Abstract
Swine influenza is a highly contagious viral infection in pigs that significantly impacts the pork industry due to weight loss and secondary infections. There is also the potential of a significant threat to public health, as was seen in 2009 when the pandemic H1N1 influenza virus strain emerged from reassortment events among avian, swine, and human influenza viruses within pigs. As classic and pandemic H1N1 strains now circulate in swine, an effective vaccine may be the best strategy to protect the pork industry and public health. Current inactivated-virus vaccines available for swine influenza protect only against viral strains closely related to the vaccine strain, and egg-based production of these vaccines is insufficient to respond to large outbreaks. DNA vaccines are a promising alternative since they can potentially induce broad-based protection with more efficient production methods. In this study we evaluated the potentials of monovalent and trivalent DNA vaccine constructs to (i) elicit both humoral and gamma interferon (IFN-γ) responses and (ii) protect pigs against viral shedding and lung disease after challenge with pandemic H1N1 or classic swine H1N1 influenza virus. We also compared the efficiency of a needle-free vaccine delivery method to that of a conventional needle/syringe injection. We report that DNA vaccination elicits robust serum antibody and cellular responses after three immunizations and confers significant protection against influenza virus challenge. Needle-free delivery elicited improved antibody responses with the same efficiency as conventional injection and should be considered for development as a practical alternative for vaccine administration.
INTRODUCTION
Swine influenza is a highly contagious viral infection in pigs and is characterized by coughing, sneezing, nasal discharge, elevated temperatures, lethargy, breathing difficulties, and depressed appetite (15). Typical pathological features of swine influenza virus (SIV) infection in pigs include changes in the cranial and ventral lung lobes, demarcation between normal and affected lung tissue, interlobular edema, hemorrhagic lymph nodes, blood-tinged fibrinous exudate in the airways, and acute respiratory distress, which can result in widespread interstitial pneumonia and hemorrhagic lymph nodes (15). The virus is spread primarily via direct contact between infected and susceptible pigs but is also capable of airborne transmission as the virus is excreted through coughing, sneezing, and nasal discharges (7, 15).
Historically, swine influenza epidemics have caused significant economic impact on the pork industry due to weight loss, increased time needed to reach market weight, and predisposition of pigs to secondary bacterial infections (7, 15). Sporadic human infections with H1 and H3 influenza virus subtypes, otherwise known as “classic” SIV, have occurred following direct contact with pigs, without any further transmission of disease. However, the emergence of the pandemic strain in 2009 highlights the potential public health threat posed by influenza infection in pigs. Molecular characterization of the pandemic viral strain revealed that it contained genes from human, classic swine, and North American avian influenza viruses (10, 11), reinforcing the possibility that pigs act as a mixing vessel (4, 12, 15, 16, 36, 53) for reassortment events that lead to the development of novel viral strains to which humans have no preexisting immunity. The pork industry was also severely impacted by the 2009 H1N1 pandemic as consumption dropped due to the “swine flu” misnomer that raised false perceptions that the disease was transmitted through eating pork (28). While the WHO has declared the pandemic to be over, the pandemic H1N1 strain continues to circulate along with other seasonal influenza viruses in humans and has been transmitted to swine in essentially all major pork-producing countries (9, 29, 49). Interestingly, reassortant viruses comprising elements of the human pandemic virus and contemporary swine viruses have already been identified (23, 25). Thus, it is important to develop swine models and vaccines that target both pandemic and classic strains of H1N1 swine flu virus; an effective pig vaccine may protect the pork industry from economic losses while curbing the development of virulent flu virus strains that may threaten public health.
Currently available commercial swine influenza vaccines are traditionally inactivated, whole-virus vaccines containing H3N2 and H1N1 subtype SIVs produced in embryonated eggs. While these vaccines are efficacious in stimulating high antibody responses, protection is afforded only when the hemagglutinin (HA) immunogen matches that of the challenge virus closely. Inactivated-virus vaccines do not effectively protect against heterovariant or heterosubtypic challenges (3, 6, 21, 42), including the pandemic H1N1 strain (13), and in some cases may even enhance disease (44). Studies have suggested that cell-mediated and/or mucosal responses, which are not stimulated by inactivated-virus vaccines, are essential to induce heterosubtypic immunity (21, 40, 41). Furthermore, the present system of production does not allow for timely responses to novel outbreaks and requires large biocontainment facilities.
DNA vaccination may offer several advantages over conventional vaccines. Since DNA vaccines can carry multiple genes from various strains and subtypes, they can offer an umbrella of broad protection by multivalent constructs and prevent escape mutations of influenza virus. Furthermore, DNA vaccines are not associated with the same risks and biosafety issues as whole-virus vaccines and have been shown to elicit both humoral and cellular responses in a variety of influenza animal challenge models, including mice, ferrets, chickens, and nonhuman primates (5, 19, 21, 26, 27, 46, 50, 52). In this study, our primary objective was to establish the proof-of-principle that DNA vaccination is immunogenic and protective against both classic SIV and the pandemic H1N1 strain in a pig challenge model. A secondary objective was to evaluate a needle-free (NF) vaccine delivery method as a potential alternative to conventional needle/syringe (NS) injection. In addition to potentially enhancing the response, NF delivery offers several advantages over NS injections, including less pain, ease of distribution, and improved vaccine acceptance (1). NF delivery may also reduce vaccination costs in high-risk, low-resource settings because of increased speed of vaccine distribution (39), reduction of safety risks and logistical problems associated with the handling of needles and syringes (1, 8), and reduction of training for health care personnel needed to perform vaccinations (1).
MATERIALS AND METHODS
Immunogen and plasmid construction.
Plasmids encoding HAs from A/swine/Ohio/2007 (classic H1N1; GenBank accession no. EU604689), A/swine/North Carolina/2008 (H3N2; GenBank accession no. ACS92895), and A/California/2009 (pandemic H1N1) were synthesized by GeneArt (Regensburg, Germany). HA genes were synthesized using mammal-preferred codons as described previously (14) and constructed in a backbone comprising the cytomegalovirus enhancer/promoter and the human T-cell leukemia virus type 1 R region (CMV/R) by GeneArt (Regensburg, Germany) as described previously (14, 31). These cytomegalovirus vectors are optimized and are the same as those approved for use in human clinical trials.
