Abstract
Serodiagnosis of amoebiasis remains the preferred method for diagnosis of amoebic liver abscess (ALA). However, the commercially available kits are problematic in areas of endemicity due to the persistently high background antibody titers. Human serum samples (n = 38) from patients with ALA who live in areas of endemicity were collected from Hospital Universiti Sains Malaysia during the period of 2008 to 2010. Western blots using excretory-secretory antigen (ESA) collected from axenically grown Entamoeba histolytica were probed with the above serum samples. Seven antigenic proteins of ESA with various reactivities were identified, i.e., 152 kDa, 131 kDa, 123 kDa, 110 kDa, 100 kDa, 82 kDa, and 76 kDa. However, only the 152-kDa and 110-kDa proteins showed sensitivities above 80% in the Western blot analysis. All the antigenic proteins showed undetectable cross-reactivity when probed with healthy human serum samples (n = 30) and serum samples from other infections (n = 33). From the matrix-assisted laser desorption ionization–two-stage time of flight (MALDI-TOF/TOF) analysis, the proteins were identified as heavy subunits of E. histolytica lectin and E. histolytica pyruvate phosphate dikinase, respectively. Use of the E. histolytica lectin for diagnosis of ALA has been well reported by researchers and is being used in commercialized kits. However, this is the first report on the potential use of pyruvate phosphate dikinase for diagnosis of ALA; thus, this molecule merits further evaluation on its diagnostic value using a larger panel of serum samples.
INTRODUCTION
Amoebiasis is caused by the enteric protozoan Entamoeba histolytica, which affects 50 million people worldwide and leads to 100,000 fatal cases annually (19, 21). Amoebic liver abscess (ALA) is the most common clinical manifestation of extraintestinal amoebiasis. It is due to the hematogenous spread of the E. histolytica trophozoites from intestine to liver through the portal vein. Patients with ALA present with hepatomegaly, right-upper-quadrant pain, tenderness of the liver, fever, jaundice, and nausea. It may lead to a fatal outcome if early diagnosis and treatment are not sought (1, 10).
Diagnosis of ALA is often initiated with radiology imaging to examine the presence of abscess in the liver. If indicated, aspiration of the sample is performed for culture, DNA detection, and/or antigen detection. The indications include large abscesses, superficial abscesses, abscesses with severe pain or marked point tenderness, abscesses with marked diaphragm elevation, a clinical picture suggesting impending abscess perforation, and left lobe abscess (7). Absence of bacterial growth in the abscess culture could rule out the possibility of pyogenic liver abscess cases. The definitive diagnosis of ALA is by microscopic observation of trophozoites in the abscess fluid, but the sensitivity of microscopic examination is low as the trophozoites are easily disintegrated, and most of them reside at the peripheral margin of the abscess. Many reports showed that DNA and antigen detection-based methods performed on the abscess sample (e.g., PCR, real-time PCR, and TechLab E. histolytica II antigen detection enzyme-linked immunosorbent assay [ELISA]) had high sensitivity (4, 11, 18).
In addition to imaging, serological testing is the preferred choice for diagnosis. The available antigen detection tests such as TechLab E. histolytica II ELISA, which detects E. histolytica lectin antigen, can be used for diagnosis of acute ALA patients who have not received treatment (23). However, often patients who are admitted to the hospital with liver abscess have received treatment prior to investigation for ALA, which significantly reduces the sensitivity of the antigen detection test. Thus, antibody detection is currently the most common serological test used to detect ALA, either by indirect hemagglutination assay (IHA) or ELISA. However, these tests mostly use amoebic lysate antigen and are problematic for diagnosis in areas of endemicity where the background antiamoebic antibody titer is high. Thus, in areas of endemicity, low specificities of these tests were reported with the low cutoff values suggested by the manufacturer (22, 24).
Comparison of crude soluble antigen (CSA) with excretory-secretory antigen (ESA) of E. histolytica showed that ESA demonstrated a higher positive detection rate when tested with sera of patients with acute amoebic dysentery and asymptomatic cyst passers and equal sensitivity for diagnosis of ALA (10, 15). Therefore, in our quest to identify new markers to improve the serodiagnosis of ALA, ESA of E. histolytica was produced and analyzed by SDS-PAGE, two-dimensional electrophoresis (2-DE), and Western blotting. The identities of the potential candidates were then identified by mass spectrometry.
