Abstract
A growing number of Acidobacteria strains have been isolated from environments worldwide, with most isolates derived from acidic samples and affiliated with subdivision 1. We recovered 18 Acidobacteria strains from an alkaline soil, among which 11 belonged to the previously uncultured subdivision 6. Various medium formulations were tested for their effects on Acidobacteria growth.
TEXT
It has been known for more than a century that the great majority of environmental microorganisms are not amenable to cultivation on/in standard laboratory media (6). Among the taxa highly recalcitrant to cultivation, the bacterial division Acidobacteria has been the focus of attention, because it is the second most abundant phylum in soils based on 16S rRNA (gene) surveys (10). Yet very little is known of the ecology and the role of its members in global biogeochemical cycles. The first Acidobacterium strain was isolated in 1991 (15). Since then a growing number of isolates have been cultured from soils (or sediments) (2, 4, 5, 11, 13, 16–18, 21, 22, 31, 32), and there are currently 212 sequences of Acidobacteria strains listed in the Ribosomal Database Project (RDP; release 10, version 21, http://rdp.cme.msu.edu/). Modified medium formulations were proposed to improve their recovery, such as solid rather than liquid media (29), polymeric growth substrates like xylan instead of easily degradable carbon sources (4, 13, 27), gellan gum instead of agar (4, 11), or culturing with air enriched in CO2 (5, 31). Notably, most sampled environments were acidic, and the great majority of those isolates (71%) belong to subdivision 1, a group favored by (slightly) acidic conditions (5, 26). Subdivision 6, which is the most phylogenetically diverse and numerous Acidobacteria subgroup (it represents 29% of all Acidobacteria sequences contained in the RDP), completely lacks any cultured representative. We present here the successful recovery from a slightly alkaline soil of Acidobacteria isolates that were mostly affiliated with subdivision 6.
Samples were obtained from a surface soil (soil UCL) at the university campus of Louvain-la-Neuve, Belgium, in 2008 (pH using water [pHH2O], 7.40; 24% sand, 64% silt, 12% clay; humidity, 8.7%; 3.22% ± 0.03% C [dry soil]; 0.24% ± 0.01% N [dry soil]). A bacterial 16S rRNA gene library of 100 clones was built from soil UCL and screened for Acidobacteria by PCR following the protocol described in reference 8 (except that we used the pDrive vector [Qiagen, Venlo, Netherlands] for library construction). Acidobacterial 16S rRNA gene sequences represented 31% of that inventory. Sixty-seven percent of acidobacterial 16S rRNA gene sequences were affiliated with Acidobacteria subdivision 6. These relative abundances were in the range of what is usually observed in soils (7, 10, 34) and more particularly in soils with a comparable pH (12). Phylogenetic inference of Acidobacteria 16S rRNA gene sequences was performed using an updated version of our ARB (http://www.arb-home.de) database (8), including all acidobacterial 16S rRNA gene sequences (>1,300 bp) available in February 2010 in the SILVA database and the sequences generated in this study. Sequences were integrated in the existing ARB alignment using the automated alignment tool of the software complemented by a manual alignment. The optimized topology of the Acidobacteria subtree (8) (in which the subdivisions were delineated according to Hugenholtz et al. [9]) was not changed. Sequences were incorporated into this tree using the ARB parsimony interactive tool.
The cultivation protocol of soil bacteria was based on a very dilute medium and long incubation times similar to those described in reference 11. Serial dilutions of soil UCL suspensions were plated on (i) dilute nutrient broth (NB) supplemented with 15 g/liter agar (Sigma-Aldrich) and 0.065 g/liter nutrient broth (Oxoid) (i.e., 200 times less than the manufacturer's instructions) (ANB 1/200) and (ii) the same medium supplemented with soil extract (ANB 1/200-SE). The soil extract was prepared by mixing 300 g of dried soil UCL with 1 liter of sterile distilled water and stirring the slurry for 1 h. The slurry was then centrifuged, and the supernatant filtered with a 0.2-μm-pore-size filter was added in a 1:1 (vol/vol) ratio to autoclaved ANB 1/200. Soil suspensions were prepared by mixing 1 g of soil UCL with 100 ml of sterile distilled water (dH2O), stirring for 2 h at 300 rpm, and settling for 1 h. The supernatant was then serially diluted and plated onto ANB 1/200 and ANB 1/200-SE plates. Plates were incubated for up to 3 months in the dark at room temperature (19 to 24°C).