Pigs and immunization.
The experimental outline is presented in Table 1. Eighty 3-week-old cross-bred pigs were obtained from a herd free of SIV and porcine reproductive and respiratory syndrome virus (PRRSV) and treated with ceftiofur crystalline-free acid according to the label directions (Pfizer Animal Health, New York, NY) to reduce bacterial contaminants prior to the start of the study. Each pig was screened for prior influenza infections, randomly assigned to one of 8 groups, and vaccinated with a prime and 2 homologous boosts at 3 and 6 weeks postpriming with 4 mg of DNA in 1 ml phosphate-buffered saline (PBS). All animals were immunized intramuscularly (i.m.) in the postauricular region of the neck by using either conventional needle and syringe injection or a needle-free 0.5-ml subcutaneous (s.c.)/i.m. injection system in accordance with the instructions of the manufacturer (PharmaJet, Inc., Golden, CO). Pigs were housed at the National Animal Disease Center (NADC), USDA (Ames, IA), in animal biosafety level 2 (ABSL-2) containment facilities during the vaccine phase of the study. On the day of challenge, pigs were transferred to an ABSL-3 containment facility for the remainder of the experiment. All procedures were approved by and were in compliance with the guidelines of the institutional animal care and use committees of the NADC and the Vaccine Research Center, NIAID, NIH (Bethesda, MD). Logistical and financial constraints prevented the inclusion of a conventional inactivated-virus vaccine treatment group to compare to DNA-vaccinated animals. Negative control groups were inoculated with an empty sham DNA plasmid not carrying influenza virus gene sequences.
Table 1.
Experimental schema evaluating immunogenicity, protection, and needle-free injectiona
| Group | DNA vaccine (delivery method) | H1N1 challenge virus |
|---|---|---|
| 1 | Control (sham DNA) (NS injection) | A/Ohio/2007 |
| 2 | Control (sham DNA) (NS injection) | A/California/2009 |
| 3 | Trivalent (NS injection) | A/Ohio/2007 |
| 4 | Trivalent (NS injection) | A/California/2009 |
| 5 | Monovalent (NS injection) | A/California/2009 |
| 6 | Trivalent (NF delivery) | A/Ohio/2007 |
| 7 | Trivalent (NF delivery) | A/California/2009 |
| 8 | Monovalent (NF delivery) | A/California/2009 |
The treatment group number, type of vaccine (and delivery method), and challenge virus are presented. The monovalent DNA vaccine encodes HA from H1N1 A/California/2009, while the trivalent DNA vaccine encodes HAs from H1N1 A/California/2009, H1N1 A/Ohio/2007, and H3N2 A/North Carolina/2008. The negative control group was inoculated with empty sham DNA. Each group contained 10 animals that received a 4-mg/ml dose of vaccine at weeks 0, 3, and 6. Animals were challenged with H1N1 virus at week 9.
HI assay.
Pig serum was collected 1 week prior to each immunization and immediately prior to challenge (at weeks −1, 2, 5, 8, and 9). For each of these time points, a hemagglutination inhibition (HI) assay was performed with homologous virus strains to assess antibody responses to vaccine treatments as described previously (42). Briefly, sera were heat inactivated at 56°C for 30 min and then treated with a 20% suspension of kaolin (Sigma-Aldrich, St. Louis, MO) and subjected to adsorption with 0.5% turkey red blood cells (RBC) to remove nonspecific hemagglutinin inhibitors and natural serum agglutinins. The HI assays were then performed using virus strains homologous to the challenge strain for each group. An additional HI assay with all three challenge strains was performed on sera collected prior to challenge to measure heterologous antibody responses. Titers were determined using 2-fold serial dilutions to detect the endpoint of HI and reported as log10 transformations.
Production of pseudotype lentiviral vectors and measurement of neutralizing antibodies.
To confirm HI assay results and evaluate levels of neutralizing antibody responses, collected sera were pooled for each time point and tested using a pseudotype lentiviral inhibition assay. Production of pseudotype lentiviral vectors for H1N1 and neutralization of pseudotype viruses were performed as described previously (47). Due to logistical constraints and the highly intensive nature of this assay, individual samples could not be analyzed.
Measurement of IFN-γ response by ELISpot assay.
To assess vaccine-induced gamma interferon (IFN-γ) responses, approximately 8 ml of blood was collected 1 week prior to challenge into a BD Vacutainer CPT tube with sodium citrate, and the peripheral blood mononuclear cell (PBMC) fraction was collected according to the tube manufacturer's recommendations. PBMCs were washed once with RPMI 1640 (Invitrogen), run over a 40-μm screen filter, washed a second time, and enumerated. An enzyme-linked immunosorbent spot (ELISpot) assay for IFN-γ-secreting cells was performed as described previously with slight modifications (54). Briefly, 96-well membrane plates (catalog no. MAIPS4510; Millipore) were prewet with 35% ethanol, washed, and coated overnight at 4°C with 6 μg/ml anti-porcine IFN-γ (P2G10; BD Biosciences). The next day, the plate was washed and blocked with complete RPMI (RPMI 1640, 10% fetal calf serum, 2 mM l-glutamine, 1% antibiotic/antimycotic [Invitrogen], and gentamicin) for 2 h at 37°C. The blocking medium was removed, and 2.5 × 105 PBMCs were plated per well. Treatment preparations were added to appropriate wells (each treatment was carried out in triplicate), and the plates were incubated for 18 h at 37°C and 5% CO2. Treatments included live influenza virus at a multiplicity of infection (MOI) of 0.5, control MDCK medium, or concanavalin A added at 5 μg/ml. After 18 h, plates were washed and incubated with 0.5 μg/ml anti-IFN-γ detection antibody (P2C11; BD Biosciences) for 2 h at 37°C. Plates were washed and developed using the ELISpot blue color module according to the recommendations of the manufacturer (R&D Systems). Plates were scanned and spots were enumerated using CTL-ImmunoSpot S5 UV analyzer and ImmunoSpot software. The number of PMBC samples analyzed for each treatment group ranged from 3 to 7.