MATERIALS AND METHODS
Maintenance of E. histolytica trophozoites.
E. histolytica axenic strain HM1:IMSS trophozoites were hermetically cultured in TYI-S-33 medium supplemented with 12.5% bovine serum (Invitrogen, New Zealand) and 1× Diamond's vitamin-Tween 80 (Sigma) at 36°C. The medium was changed every 48 to 72 h (2).
Collection and preparation of ESA.
Mass cultures of E. histolytica trophozoites were collected at log phase and washed three time with RPMI medium supplemented with 0.1% l-cysteine and 0.02% ascorbic acid (RPMI-C-A medium) with centrifugation at 22 × g for 2 min at room temperature (RT). Subsequently, the cell density was determined via the trypan blue exclusion method. Trophozoites were seeded into a culture tube 80% filled with RPMI-C-A medium at a cell density of 0.8 × 106 cells per ml and incubated at 36°C for 6 h. Using this method, we have previously shown that ∼95% trophozoite viability can be maintained throughout the incubation period (20). Upon completion, culture tubes were subjected to centrifugation at 22 × g for 2 min at 4°C. The supernatants in the culture tubes were collected and mixed with 0.5 M iodoacetamide to a final concentration of 1 mM. Next, the supernatant was again subjected to centrifugation at 10, 000 × g for 5 min at 4°C and filtered through 0.2-μm-pore-size membrane. Subsequently, ESA in the supernatant was concentrated 1,000 times using a U-tube concentrator with a molecular-weight cutoff (MWCO) of 10 kDa. Cocktail protease inhibitor (Roche Diagnostic, Germany) was then added to the concentrated ESA. The protein concentration of the ESA was estimated using a Bradford protein assay (10, 15, 20).
Serum samples and ethical approval.
Serum samples in the current study were collected from Hospital Universiti Sains Malaysia (USM) during the period of 2008 to 2010. The procedures of collecting and handling the serum samples were approved by USM Human Ethical Committee (reference number USMKK/PPP/JEPem[213.3(10)]). Human serum samples included in this study were divided into four groups: (i) group A, human ALA serum samples (n = 24) with consistent clinical symptoms (i.e., fever, abdominal/right hepatic chest pain, hepatomegaly, and jaundice) and radiological image and IHA-positive results; (ii) group B, human ALA serum samples (n = 14) from patients whose abscesses were positive by real-time PCR for E. histolytica DNA and negative by bacterial culture (the primers and probe sequences were as reported previously [9]); (iii) group C, healthy blood donor serum samples which were negative by IHA (n = 30); (iv) group D, serum samples from patients with other infections (n = 33), i.e., salmonellosis (n = 5), shigellosis (n = 1), Escherichia coli septicemia (n = 2), Staphylococcus sp. septicemia (n = 2), Helicobacter pylori (n = 6), pyogenic liver abscess (n = 4), Stenotrophomonas maltophilia septicemia (n = 1), enteropathogenic E. coli (n = 1), Ascaris lumbricoides (n = 1), Klebsiella pneumoniae (n = 1), and toxoplasmosis (n = 9). These sera were negative by IHA for amoebiasis.
SDS-PAGE.
Protein samples were electrophoretically separated via SDS-PAGE using a Bio-Rad Mini-Protean III Electrophoresis Cell and Protean II xi Cell according to the Laemmli (8) protocol, with modifications. Prior to SDS-PAGE, ESA was mixed with 2× Laemmli sample buffer and boiled for 5 min. Subsequently, ESA was separated using a 6% SDS-PAGE gel at a constant current of 25 mA per gel for about 1 h.
Western blotting.