The numbers of colonies that were visible on plates progressively increased over 3 months of incubation. After 91 days of incubation, CFU numbers had reached 1.55 × 107 ± 0.14 × 107 and 1.82 × 107 ± 0.16 × 107 CFU per gram of dry soil on ANB and ANB-SE plates, respectively. Cultured Acidobacteria were not detected on the plates by plate wash PCR (using primers Acid31F [1] and 518R-GC [20]) after 21 days of incubation, whereas they were detected on all the plates analyzed after 35, 63, and 91 days of incubation. This delay (one month), needed to detect cultured Acidobacteria, illustrates their slow growth, as reported in another study (4). Denaturing gradient gel electrophoresis (DGGE) analysis of the PCR products and subsequent identification of the DGGE bands revealed an increase in cultured Acidobacteria diversity over the incubation time and the predominance of subdivision 6 members in the cultured Acidobacteria community (data not shown). The PCR mix, PCR cycle, and DGGE conditions were those described in reference 8.
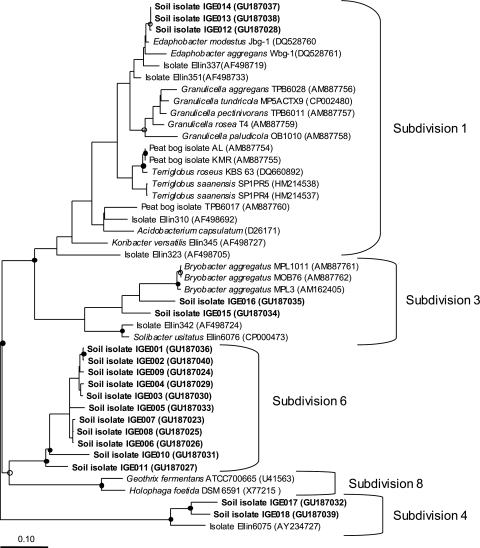
A protocol similar to that described in reference 31 was used to localize Acidobacteria on the plates after 3 months of incubation. Three thousand nine hundred colonies from ANB 1/200 or ANB 1/200-SE plates were subcultured onto fresh ANB 1/200 plates using toothpicks. After 1 month of incubation, an Acidobacteria colony PCR (with an annealing temperature at 58°C) was performed on each individual colony using primers Acid31F and 1492R. Thirty-six colonies gave a positive PCR amplicon (0.9%), among which 18 could be recovered as pure cultures and repeatedly subcultured. The medium ANB 1/200-SE proved to be superior to ANB 1/200 for isolation of Acidobacteria from the soil environment, as 12 out of the 18 isolates (IGE002, IGE003, IGE004, IGE006, IGE007, IGE008, IGE009, IGE011, IGE013, IGE014, IGE017, IGE018) were initially isolated on the former. Three isolates belonged to subdivision 1, two to subdivision 3, two to subdivision 4, and 11 (61%) to subdivision 6 (Fig. 1). Clearly, subdivision 6 predominated within these isolates, which was remarkable considering that this was the first time that members of this subdivision had been isolated under laboratory conditions. A neighbor-joining (NJ) distance matrix was generated for the sequences presented in Fig. 1 using ARB, and it was used as an input file for the program DOTUR (28). DOTUR grouped isolates IGE012 to IGE014 (subdivision 1), Edaphobacter modestus Jbg-1, and Edaphobacter aggregans Wbg-1 as a single operational taxonomic unit (OTU) at the 0.97 identity level (OTU0.03). All the other strains represented novel Acidobacteria strains. Isolates IGE001 to IGE009 (subdivision 6) were grouped as a single OTU0.03. Other isolates (IGE010, IGE011, IGE015, IGE016, IGE017, IGE018) represented a unique OTU0.03. Therefore, our isolates represented multiple OTU0.03 in the subdivisions 3, 4, and 6.