H1N1 influenza virus challenge.
Three weeks after the final boost, all pigs were challenged intranasally with 2 × 106 50% tissue culture infective doses (TCID50)/pig of either A/Ohio/2007 H1N1 or pandemic A/California/2009 H1N1 virus prepared in MDCK cells. As indicated in Table 1, pigs that received monovalent and trivalent DNA vaccines were challenged with A/California/2009 H1N1, while only the trivalent vaccine was tested against challenge with A/Ohio/2007 H1N1. Pigs were observed daily for clinical signs of disease. Nasal swab samples were taken with Fisherbrand Dacron swabs (Fisher Scientific, Pittsburg, PA) at 0, 3, 5, and 7 days postchallenge (dpc) to evaluate nasal virus shedding by wetting the swab in minimal essential medium (MEM) and inserting the swab approximately 2.5 cm into each nare. Samples were stored at −80°C until testing. Five pigs per group were humanely euthanized with a lethal dose of pentobarbital (Sleepaway; Fort Dodge Animal Health, Fort Dodge, IA) at 5 dpc to evaluate lung lesions and viral replication in the lung. The remaining challenged pigs were euthanized at 12 dpc, and the same types of samples were collected.
Viral shedding and quantitation.
A real-time PCR assay developed to detect avian influenza A viruses (37) was used for detecting the swine viruses, and this assay was modified to detect the pandemic H1N1 virus (20). For virus isolation, samples were thawed, subjected to a vortex for 15 s, and centrifuged for 10 min at 640 × g and the supernatant was passed through 0.45-μm filters to reduce bacterial contaminants. Nasal swabs (200 μl) were placed on confluent MDCK cells in 24-well plates to incubate for 1 h, after which the sample was removed and 400 μl MEM with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin was added. The plate was checked at 24 and 48 h for cytopathic effects (CPE). After 48 h, the plate was frozen and thawed one time and all samples (200 μl) were blindly passed onto a confluent 48-well plate. After 48 h, the plates were evaluated for CPE and samples were fixed with 4% phosphate-buffered formalin and stained with an anti-influenza A virus nucleoprotein monoclonal antibody as described previously (43, 45) to detect virus antigen. Filtered samples were thawed and placed on MDCK cells in 96-well plates. Tenfold serial dilutions in serum-free MEM supplemented with TPCK trypsin and antibiotics were made with each positive nasal swab sample. Each dilution was plated in triplicate in 100 μl volumes onto PBS-washed confluent MDCK cells in 96-well plates. Plates were evaluated for CPE between 48 and 72 h postinfection. At 72 h, plates were fixed and stained using immunocytochemistry as described above. A TCID50 titer was calculated for each sample by the Reed and Muench method (38).
Histopathology and immunohistochemistry.
At 5 dpc, 5 pigs/group were euthanized and lung samples were collected for histopathologic evaluation. Tissues were fixed by immersion for 24 h in 10% neutral buffered formalin, processed routinely in an automated tissue processor, embedded in paraffin, sectioned at 6 μm, and stained with hematoxylin and eosin. Sections were cut at 3 μm, mounted onto poly-l-lysine-coated slides, and processed for immunohistochemical (IHC) evaluation of the presence of influenza A virus-specific antigen using a previously described method (45). IHC staining was applied to lung sections from all pigs. Tissues were evaluated by a board-certified pathologist for histopathologic lesions characteristic of influenza virus infection.
Statistical analysis.
For HI antibody responses, NF-delivery groups were compared to NS injection groups by using a Mann-Whitney t test with GraphPad Prism software (GraphPad, San Diego, CA). For IFN-γ responses and viral load titers, each vaccinated group was compared to controls by using the same test and software. Bonferroni's adjustment was applied to account for multiple comparisons; thus, differences with P values of less than 0.05, divided by the number of comparisons, were considered significant.
RESULTS
Antibody responses in immunized pigs.
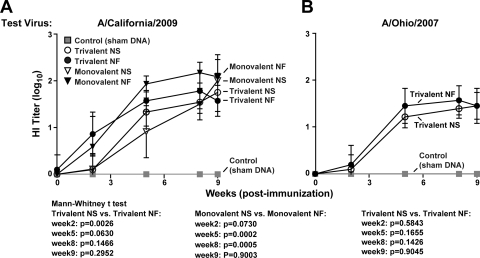
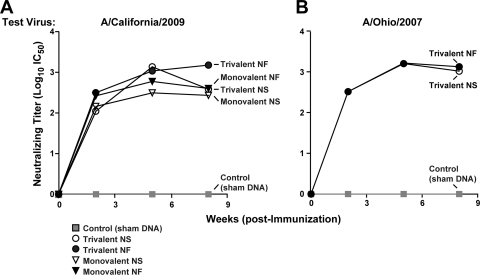
HI antibody assays demonstrated that DNA vaccination elicited robust levels of antibody response against both A/California/2009 (Fig. 1 A) and A/Ohio/2007 (Fig. 1B). Against A/California/2009, NF trivalent vaccine groups showed significantly higher responses than NS-injected trivalent vaccine groups after the first immunization (P = 0.0026) but not after the second immunization (Fig. 1A). NF monovalent vaccine groups showed significantly higher antibody titers than NS-injected monovalent vaccine groups after the second and third immunizations (P = 0.0002 and 0.0005, respectively). Immediately prior to challenge, both NS injection and NF-delivery animals showed robust HI titers that were not statistically different from each other. Against A/Ohio/2007 (Fig. 1B), the two trivalent vaccines stimulated similar levels of antibody responses, which were enhanced by the first boost and sustained by the second. A follow-up lentiviral pseudotype inhibition assay supported this finding, and while results for treatment groups did not agree with the HI assay in terms of relative immunogenicity, all vaccinated groups showed robust levels of neutralizing antibodies (Fig. 2). NS and NF delivery elicited similar levels of neutralizing antibody responses, although statistical differences could not be determined since sera were pooled for this assay.
Fig. 1.