Upon completion of SDS-PAGE, proteins in the gel were electrophoretically transferred onto a 0.45-μm-pore-size nitrocellulose paper (NCP) membrane using a semidry transblot apparatus (Bio-Rad) at a constant voltage of 15 V for 30 min. The NCP was blocked for 1 h at RT with 5% skim milk prepared in 10 mM Tris-buffered saline (TBS), pH 7.2. Subsequently, the NCP was washed (three times for 5 min each time) with TBS containing 0.1% Tween 20 (TBS-T). Then, the NCP was cut into multiple strips and incubated with human serum at a dilution of 1:200 (in TBS-T) for 2 h at RT. The NCP strips were then washed three times with TBS-T and then incubated with monoclonal mouse anti-human IgG conjugated with horseradish peroxides (HRP) at a dilution of 1:6,000 for 1 h. Subsequently, the NCP strips were again washed (three times for 5 min each time) with TBS-T. Western blot substrates, i.e., enhanced chemiluminescence (ECL) blotting reagent (Roche Diagnostics, Germany) or tetramethylbenzidine (TMB) substrate for membrane (Sigma), were used as substrates. The Western blot signal was captured using a camera (Lumix, Germany).
2-DE and Western blotting.
Selected protein bands which showed potential diagnostic value were further analyzed using 2-DE to ensure that the bands were well separated. An Offgel Fractionator 3100 (Agilent Technologies, Germany) was used to separate the proteins by isoelectric point (pI) followed by SDS-PAGE and Western blotting. An Agilent Offgel kit, pH 3 to 10, with a 12-well setup frame was used. Sample was prepared by mixing 1,600 μl of the ready stock solution (1.25×) with 400 μl of the sample with a total protein amount of 2 mg and then gently vortexed. Forty microliters of immobilized pH gradient (IPG) strip rehydration buffer was added into each well to swell the gel for 15 min. Wetted electrode pads were placed at the cathode and anode ends of the IPG strip gel surface. After reswelling of the gel, 150 μl of protein sample was loaded into each well. Ten microliters of rehydration buffer was reapplied onto the electrode pads at each of the IPG gel ends. Cover fluid (mineral oil) was pipetted onto the gel strip ends. Subsequently, the sample was focused with a maximum power of 200 mW, maximum current of 50 mA, and typical voltages ranging from 500 to 4,500 V until 50 kVh was reached after 24 h. Upon completion, each of the 12 fractionated ESA samples was separately mixed with 5× Laemmli sample buffer without boiling and electrophoretically separated via SDS-PAGE. Western blotting was performed using pooled and individual human serum samples to identify the selected antigenic proteins.
Mass spectrometry analysis and protein identification.
The selected proteins were excised from a 2-D SDS-PAGE gel and sent for matrix-assisted laser desorption ionization–two-stage time of flight ([MALDI-TOF/TOF] ABI 4800 instrument) analysis at the Proteomic Laboratory Service Center, Australia, and searched with a Swiss-Prot protein database.
RESULTS
IgG blots of ESA.
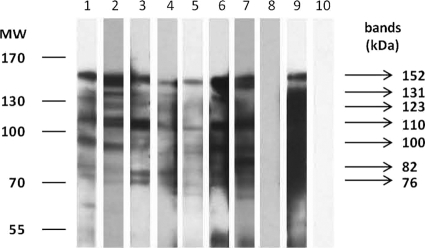
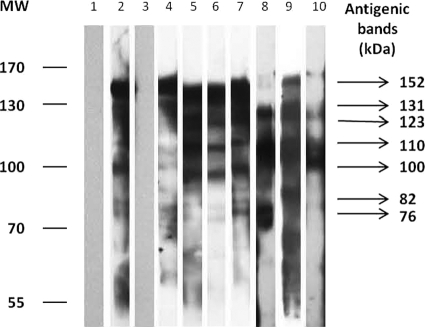
IgG blots of ESA probed with human ALA serum samples from group A showed seven antigenic bands with consistent reactivities (Fig. 1). In addition, these antigenic proteins were also similar to the bands present in the IgG blots probed with human ALA serum samples from group B (Fig. 2). However, mean sensitivities of the bands to detect ALA varied from 16% to 84%. Only two of the antigenic bands, i.e., 152-kDa and 110-kDa bands, showed high sensitivities of above 80% (Table 1). Neither of these two antigenic bands showed reactivity in IgG blots probed with serum samples from groups C and D, thus showing 100% specificity. In this study, sensitivity is defined as the number of serum samples which reacted with the band out of the total number of samples from ALA patients, i.e., group A and B sera. Meanwhile, specificity is defined as the number of serum samples which do not react with the band out of the total number of samples from individuals without ALA, i.e., group C and D sera.