Fig. 1.
Phylogenetic affiliation of Acidobacteria isolates recovered from soil UCL and all other published terrestrial isolates based on comparative analysis of 16S rRNA gene sequence data. Isolates obtained in this study are in boldface. Sequences were inserted into the maximum parsimony dendrogram using the parsimony insertion tool of ARB. Bootstrap values of >90% and >70% are indicated in the figure as filled and open circles, respectively. The scale bar represents a change of 0.10 per nucleotide.
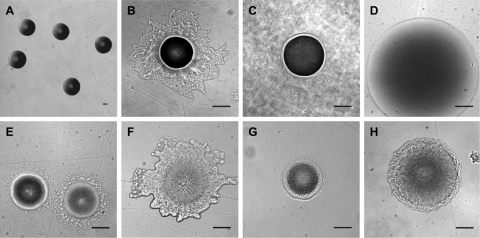
On ANB 1/200 plates, colonies of Acidobacteria took 3 to 4 days (subdivision 1), 1 to 2 weeks (subdivisions 3 and 4), or 3 to 4 weeks (subdivision 6) to form colonies with a diameter of >20 μm. After 3 months of incubation, colonies generally reached 1 to 2 mm in diameter, except subdivision 1 and subdivision 4 colonies, which were bigger (3 to 5 mm in diameter). Colony size kept increasing slowly with increasing incubation times (up to 1.5 year) except for subdivision 1 isolates. Colonies were white except subdivision 1 colonies, which were all pale pink, subdivision 6 colonies, which were all yellow, and strain IGE017 (subdivision 4), which was pink. Subdivision 1 and 4 isolates formed smooth, circular, semitransparent colonies, whereas subdivision 3 and 6 isolates formed dense, rubbery colonies, generally with an irregular margin at a later stage of development (>2 months of incubation) (Fig. 2). Subdivision 6 colonies were notably cohesive and partly grown into the agar after 3 months of incubation (except on ANB 1/200-SE), which hampered the collection of colony material by using toothpicks or even a scalpel. This feature might reflect a competitive advantage in the soil environment via the production of as-yet-uncharacterized extrapolymeric substances that promote adherence to soil particles, promote resistance to desiccation, and/or help to retain nutrients (25, 33). Their production has previously been observed for subdivision 1 and 3 isolates (5, 15, 16, 21), and full or incomplete cellulose synthesis operons have been identified in the genomes of strains belonging to those subdivisions (33).
Fig. 2.
Examples of Acidobacteria colonies recovered from soil UCL after 3 months of incubation (2 months for strain IGE012) on ANB. A, strain IGE012 (subdivision 1); B, strain IGE015 (subdivision 3); C, strain IGE016 (subdivision 3); D, strain IGE017 (subdivision 4); E, strain IGE001 (subdivision 6); F, strain IGE007 (subdivision 6); G, strain IGE005 (subdivision 6); H, strain IGE010 (subdivision 6). Bar, 100 μm. All pictures were taken from the bottom of the plate with a 10× objective, except picture A, which was taken with a 2.5× objective.