DNA vaccination elicits anti-H1N1 antibody responses detectable by an HI assay. Antibody titers against A/California/2009 and A/Ohio/2007 in vaccinated and control (sham DNA-vaccinated) pigs were determined by an HI assay, with 10 pigs/group. Log10 transformations of the data are plotted with error bars representing standard deviations (SD). For A/California/2009, data for five treatment groups are presented: animals receiving a monovalent NS vaccine, a monovalent NF vaccine, a trivalent NS vaccine, or a trivalent NF vaccine and controls. The same symbols are used for the HI tests against A/Ohio/2007, excluding the monovalent vaccine groups.
Fig. 2.
A pseudotype lentiviral inhibition assay confirms anti-H1N1 neutralizing antibody responses in pooled sera. Neutralizing antibody responses against A/California/2009 and A/Ohio/2007 were confirmed by a pseudotype inhibition assay performed on pooled sera from each treatment group, with 10 pigs/group. Neutralizing antibody titers are presented as log10 transformations of half-maximal inhibitory concentrations (IC50). Data points are plotted for each treatment group with the same symbols used in Fig. 1.
IFN-γ responses in immunized pigs.
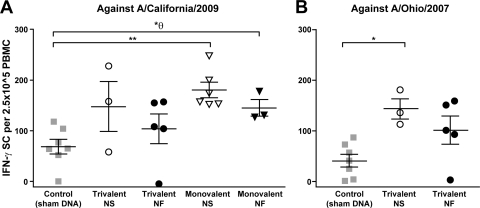
All immunized animals tested showed IFN-γ responses against both A/California/2009 and A/Ohio/2007, as IFN-γ-secreting cells (ISCs) were detected in PBMC samples (Fig. 3). Against A/California/2009, only the NS-injected monovalent vaccine group showed a significant difference compared to controls (P = 0.0012) (Fig. 3A). The NF monovalent vaccine group initially showed a significant difference by the Mann-Whitney t test (P = 0.017); however, only P values of less than 0.0125 were considered significant with Bonferroni's adjustment applied. Against A/Ohio/2007, both groups of immunized animals elicited cellular responses, although only the NS-injected trivalent vaccine group showed statistically higher responses than controls (P = 0.017) (Fig. 3B). NF-delivery groups did not statistically differ from NS injection groups, regardless of the use of trivalent (P = 0.4341) or monovalent (P = 0.3502) constructs. Due to technical issues with blood collection, the number of samples tested for some of the vaccination groups was quite small. Control animals showed relatively high background levels of IFN-γ responses that are not uncommon given the high percentage of circulating gamma-delta T cells in this species as well as sensitivity to the PBMC isolation method, as described previously (2).
Fig. 3.
DNA vaccination elicits IFN-γ responses measured by an ELISpot assay with PBMCs collected 1 week prior to H1N1 challenge. An ELISpot assay was used to detect cells secreting IFN-γ against A/California/2009 and A/Ohio/2007 in vaccinated animals. Results are presented as single data points in a group, with lines indicating the mean and standard error of the mean (SEM) for each group. A Mann-Whitney t test was used for statistical analysis comparing values between immunized and nonimmunized control animals. P values of less than 0.05 are indicated: * represents a P value between 0.05 and 0.001, while ** indicates a P value of ≤0.001. θ indicates a value that is nonsignificant when Bonferroni's adjustment is applied.
Postchallenge viral shedding.
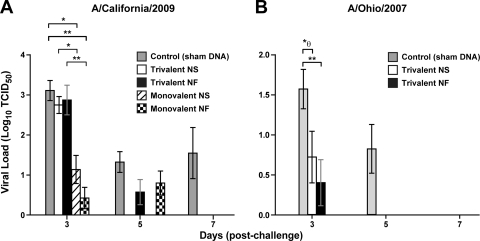
In animals challenged with A/California/2009, plaque assays demonstrated a significant reduction in viral loads by 3 dpc in both NS-injected monovalent vaccine animals (P = 0.002) and NF monovalent vaccine animals (P = 0.0004) compared to controls (Fig. 4 A). Interestingly, despite similar HI titers, monovalent vaccine groups showed significant protection compared to trivalent vaccine groups, among both NS-injected animals (P = 0.0031) and NF-delivery animals (P = 0.0009). Animals vaccinated with the trivalent vaccine had viral loads similar to those of controls at 3 dpc, but the NS-immunized group showed full reduction by 5 dpc and the NF-delivery group showed full reduction by 7 dpc, while A/California/2009 virus persisted in sham-vaccinated controls (Fig. 4A). For animals challenged with A/Ohio/2007, compared to nonvaccinated controls, a significant decrease in viral load was detected 3 dpc in NF trivalent vaccine pigs (P = 0.004) (Fig. 4B). Initially, the NS-injected trivalent vaccine group showed a significant difference by the Mann-Whitney test (P = 0.047), but the difference was considered insignificant after Bonferroni's adjustment. At 5 dpc, both vaccinated groups cleared A/Ohio/2007 virus to undetectable levels while viral loads persisted in controls.
Fig. 4.
DNA vaccination reduces viral load and protects against H1N1 influenza virus challenge. Immunized and control animals were challenged with either H1N1 A/California/2009 or A/Ohio/2007. Postchallenge viral loads were assessed for up to 7 days using a plaque reduction assay. Viral titers are presented as the log of the TCID50 for each animal, with bars indicating means and error bars indicating SEMs. Differences between immunized animals and controls were analyzed for statistical significance using a Mann-Whitney t test. P values of less than 0.05 are indicated: * represents a P value between 0.05 and 0.001, while ** indicates a value of ≤0.001. θ indicates a value that is nonsignificant when Bonferroni's adjustment is applied.
Histopathology and IHC detection of influenza virus antigen in the lungs of H1N1-challenged pigs.