Fig. 1.
Representative IgG blot of ESA probed with human serum samples. Lanes 1 to 7, individual ALA serum samples from group A; lane 8, pooled IHA-negative serum sample (group C); lane 9, pooled ALA serum sample (positive control); lane 10, TBS. MW, molecular weights in thousands.
Fig. 2.
Representative IgG blot of ESA when probed with human serum samples. Lane 1, TBS; lane 2, pooled ALA serum sample (positive control); lane 3, pooled IHA-negative serum sample (group D); lanes 4 to 10, individual ALA serum samples from group B. MW, molecular weights in thousands.
Table 1.
Sensitivity and specificity of antigenic bands of ESA from IgG blots
| Antigenic band (kDa)c | Sensitivity (no. of positive tests [%])a | Specificity (no. of negative tests [%])b |
|---|---|---|
| 152 | 32 (84) | 100 |
| 131 | 17 (45) | 100 |
| 123 | 6 (16) | 100 |
| 110 | 31 (82) | 100 |
| 100 | 7 (18) | 100 |
| 82 | 15 (39) | 100 |
| 76 | 15 (39) | 100 |
n = 38, total number of tests within the groups with ALA (groups A and B).
n = 63, total number of tests within the groups without ALA (groups C and D).
The bands with sensitivities above 80% are shown in boldface.
2-DE Western blotting and protein identification.
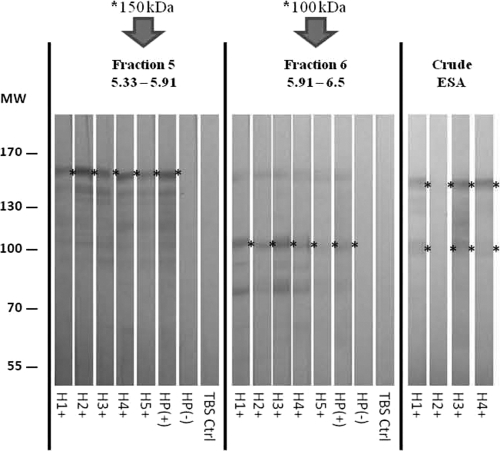
IgG blotting of 12 ESA fractions with pooled serum samples revealed that the 152-kDa and 110-kDa proteins were located in fraction 5 (pI 5.33 to 5.91) and fraction 6 (pI 5.91 to 6.5), respectively (Fig. 3). Further IgG blot analysis of these ESA fractions with individual serum samples (n = 5) confirmed the location of these antigenic proteins (Fig. 4). These protein bands were excised and sent for MALDI-TOF/TOF analysis. According to the Mascot search result from the mass spectrometry database (MSDB) search engine, the 152-kDa protein matched the E. histolytica lectin protein (C4LTMO) with a protein score of 273. A score of >55 indicates identity or extensive homology at a significant level (P < 0.05). Seven peptides matched to the Gal/GalNAc lectin heavy subunit. The 110-kDa protein matched pyruvate phosphate dikinase (EHI_009530) with a protein score of 544, with nine matched peptides. Since pyruvate phosphate dikinase has not been reported as a diagnostic protein, the 110-kDa bands from separate gels were sent three times for the MALDI-TOF/TOF analysis, and same results were obtained each time.
Fig. 3.
IgG blot of ESA separated by 2-DE and probed with human serum samples. H+, pooled positive ALA serum sample from group B; H−, pooled IHA-negative serum sample (group C). MW, molecular weights in thousands. pI values are indicated for each fraction.
Fig. 4.
IgG blot of ESA separated by 2-DE and probed with human serum samples. H1+ to H5+, individual ALA serum samples from group B; HP+, pooled ALA serum sample (positive control); HP−, pooled IHA-negative serum sample (group D). Ctrl, control; MW, molecular weights in thousands. pI values are indicated for the fractions.
DISCUSSION
Current diagnosis of ALA still depends on the results of clinical manifestations, radiology imaging, and serology since stool examination is inapplicable for diagnosis of extraintestinal amoebiasis and abscess aspiration is performed only when indicated.