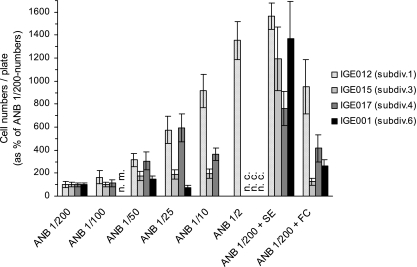
In order to improve the growth of Acidobacteria in laboratory conditions, we tested different growth media, i.e., agar (15 g/liter, pH 5.5) supplemented with increasing concentrations of NB (0.065 g/liter [ANB 1/200], 0.130 g/liter [ANB 1/100], 0.260 g/liter [ANB 1/50], 0.510 g/liter [ANB 1/25], 1.300 g/liter [ANB 1/10], or 6.500 g/liter [ANB 1/2]), ANB-SE, and ANB 1/200-FC (same medium as ANB 1/200, where a solution of 2,000 U of catalase [bovine liver; Sigma-Aldrich] was filtered with a 0.2-μm-pore-size filter and spread onto each plate). Aliquots of the same suspension of strains IGE012 (subdivision 1), IGE015 (subdivision 3), IGE017 (subdivision 4), or IGE001 (subdivision 6) were plated on each medium. After 3 months of incubation, cells were collected from triplicate plates using a glass spreader and/or a razor blade and sterile dH2O. They were stained with 4′,6-diaminido-2-phenylindole (DAPI) (1 μg/ml) for enumeration by microscopy. A total of 5.72 × 108 ± 1.62 × 108, 8.42 × 107 ± 1.88 × 107, 2.68 × 107 ± 0.42 × 107, and 1.90 × 108 ± 0.30 × 108 cells were recovered per plate of standard medium for each strain, respectively. None of the strains could grow on agar without nutrients (data not shown). The soil extract-amended medium was the best growth medium among the media tested for the four Acidobacteria strains and resulted in a cell number increase of 7.6- to 15.6-fold (Fig. 3). This result corroborates the beneficial effect of this undefined medium amendment on the initial isolation of Acidobacteria from the soil environment and on the growth of fastidious soil bacteria in general (4). However, it is probable that this positive effect was overestimated for subdivision 6 isolates, as collection of colony material was notably easier on that medium. The addition of filtered catalase resulted in an increase in cell numbers as well, although more moderate (1.2- to 9.5-fold). This result confirms the general sensitivity of Acidobacteria to damage by reactive oxygen species (5, 31), a phenomenon well studied in Escherichia coli (30). Finally, the four strains exhibited a different response to increasing nutrient concentrations which likely reflected different lifestyle strategies among members of this broad phylum. Acidobacteria are known to be more abundant in environments with low C availability (7) and seem to prefer bulk soil to nutrient-rich rhizosphere (3, 14, 19), but such intraphylum differences regarding nutrient concentrations have not been previously reported. Strain IGE012 (subdivision 1) showed a constant increase in cell numbers with increasing nutrient concentrations, to reach a 13.5-fold increase on ANB 1/2. This response was not surprising considering that subdivision 1 is the most readily cultured group of Acidobacteria (4, 23, 26, 27). For the 3 other strains, the medium ANB 1/2 inhibited growth, as no colonies were observed on the plates. The nutrient concentrations at which the largest number of bacterial cells was achieved for each Acidobacteria strain, relative to ANB 1/200, were ANB 1/10 for strain IGE015 (2-fold increase in cell numbers), ANB 1/25 for strain IGE017 (5.9-fold increase), and ANB 1/50 for strain IGE001 (1.5-fold increase). The optimal nutrient concentration for strain IGE001 could actually be lower, as we have no data on ANB 1/100 due to a problem in the preparation of the medium. For each strain, cell numbers on the medium with the highest cell count (ANB-SE and ANB-FC excluded) were statistically different from those on standard medium (one-tailed Student's t test, P = 0 × 10−6 for strain IGE012, IGE015, or IGE017, and P = 12 × 10−6 for strain IGE001). To isolate Acidobacteria strains, some other research groups used richer media than our standard 200-fold diluted NB, e.g., 10-fold diluted Trypticase soy agar (TSA) (4, 13). However, in light of our results, such media are suboptimal for the recovery of subdivision 6 strains, as the number of cells collected on ANB 1/10 represented only 2% of the cells collected on ANB 1/200 (Fig. 3). Assessment of the nutrient concentrations based on colony diameter measured after 3 months of incubation led to similar conclusions (data not shown). The sole exception was strain IGE017, which developed very-large-diameter (but scarce) bright-pink-colored colonies on ANB 1/10 (data not shown). This strong coloration is reminiscent of the pink coloration observed for some Acidobacteria subdivision 1 strains under a higher oxygen concentration (5) and may be due to a stress response linked to higher nutrient levels. We are presently characterizing the physiology of our strains. Moreover, some of these isolates have been found to contain domains predicted to function within polyketide synthase pathways (24), and it will be of interest to establish the diversity of secondary metabolites expressed by these Acidobacteria strains.
Fig. 3.