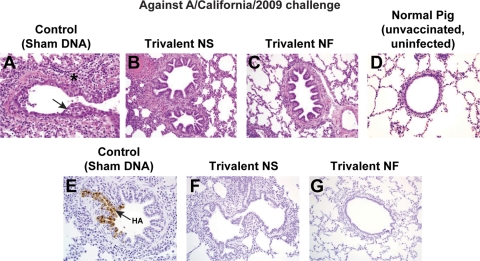
Necrotizing bronchiolitis with peribronchiolar lymphocytic cuffing and mild interstitial pneumonia consistent with influenza infection were evident in 2 of 5 sham-vaccinated pigs challenged with A/California/2009 (Fig. 5) and in 3 of 5 sham-vaccinated pigs challenged with A/Ohio/2007 (Fig. 6) that were euthanized at 5 dpc. In a representative control (sham-vaccinated) pig challenged with A/California/2009 (Fig. 5), the epithelium lining the bronchiole is irregular (arrow) due to necrosis and sloughing of infected cells and reactive flattening of the remaining cells to cover the defect. Prominent peribronchiolar lymphocyte infiltration (*) is evident. In the vaccinated pigs, a normal epithelial layer of well-ordered cuboidal to columnar cells lines the airway, and a few infiltrating lymphocytes can be seen around the airways in some of the vaccinated pigs, particularly in the NS-injected trivalent vaccine group. In the control IHC photomicrograph, virus antigen in bronchiolar epithelial cells (arrow) appears as brown coloration of the nucleus and cytoplasm. Virus infection in this airway has not yet resulted in epithelial cell necrosis. Influenza virus antigen was detected by IHC analysis in the lungs of all 5 pigs challenged with A/California/2009, while no virus antigen was detected in the bronchioles from vaccinated pigs.
Fig. 5.
DNA vaccination elicits protection from lung disease induced by A/California/2009 challenge and prevents viral replication in the lung. Lungs were collected from 5 pigs per group at 5 dpc with A/California/2009. Hematoxylin and eosin staining was performed to assess histopathology, and immunohistochemistry analysis was performed to detect influenza virus antigens in the lung. Micrographs are representative for each treatment group (including monovalent NS and NF vaccine groups) at a magnification of ×20. See Results for lesion description. In panel A, the epithelium lining the bronchiole is irregular (arrow), and prominent peribronchiolar lymphocyte infiltration (*) is evident. In the control IHC photomicrograph in panel E, virus antigen in bronchiolar epithelial cells (arrow) appears as brown coloration of the nucleus and cytoplasm.
Fig. 6.
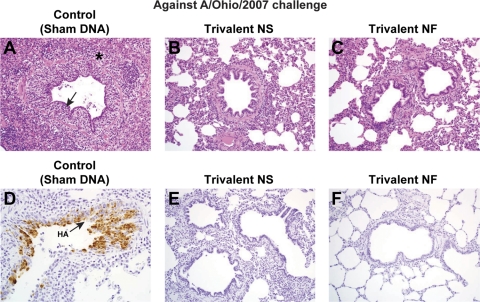
DNA vaccination elicits protection from lung disease induced by A/Ohio/2007 challenge and prevents viral replication in the lung. Lungs were collected from 5 pigs per group at 5 dpc with A/Ohio/2007. Hematoxylin and eosin staining was performed to assess histopathology, and immunohistochemistry analysis was performed to detect influenza virus antigens in the lung. Micrographs are representative for each treatment group at a magnification of ×20, except for one animal immunized with a trivalent NF vaccine in which virus was detected. See Results for lesion description. In panel A, the epithelium lining the bronchiole is irregular (arrow), and prominent peribronchiolar lymphocyte infiltration (*) is evident. In the control IHC photomicrograph in panel D, virus antigen in bronchiolar epithelial cells (arrow) appears as brown coloration of the nucleus and cytoplasm.
In a representative control (sham-vaccinated) pig challenged with A/Ohio/2007 (Fig. 6), the epithelium lining the bronchiole is severely attenuated (arrow) due to necrosis and sloughing of infected cells and reactive flattening of the remaining cells to cover the defect. Prominent subepithelial and peribronchiolar lymphocyte infiltration (*) is evident. In the vaccinated pigs, a normal epithelial layer of well-ordered cuboidal to columnar cells lines the airway and minimal or no lymphocyte infiltration is evident. In the control IHC photomicrograph, virus antigen in disorganized bronchiolar epithelial cells (arrow) appears as brown coloration of the nucleus and cytoplasm. Influenza virus antigen was detected by IHC analysis in the lungs of 3 of 5 pigs challenged with A/Ohio/2007 (Fig. 6), while no virus antigen was detected in the bronchioles from the vaccinated pigs.
DISCUSSION
Current available swine influenza vaccines are whole, inactivated viruses that have limited efficacy against heterologous strains, including new emerging strains. This can result in substantial economic losses to the farmer and an increased likelihood of the production of novel viruses. Ideally, the next generation of influenza virus vaccines will induce a balanced immune response that has broad cross-protection with production that is more efficient than that of inactivated virus vaccines that are prepared in embryonated chicken eggs. DNA vaccines have been shown to meet these criteria (5, 19, 21, 26, 27, 46, 50, 52). Here, we tested the abilities of a DNA vaccine to elicit humoral and cellular immune responses and protect against challenge with classic swine or pandemic H1N1 influenza virus.
In all vaccinated animals, robust humoral responses against both A/California/2009 and A/Ohio/2007 were detected by an HI assay, and neutralization was confirmed by a pseudotype lentiviral inhibition assay. This pseudotype assay has been shown to be more sensitive than microneutralization assays and does not require the use of BSL-2 or BSL-3 conditions (14, 47, 48, 51). The induction of prechallenge antibody responses, with a marked boosting effect from the second immunization, is consistent with results from previous DNA vaccine studies with pigs (17, 22). Interestingly, the trivalent DNA vaccine elicited titers against A/California/2009 similar to those elicited by the monovalent vaccine, despite containing only a third of the dose of A/California/2009 HA immunogen (1.33 mg in the trivalent vaccine compared to 4 mg in the monovalent vaccine). While this may be due to cross-reactivity with the conserved epitopes shared between the three HA genes, it supports the hypothesis that DNA vaccines can encode multiple influenza immunogens to enhance the breadth of responses without sacrificing the magnitude of response. This is further supported by our detection of HI antibody responses against and reduction of the load of A/Illinois/2009 H3N2, another strain that contributes to classic swine influenza (see Fig. S1 in the supplemental material). Due to limited resources, only one vaccinated group was challenged with H3N2 to demonstrate immunogenicity and protection against all three HAs included in our trivalent construct. The A/Illinois/2009 strain is 98% homologous to A/North Carolina/2008, which was encoded in our vaccine, with 100% identity in the regions believed to encode the antigenic HA protein, and was used due to limited availability of A/North Carolina/2008.