E. histolytica ESA contains proteins shed from trophozoites during active multiplication and metabolites released by trophozoites during incubation in RPMI-C-A medium. Although great care was taken to produce good quality ESA, there were probably still some partial proteins released from lysed trophozoites. In this study, the ESA antigenic bands that ranged from 97.2 to 158 kDa consistently showed reactivities when incubated with human ALA serum samples (n = 7). Analysis of the IgG blots showed that 152-kDa and 110-kDa proteins had higher association with serum samples from patients with PCR-positive abscess, i.e., 13/14 (93%) and 12/14 (86%), respectively, than serum samples from patients with unknown PCR results, i.e., 19/24 (79%) for both bands. In addition, the sensitivities of both the 152-kDa and 110-kDa bands were similar, i.e., 32/38 (84%) and 31/38 (82%), respectively. Both antigens showed high specificity as there were undetectable reactivities in the IgG blot of ESA probed with serum samples from healthy individuals and from those with other infections.
The protein components of ESA in the current study were different from those reported by Sengupta et al. (15). In the latter study, the ESA proteins ranged between 200 to 20 kDa, with the predominant protein bands below 66 kDa (e.g., 45 kDa and 29 kDa), while the high-molecular-weight proteins (>100 kDa) were faint. This may be due to the differences in antigen preparations. The ESA in this study was concentrated 1,000 times instead of 10 times to enrich the low-abundance proteins. In addition, to enhance the reproducibility of ESA, protein-free defined RPMI-C-A medium was used in this study instead of serum- and vitamin-free TYI-S-33 medium. In addition, iodoacetamide was added to the RPMI-C-A medium containing the ESA upon its collection in order to protect the protein from degradation by proteases (3, 13, 15). Although we have previously optimized the growth conditions of E. histolytica trophozoites so that ∼ 95% of the parasites were alive when ESA was collected (20), nevertheless it is still possible that some of the antigenic bands may have come from ruptured trophozoites.
2-DE protein separation via an Agilent 3100 Offgel Fractionator followed by SDS-PAGE allowed only the selected ESA fractions to be tested with serum samples. The protein bands excised from the 2-DE gels were well separated, thus avoiding the presence of multiple proteins in each band. In this study, the 152-kDa protein was identified as an E. histolytica lectin protein, which has been reported to be sensitive for diagnosis of invasive amoebiasis (5, 6). Specific monoclonal antibody against this protein has been used for antigen detection in a TechLab Entamoeba histolytica II kit (TechLab Inc.). The 110-kDa protein, identified as the E. histolytica pyruvate phosphate dikinase, was also found to show similarly high sensitivity for diagnosis of ALA. This protein was reported to be a key enzyme in the anaerobic metabolism via pyrophosphate-dependent glycolysis and has no counterpart with proteins in human metabolism (14). Molecular modeling of this enzyme had been reported, and specific inhibitors to it for therapeutic purpose have been studied (17). The protein sequence of pyruvate phosphate dikinase showed high similarity with a closely related pathogenic intestinal anaerobic protozoon, Giardia lamblia. This suggests the possibility of producing specific antibody to pyruvate phosphate dikinase for simultaneous detection of both E. histolytica and G. lamblia species in fecal samples (14).
To date, there is no report on the application of pyruvate phosphate kinase for diagnosis of amoebiasis. Nevertheless, to confirm the diagnostic value of pyruvate phosphate dikinase, it would be necessary to produce the recombinant form of the protein and then demonstrate that it is recognized by patient serum. This is indeed our aim for future research.
ACKNOWLEDGMENTS
This study was funded by FRGS grant number 203/CIPPM/6711122, USM-RU grant number 1001/PPSK/813009, and USM-RU-PGRS grant number 1001/INFORMM/8032030. W.W.K. is a recipient of a USM Fellowship, and N.O. received financial support from the Vice Chancellor's Award.