Cell numbers recovered by plate washing after incubation of four Acidobacteria strains for 91 days. Cell suspensions were plated on agar with increasing concentrations of nutrient broth (ANB 1/200, ANB 1/100, ANB 1/50, ANB 1/25, ANB 1/10, ANB 1/2) or supplemented with soil extract (ANB-SE) or filtered catalase (ANB-FC). Each solid bar represents standard errors of triplicates. n.m., not measured; n.c., no colony.
Due to their small size, slow growth, adaptation to very low nutrient concentrations, and attachment to the agar substrate, it is likely that subdivision 6 Acidobacteria were overlooked in previous cultivation attempts or that the alkaline soil used in this study was more conducive to subdivision 6 cultivation. This study illustrates that there is still great potential to isolate novel environmental strains using simple and inexpensive cultivation strategies.
Nucleotide sequence accession numbers.
Sequence data have been deposited in the GenBank database under the following accession numbers: JF521744 to JF521838 for the Acidobacteria 16S rRNA gene sequences of soil UCL and GU187023 to GU187040 for the 16S rRNA gene sequences of the Acidobacteria isolates.
Acknowledgments
Isabelle F. George benefited from a grant of “Chargée de Recherches” of the National Fund for Scientific Research of Belgium (FNRS). Her visits to the National Oceanography Centre, Southampton, United Kingdom, were supported by travel grants from the FNRS and the British Council/FNRS-CWBI Partnership Programme in Science.
Laurence Pesché and Benjamin Vidick are acknowledged for their help.
Footnotes
Published ahead of print on 23 September 2011.
REFERENCES
- 1. Barns S. M., Takala S. L., Kuske C. R. 1999. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 65:1731–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Coates J. D., Ellis D. J., Gaw C. V., Lovley D. R. 1999. Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int. J. Syst. Bacteriol. 49(Pt. 4):1615–1622 [DOI] [PubMed] [Google Scholar]
- 3. da Rocha U. N., van Overbeek L., van Elsas J. D. 2009. Exploration of hitherto-uncultured bacteria from the rhizosphere. FEMS Microbiol. Ecol. 69:313–328 [DOI] [PubMed] [Google Scholar]
- 4. Davis K. E., Joseph S. J., Janssen P. H. 2005. Effects of growth medium, inoculum size, and incubation time on culturability and isolation of soil bacteria. Appl. Environ. Microbiol. 71:826–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eichorst S. A., Breznak J. A., Schmidt T. M. 2007. Isolation and characterization of soil bacteria that define Terriglobus gen. nov., in the phylum Acidobacteria. Appl. Environ. Microbiol. 73:2708–2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Epstein S. S. 2009. Uncultivated microorganisms. Springer-Verlag, New York, NY [Google Scholar]
- 7. Fierer N., Bradford M. A., Jackson R. B. 2007. Toward an ecological classification of soil bacteria. Ecology 88:1354–1364 [DOI] [PubMed] [Google Scholar]
- 8. George I. F., et al. 2009. Changes in soil Acidobacteria communities after 2,4,6-trinitrotoluene contamination. FEMS Microbiol. Lett. 296:159–166 [DOI] [PubMed] [Google Scholar]
- 9. Hugenholtz P., Goebel B. M., Pace N. R. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Janssen P. H. 2006. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 72:1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Janssen P. H., Yates P. S., Grinton B. E., Taylor P. M., Sait M. 2002. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Environ. Microbiol. 68:2391–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones R. T., et al. 2009. A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. ISME J. 3:442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Joseph S. J., Hugenholtz P., Sangwan P., Osborne C. A., Janssen P. H. 2003. Laboratory cultivation of widespread and previously uncultured soil bacteria. Appl. Environ. Microbiol. 69:7210–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kielak A., Pijl A. S., van Veen J. A., Kowalchuk G. A. 2008. Differences in vegetation composition and plant species identity lead to only minor changes in soil-borne microbial communities in a former arable field. FEMS Microbiol. Ecol. 63:372–382 [DOI] [PubMed] [Google Scholar]