The significant detection of influenza virus-specific ISCs in PBMCs in DNA-vaccinated pigs is consistent with previous experiments in which DNA vaccines induced both humoral and cellular immune responses in other animal models (19, 26, 50). However, in previous DNA vaccine studies with pigs, cellular immune responses have been detected only at low levels in the spleen and in pigs covaccinated with killed vaccines (17). Studies have also suggested that such cell-mediated responses may be essential to induce heterosubtypic immunity against influenza (21, 40, 41). Further experiments with more robust sample sizes may be needed to confirm our findings and to delineate between CD4-specific and CD8-specific cellular responses.
In previous challenge experiments with pigs, DNA vaccines have exhibited only partial protection against classic swine influenza virus strains with significant reduction of viral loads at 5 dpc or later (17, 18, 22). However, significant viral load reduction at 3 dpc was observed previously when a DNA prime was boosted by a conventional killed vaccine (17). Commercial inactivated viruses also provide only partial protection against pandemic H1N1 with viral load reduction at 5 dpc (42). In our study, pigs immunized with the monovalent DNA vaccine showed significant reduction in A/California/2009 viral loads by 3 dpc, similar to the significant protection demonstrated by the DNA prime, killed-vaccine boost regimen (17). While similar protection was also conferred by our trivalent vaccine, it is interesting that monovalent constructs reduced viral titers earlier than trivalent constructs against the pandemic strain, despite similarities in prechallenge antibody responses. It is possible that homologous antibody responses, rather than broad stimulation, may be preferable to confer protection against a single strain. This could be due to more specific stimulation of higher-quantity and higher-quality neutralizing antibody responses in the monovalent vaccine groups, as well as other responses that may augment the activity of T cells. This could also be the result of antibody responses against unmatched HA antigens in trivalent vaccine groups interfering with the protective activity of the HA matched to the challenge strain. Another possible explanation is that trivalent vaccines stimulated antibody responses that, while comparable to those stimulated by the monovalent vaccines, were just below the threshold of protection, whereas monovalent vaccine groups reached a titer just above the threshold, as discussed previously (24). Nevertheless, all monovalent and trivalent vaccine groups cleared A/California/2009 infection by 7 dpc and A/Ohio/2007 infection by 5 dpc; thus, considerable protection is afforded by either construct.
While influenza virus antigen and lung lesions characteristic of influenza virus infection were detected in the lungs of sham-vaccinated control animals infected with A/Ohio/2007, there was nearly complete blockage of lesion development and almost total elimination of virus-infected cells detectable by IHC analysis in the immunized groups, indicating a protective effect by both the trivalent and monovalent DNA vaccines. Sham-vaccinated pigs infected with A/California/2009 developed lung disease that was less severe than that in pigs challenged with A/Ohio/2007, but all vaccinated pigs were also noticeably protected against viral replication in the lung and the development of lesions. While some lymphocyte infiltration was observed in vaccinated groups, particularly NS-injected trivalent vaccine animals, this was considered by board-certified pathologists (with extensive swine experience) to be within normal limits. This suggests a potential effect of either circulating neutralizing antibodies or T lymphocytes at the site of viral replication, although more studies are required to characterize this observation.
The higher levels of protection observed in this study than in previous DNA studies may be attributed to a number of variables, including differences in codon optimization, encoded immunogens, dose, and methods of inoculation, as well as a three-immunization regimen compared to a two-immunization regimen. Interestingly, vaccine groups that showed significant IFN-γ responses also had significant protection against A/California/2009 at 3 dpc compared to groups that did not show significant IFN-γ responses. This was similarly demonstrated for a classic SIV strain by Larsen et al. (17), suggesting that while HA-specific antibody responses may be indicated as a primary factor in protection (3, 17, 30, 34, 35), cellular immune responses may also be associated with enhanced protection.
Immunogenicity data and postchallenge results suggest that NF delivery may be advantageous compared to conventional NS injection in efficiency and protection. NF delivery elicited slightly higher levels of antibody responses, comparable IFN-γ immune responses, and similar protection against viral replication and lung lesions. Previous studies have indicated that NF delivery particularly enhances DNA vaccination against a variety of diseases, including influenza, HIV infection, dengue fever, and others (1, 32, 33). Given that there are no disadvantages in efficiency and protection compared to conventional parenteral injection, NF delivery should be considered a practical alternative method for vaccine administration.
By reducing and clearing influenza virus earlier in the infection period and abrogating viral replication in the lung, DNA vaccination may prevent the development of clinical disease, the spread of virus to other animals or humans, and the formation of novel virulent strains. However, while the protection of experimental pigs against classic and pandemic influenza is promising, more effort will be required to develop DNA vaccines into a viable and practical alternative. Future studies will focus on optimizing DNA vaccine efficacy and cost-efficiency by evaluating variables such as dose and coadministration of adjuvants, while also continuing to shed light on precise mechanisms of protection.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIH Vaccine Research Center.
We thank Judy Stein for assistance in coordinating material transfers and managing regulatory issues, Amanda Burow and Deborah Adolphson for technical assistance, Jason Huegel and Brian Pottebaum for animal care, Martha Nason for assistance in statistical analysis, Mythreyi Shastri and Brenda Hartman for assistance in manuscript preparation, and Linda Bessacque for administrative support.
Gary J. Nabel, Srinivas S. Rao, Wing-Pui Kong, and Chih-Jen Wei are each listed on a patent filing for our DNA vaccine technology, U.S. patent application 2/838,292, which is an adjunct to an existing patent, “Influenza DNA Vaccination and Methods of Use Thereof,” U.S. patent 61/023,341.
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. The USDA is an equal-opportunity provider and employer.