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Akgun Y., Tacyildiz I. H., Celik Y. 1999. Amebic liver abscess: changing trends over 20 years. World J. Surg. 23:102–106 [DOI] [PubMed] [Google Scholar]
- 2. Diamond L. S., Harlow D. R., Cunnick C. C. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72:431–432 [DOI] [PubMed] [Google Scholar]
- 3. Flores M. S., et al. 2005. Preparation of Entamoeba histolytica antigens without enzymatic inhibitors. Parasitology 131:231–236 [DOI] [PubMed] [Google Scholar]
- 4. Fotedar R., et al. 2007. Laboratory diagnostic techniques for Entamoeba species. Clin. Microbiol. Rev. 20:511–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haque R., et al. 1997. Entamoeba histolytica and Entamoeba dispar infection in children in Bangladesh. J. Infect. Dis. 175:734–736 [DOI] [PubMed] [Google Scholar]
- 6. Haque R., Petri W. A., Jr 2006. Diagnosis of amebiasis in Bangladesh. Arch. Med. Res. 37:273–276 [DOI] [PubMed] [Google Scholar]
- 7.Kapoor O. P.Amoebic liver abscess. S. S. Publishers, Bombay, India. 1979. http://www.bhj.org/books/liver/s7c02.htm.
- 8. Laemmli U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 9. Othman N., et al. 2010. Application of real-time polymerase chain reaction in detection of Entamoeba histolytica in pus aspirates of amoebic liver abscess patients. Foodborne Pathog. Dis. 7:637–641 [DOI] [PubMed] [Google Scholar]
- 10. Pal S., et al. 1996. Comparative evaluation of somatic and excretory-secretory antigens of Entamoeba histolytica in serodiagnosis of human amoebiasis by ELISA. Indian J. Med. Res. 104:152–156 [PubMed] [Google Scholar]
- 11. Paul J., Srivastava S., Bhattacharya S. 2007. Molecular methods for diagnosis of Entamoeba histolytica in a clinical setting: an overview. Exp. Parasitol. 116:35–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reference deleted.
- 13. Que X., Reed S. L. 2000. Cysteine proteinases and the pathogenesis of amebiasis. Clin. Microbiol. Rev. 13:196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saavedra-Lira E., Ramirez-Silva L., Perez-Montfort R. 1998. Expression and characterization of recombinant pyruvate phosphate dikinase from Entamoeba histolytica. Biochim. Biophys. Acta 1382:47–54 [DOI] [PubMed] [Google Scholar]
- 15. Sengupta S., et al. 2000. Role of excretory-secretory products of Entamoeba histolytica in human amebiasis. Arch. Med. Res. 31:S226–S228 [DOI] [PubMed] [Google Scholar]
- 16.Reference deleted.
- 17. Stephen P., et al. 2008. Molecular modeling on pyruvate phosphate dikinase of Entamoeba histolytica and in silico virtual screening for novel inhibitors. J. Comput. Aided Mol. Des. 22:647–660 [DOI] [PubMed] [Google Scholar]
- 18. Tanyuksel M., Petri W. A., Jr 2003. Laboratory diagnosis of amebiasis. Clin. Microbiol. Rev. 16:713–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Walsh J. A. 1986. Problems in recognition and diagnosis of amebiasis: estimation of the global magnitude of morbidity and mortality. Rev. Infect. Dis. 8:228–238 [DOI] [PubMed] [Google Scholar]
- 20. Wong W. K., et al. 2011. Comparison of protein-free defined media, and effect of l-cysteine and ascorbic acid supplementation on viability of axenic Entamoeba histolytica. Parasitol. Res. 108:425–430 [DOI] [PubMed] [Google Scholar]
- 21. World Health Organization. 1997. WHO/PAHO/UNESCO report. A consultation with experts on amoebiasis. Mexico City, Mexico 28–29 January, 1997. Epidemiol. Bull. 18:13–14 [PubMed] [Google Scholar]
- 22. Zambrano-Villa S., et al. 2002. How protozoan parasites evade the immune response. Trends Parasitol. 18:272–278 [DOI] [PubMed] [Google Scholar]
- 23. Zeehaida M., et al. 2008. A study on the usefulness of TechLab Entamoeba histolytica II antigen detection ELISA in the diagnosis of amoebic liver abscess (ALA) at Hospital Universiti Sains Malaysia (HUSM), Kelantan, Malaysia. Trop. Biomed. 25:209–216 [PubMed] [Google Scholar]
- 24. Zeehaida M., et al. 2009. Analysis of indirect hemagglutination assay results among patients with amoebic liver abscess. Int. Med. J. 16:195–199 [Google Scholar]