- 15. Kishimoto N., Kosako Y., Tano T. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Microbiol. 22:1–7 [DOI] [PubMed] [Google Scholar]
- 16. Koch I. H., Gich F., Dunfield P. F., Overmann J. 2008. Edaphobacter modestus gen. nov., sp. nov., and Edaphobacter aggregans sp. nov., acidobacteria isolated from alpine and forest soils. Int. J. Syst. Evol. Microbiol. 58:1114–1122 [DOI] [PubMed] [Google Scholar]
- 17. Liesack W., Bak F., Kreft J. U., Stackebrandt E. 1994. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 162:85–90 [DOI] [PubMed] [Google Scholar]
- 18. Mannisto M. K., Rawat S., Starovoytov V., Haggblom M. M. 2011. Terriglobus saanensis sp. nov., a novel Acidobacterium isolated from tundra soil of Northern Finland. Int. J. Syst. Evol. Microbiol. 61(Pt. 8):1823–1828 [DOI] [PubMed] [Google Scholar]
- 19. Marilley L., Aragno M. 1999. Phylogenetic diversity of bacterial communities differing in degree of proximity of Lolium perenne and Trifolium repens roots. Appl. Soil Ecol. 13:127–136 [Google Scholar]
- 20. Muyzer G., de Waal E. C., Uitterlinden A. G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pankratov T. A., Dedysh S. N. 2010. Granulicella paludicola gen. nov., sp. nov., G. pectinivorans sp. nov., G. aggregans sp. nov. and G. rosea sp. nov., novel acidophilic, polymer-degrading acidobacteria from Sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 60:2951–2959 [DOI] [PubMed] [Google Scholar]
- 22. Pankratov T. A., Kirsanova L. A., Kaparullina E. N., Kevbrin V. V., Dedysh S. N. 2011. Telmatobacter bradus gen. nov., sp. nov., a cellulolytic facultative anaerobe from subdivision 1 of the Acidobacteria and emended description of Acidobacterium capsulatum Kishimoto et al. 1991. Int. J. Syst. Evol. Microbiol. [Epub ahead of print.] doi:10.1099/ijs.0.029629-0 [DOI] [PubMed] [Google Scholar]
- 23. Pankratov T. A., Serkebaeva Y. M., Kulichevskaya I. S., Liesack W., Dedysh S. N. 2008. Substrate-induced growth and isolation of acidobacteria from acidic Sphagnum peat. ISME J. 2:551–560 [DOI] [PubMed] [Google Scholar]
- 24. Parsley L. C., et al. 2011. Polyketide synthase pathways identified from a metagenomic library are derived from soil acidobacteria. FEMS Microbiol. Ecol. 78:176–187 [DOI] [PubMed] [Google Scholar]
- 25. Roberts I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50:285–315 [DOI] [PubMed] [Google Scholar]
- 26. Sait M., Davis K. E., Janssen P. H. 2006. Effect of pH on isolation and distribution of members of subdivision 1 of the phylum acidobacteria occurring in soil. Appl. Environ. Microbiol. 72:1852–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sait M., Hugenholtz P., Janssen P. H. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654–666 [DOI] [PubMed] [Google Scholar]
- 28. Schloss P. D., Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schoenborn L., Yates P. S., Grinton B. E., Hugenholtz P., Janssen P. H. 2004. Liquid serial dilution is inferior to solid media for isolation of cultures representative of the phylum-level diversity of soil bacteria. Appl. Environ. Microbiol. 70:4363–4366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Speck M. L., Ray B., Read R. B., Jr 1975. Repair and enumeration of injured coliforms by a plating procedure. Appl. Microbiol. 29:549–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stevenson B. S., Eichorst S. A., Wertz J. T., Schmidt T. M., Breznak J. A. 2004. New strategies for cultivation and detection of previously uncultured microbes. Appl. Environ. Microbiol. 70:4748–4755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stott M. B., et al. 2008. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ. Microbiol. 10:2030–2041 [DOI] [PubMed] [Google Scholar]
- 33. Ward N. L., et al. 2009. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl. Environ. Microbiol. 75:2046–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Youssef N. H., Elshahed M. S. 2009. Diversity rankings among bacterial lineages in soil. ISME J. 3:305–313 [DOI] [PubMed] [Google Scholar]