Footnotes
Supplemental material for this article may be found at http://cvi.asm.org/.
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Amorij J. P., Hinrichs W. L., Frijlink H. W., Wilschut J. C., Huckriede A. 2010. Needle-free influenza vaccination. Lancet Infect. Dis. 10:699–711 [DOI] [PubMed] [Google Scholar]
- 2. Armengol E., et al. 2002. Identification of T-cell epitopes in the structural and non-structural proteins of classical swine fever virus. J. Gen. Virol. 83:551–560 [DOI] [PubMed] [Google Scholar]
- 3. Bikour M. H., Cornaglia E., Elazhary Y. 1996. Evaluation of a protective immunity induced by an inactivated influenza H3N2 vaccine after an intratracheal challenge of pigs. Can. J. Vet. Res. 60:312–314 [PMC free article] [PubMed] [Google Scholar]
- 4. Castrucci M. R., et al. 1994. Antigenic and sequence analysis of H3 influenza virus haemagglutinins from pigs in Italy. J. Gen. Virol. 75(Pt. 2):371–379 [DOI] [PubMed] [Google Scholar]
- 5. Chen M. W., et al. 2008. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 105:13538–13543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Durrwald R., et al. 2010. Swine influenza A vaccines, pandemic (H1N1) 2009 virus, and cross-reactivity. Emerg. Infect. Dis. 16:1029–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Easterday B., Van Reeth K. 1999. Swine influenza, p. 277–290 In Straw B., D'Allaire S., Mengeling W., Taylor D. (ed.), Diseases of swine. Iowa State University Press, Ames, IA [Google Scholar]
- 8. Ekwueme D. U., Weniger B. G., Chen R. T. 2002. Model-based estimates of risks of disease transmission and economic costs of seven injection devices in sub-Saharan Africa. Bull. World Health Organ. 80:859–870 [PMC free article] [PubMed] [Google Scholar]
- 9. Forgie S. E., et al. 2011. Swine outbreak of pandemic influenza A virus on a Canadian research farm supports human-to-swine transmission. Clin. Infect. Dis. 52:10–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Furuse Y., Suzuki A., Oshitani H. 2010. Reassortment between swine influenza A viruses increased their adaptation to humans in pandemic H1N1/09. Infect. Genet. Evol. 10:569–574 [DOI] [PubMed] [Google Scholar]
- 11. Girard M. P., Tam J. S., Assossou O. M., Kieny M. P. 2010. The 2009 A (H1N1) influenza virus pandemic: a review. Vaccine 28:4895–4902 [DOI] [PubMed] [Google Scholar]
- 12. Kida H., et al. 1994. Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 75(Pt. 9):2183–2188 [DOI] [PubMed] [Google Scholar]
- 13. Kobinger G. P., et al. 2010. Assessment of the efficacy of commercially available and candidate vaccines against a pandemic H1N1 2009 virus. J. Infect. Dis. 201:1000–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kong W. P., et al. 2006. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proc. Natl. Acad. Sci. U. S. A. 103:15987–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kothalawala H., Toussaint M. J., Gruys E. 2006. An overview of swine influenza. Vet. Q. 28:46–53 [PubMed] [Google Scholar]
- 16. Kundin W. D. 1970. Hong Kong A-2 influenza virus infection among swine during a human epidemic in Taiwan. Nature 228:857. [DOI] [PubMed] [Google Scholar]
- 17. Larsen D. L., Karasin A., Olsen C. W. 2001. Immunization of pigs against influenza virus infection by DNA vaccine priming followed by killed-virus vaccine boosting. Vaccine 19:2842–2853 [DOI] [PubMed] [Google Scholar]
- 18. Larsen D. L., Olsen C. W. 2002. Effects of DNA dose, route of vaccination, and coadministration of porcine interleukin-6 DNA on results of DNA vaccination against influenza virus infection in pigs. Am. J. Vet. Res. 63:653–659 [DOI] [PubMed] [Google Scholar]
- 19. Liu M. A., McClements W., Ulmer J. B., Shiver J., Donnelly J. 1997. Immunization of non-human primates with DNA vaccines. Vaccine 15:909–912 [DOI] [PubMed] [Google Scholar]
- 20. Lorusso A., Faaberg K. S., Killian M. L., Koster L., Vincent A. L. 2010. One-step real-time RT-PCR for pandemic influenza A virus (H1N1) 2009 matrix gene detection in swine samples. J. Virol. Methods 164:83–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma W., Richt J. A. 2010. Swine influenza vaccines: current status and future perspectives. Anim. Health Res. Rev. 11:81–96 [DOI] [PubMed] [Google Scholar]
- 22. Macklin M. D., et al. 1998. Immunization of pigs with a particle-mediated DNA vaccine to influenza A virus protects against challenge with homologous virus. J. Virol. 72:1491–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moreno A., et al. 2011. Novel H1N2 swine influenza reassortant strain in pigs derived from the pandemic H1N1/2009 virus. Vet. Microbiol. 149:472–477 [DOI] [PubMed] [Google Scholar]
- 24. Nauta J. J., Beyer W. E., Osterhaus A. D. 2009. On the relationship between mean antibody level, seroprotection and clinical protection from influenza. Biologicals 37:216–221 [DOI] [PubMed] [Google Scholar]
- 25. Octaviani C. P., Li C., Noda T., Kawaoka Y. 2011. Reassortment between seasonal and swine-origin H1N1 influenza viruses generates viruses with enhanced growth capability in cell culture. Virus Res. 156:147–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Okuda K., et al. 2001. Protective immunity against influenza A virus induced by immunization with DNA plasmid containing influenza M gene. Vaccine 19:3681–3691 [DOI] [PubMed] [Google Scholar]
- 27. Oveissi S., Omar A. R., Yusoff K., Jahanshiri F., Hassan S. S. 2010. DNA vaccine encoding avian influenza virus H5 and Esat-6 of Mycobacterium tuberculosis improved antibody responses against AIV in chickens. Comp. Immunol. Microbiol. Infect. Dis. 33:491–503 [DOI] [PubMed] [Google Scholar]
- 28. Pappaioanou M., Gramer M. 2010. Lessons from pandemic H1N1 2009 to improve prevention, detection, and response to influenza pandemics from a One Health perspective. ILAR J. 51:268–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pasma T., Joseph T. 2010. Pandemic (H1N1) 2009 infection in swine herds, Manitoba, Canada. Emerg. Infect. Dis. 16:706–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Potter C. W., Oxford J. S. 1979. Determinants of immunity to influenza infection in man. Br. Med. Bull. 35:69–75 [DOI] [PubMed] [Google Scholar]
- 31. Rao S., et al. 2008. Multivalent HA DNA vaccination protects against highly pathogenic H5N1 avian influenza infection in chickens and mice. PLoS One 3:e2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Raviprakash K., et al. 2003. Needle-free Biojector injection of a dengue virus type 1 DNA vaccine with human immunostimulatory sequences and the GM-CSF gene increases immunogenicity and protection from virus challenge in Aotus monkeys. Virology 315:345–352 [DOI] [PubMed] [Google Scholar]
- 33. Raviprakash K., Porter K. R. 2006. Needle-free injection of DNA vaccines: a brief overview and methodology. Methods Mol. Med. 127:83–89 [DOI] [PubMed] [Google Scholar]
- 34. Ruben F. L., Akers L. W., Stanley E. D., Jackson G. G. 1973. Protection with split and whole virus vaccines against influenza. Arch. Intern. Med. 132:568–571 [PubMed] [Google Scholar]
- 35. Scherle P. A., Palladino G., Gerhard W. 1992. Mice can recover from pulmonary influenza virus infection in the absence of class I-restricted cytotoxic T cells. J. Immunol. 148:212–217 [PubMed] [Google Scholar]
- 36. Schultz U., Fitch W. M., Ludwig S., Mandler J., Scholtissek C. 1991. Evolution of pig influenza viruses. Virology 183:61–73 [DOI] [PubMed] [Google Scholar]
- 37. Spackman E., Suarez D. L. 2008. Type A influenza virus detection and quantitation by real-time RT-PCR. Methods Mol. Biol. 436:19–26 [DOI] [PubMed] [Google Scholar]
- 38. Stanic M. 1963. A simplification of the estimation of the 50 percent endpoints according to the Reed and Muench method. Pathol. Microbiol. (Basel) 26:298–302(In German.) [PubMed] [Google Scholar]
- 39. Sugimura T., et al. 2008. Improved antibody responses in infants less than 1 year old using intradermal influenza vaccination. Vaccine 26:2700–2705 [DOI] [PubMed] [Google Scholar]
- 40. Van Reeth K., Brown I., Essen S., Pensaert M. 2004. Genetic relationships, serological cross-reaction and cross-protection between H1N2 and other influenza A virus subtypes endemic in European pigs. Virus Res. 103:115–124 [DOI] [PubMed] [Google Scholar]
- 41. Van Reeth K., Gregory V., Hay A., Pensaert M. 2003. Protection against a European H1N2 swine influenza virus in pigs previously infected with H1N1 and/or H3N2 subtypes. Vaccine 21:1375–1381 [DOI] [PubMed] [Google Scholar]
- 42. Vincent A. L., et al. 2010. Efficacy of inactivated swine influenza virus vaccines against the 2009 A/H1N1 influenza virus in pigs. Vaccine 28:2782–2787 [DOI] [PubMed] [Google Scholar]
- 43. Vincent A. L., et al. 2010. Experimental inoculation of pigs with pandemic H1N1 2009 virus and HI cross-reactivity with contemporary swine influenza virus antisera. Influenza Other Respir. Viruses 4:53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vincent A. L., Lager K. M., Janke B. H., Gramer M. R., Richt J. A. 2008. Failure of protection and enhanced pneumonia with a US H1N2 swine influenza virus in pigs vaccinated with an inactivated classical swine H1N1 vaccine. Vet. Microbiol. 126:310–323 [DOI] [PubMed] [Google Scholar]
- 45. Vincent L. L., Janke B. H., Paul P. S., Halbur P. G. 1997. A monoclonal-antibody-based immunohistochemical method for the detection of swine influenza virus in formalin-fixed, paraffin-embedded tissues. J. Vet. Diagn. Invest. 9:191–195 [DOI] [PubMed] [Google Scholar]
- 46. Webster R. G., Fynan E. F., Santoro J. C., Robinson H. 1994. Protection of ferrets against influenza challenge with a DNA vaccine to the haemagglutinin. Vaccine 12:1495–1498 [DOI] [PubMed] [Google Scholar]
- 47. Wei C. J., et al. 2010. Cross-neutralization of 1918 and 2009 influenza viruses: role of glycans in viral evolution and vaccine design. Sci. Transl. Med. 2:24ra21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wei C. J., et al. 2010. Induction of broadly neutralizing H1N1 influenza antibodies by vaccination. Science 329:1060–1064 [DOI] [PubMed] [Google Scholar]
- 49. Welsh M. D., et al. 2010. Initial incursion of pandemic (H1N1) 2009 influenza A virus into European pigs. Vet. Rec. 166:642–645 [DOI] [PubMed] [Google Scholar]
- 50. Yager E. J., Dean H. J., Fuller D. H. 2009. Prospects for developing an effective particle-mediated DNA vaccine against influenza. Expert Rev. Vaccines 8:1205–1220 [DOI] [PubMed] [Google Scholar]
- 51. Yang Z. Y., et al. 2007. Immunization by avian H5 influenza hemagglutinin mutants with altered receptor binding specificity. Science 317:825–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhang F., et al. 2005. Maternal immunization with both hemagglutinin- and neuraminidase-expressing DNAs provides an enhanced protection against a lethal influenza virus challenge in infant and adult mice. DNA Cell Biol. 24:758–765 [DOI] [PubMed] [Google Scholar]
- 53. Zhou N. N., et al. 1999. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J. Virol. 73:8851–8856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zuckermann F. A., et al. 1998. Interleukin-12 enhances the virus-specific interferon gamma response of pigs to an inactivated pseudorabies virus vaccine. Vet. Immunol. Immunopathol. 63:57–67 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.