Abstract
Cryptosporidiosis is a ubiquitous infectious disease, caused by the protozoan parasites Cryptosporidium hominis and Cryptosporidium parvum, leading to acute, persistent, and chronic diarrhea with life-threatening consequences in immunocompromised individuals. In developing countries, cryptosporidiosis in early childhood has been associated with subsequent significant impairment in growth, physical fitness, and intellectual abilities. Currently, vaccines are unavailable and chemotherapeutics are toxic and impractical, and agents for immunoprophylaxis or treatment of cryptosporidiosis are a high priority. Availability of the genome sequences for C. hominis and C. parvum provides new opportunities to procure and examine novel vaccine candidates. Using the novel approach of “reverse vaccinology,” we identified several new potential vaccine candidates. Three of these antigens—Cp15, profilin, and a Cryptosporidium apyrase—were delivered in heterologous prime-boost regimens as fusions with cytolysin A (ClyA) in a Salmonella live vaccine vector and as purified recombinant antigens, and they were found to induce specific and potent humoral and cellular immune responses, suggesting their potential as new vaccinogens against Cryptosporidium infection.
INTRODUCTION
Cryptosporidiosis is a ubiquitous infectious disease caused primarily by the protozoan parasites Cryptosporidium hominis and Cryptosporidium parvum (21, 38). Transmission occurs by ingestion of infective oocysts, from which invasive sporozoites emerge and invade the intestinal epithelium (35). Proliferation of the parasite in the gastrointestinal (GI) tract often results in acute, persistent, and chronic diarrhea. In the United States, infection rates vary between 1.0 and 1.3 infections per 100,000 persons per year (17). Infections are significantly more common among HIV patients (22). The severity of the clinical manifestations in HIV patients is associated with low CD4+-cell counts (15). Furthermore, the persistent diarrhea caused by Cryptosporidium infection in AIDS patients is potentially life-threatening and was recently recognized as one of the reasons for impairment of antiviral drug adsorption and failure of treatment (4). The current use of more active antiretroviral combination therapy has greatly reduced the incidence of cryptosporidiosis among HIV/AIDS patients (15).
In developing countries, cryptosporidiosis in early childhood has been reported to be associated with subsequent impairment in growth, physical fitness, and intellectual capacity (14). In addition to the prevalence of cryptosporidiosis caused by accidental infection, there is now the increased threat of bioterrorism and deliberate contamination of the water supply with Cryptosporidium. The ease with which Cryptosporidium oocysts can survive processes used at water treatment facilities (including disinfection procedures) combined with the large number of persons that would be at risk from such an attack underscores this possibility. For instance, in 1993 a malfunction in a water treatment facility in Milwaukee, WI, led to an outbreak of Cryptosporidium infection that affected roughly 500,000 persons. Currently, there are no safe and effective vaccines, and new therapeutics for immunocompromised patients are urgently required. Thus, development of alternative therapeutic agents and vaccines to control and/or prevent this disease are a high priority for future public safety and health.
Resistance to and control of C. parvum infection involve a broad activation of the immune system (26). Both innate and adaptive immune mechanisms are triggered during the infection of the intestinal epithelial cells with Cryptosporidium, although final clearance of the parasite requires adaptive immunity. The same adaptive immune response would also likely be required for vaccine-induced immunity.
Invasion of enterocytes by sporozoites activates NF-κB and induces production of interleukin-1β (IL-1β) and inducible nitric oxide synthase (iNOS), leading to protection of these cells (13, 31). Several Toll-like receptors (TLRs) are associated with protection against infection (2, 24, 25, 41). The adaptive immune response to Cryptosporidium is characterized as a T-helper 1 (Th1) response with significant contribution of IL-12, IL-18, IL-23, and gamma interferon (IFN-γ) to the clearance of infection (8). In particular, CD4+ T lymphocytes located in the lamina propria are key components of the immune mucosal response against Cryptosporidium. Although it has been shown that CD8+ T cells produce IFN-γ in response to Cryptosporidium antigens, their role in protection and clearance of the parasite remains undefined (8). Preidis et al. (28) described the production of IFN-γ by peripheral blood mononuclear cells of seropositive but not seronegative individuals in response to ex vivo stimulation with recombinant Cryptosporidium hominis gp15.
Several Cryptosporidium sporozoite antigens have been identified as potential vaccine candidates using traditional methods such as analysis of serum specificities after Cryptosporidium infection. However, despite the considerable amount of structural and immunological data obtained from characterizations of multiple sporozoite surface antigens, a vaccine is not yet available (9, 16, 18).
A “reverse vaccinology” strategy using in silico analyses based on the genome sequence information of the organism represents a novel approach to identifying vaccinogens. This approach is particularly useful in organisms that, like Cryptosporidium, are difficult to cultivate continuously in the laboratory. The strategy is based on the ability to predict proteins that are associated with the parasite surface, and therefore have the potential for interaction with the host immune mechanisms, by in silico screening for signal peptides, glycosylphosphatidylinositol (GPI) signal anchors, and similarities with known pathogenic factors. Recently the genomes of C. hominis, which is primarily a human pathogen, and C. parvum, which exhibits a relatively broad mammalian host range, were completed (1, 40), providing the opportunity to apply a reverse vaccinology strategy to identify new candidates for vaccines against Cryptosporidium. By using a reverse vaccinology strategy, we have identified three promising Cryptosporidium vaccinogens that induce strong humoral and cellular immune responses, suggesting that they could be used as components of a vaccine against Cryptosporidium infection.
MATERIALS AND METHODS
Parasites, DNA, and mice.
Iowa strain C. parvum oocysts used in this study were purchased from the Sterling Parasitology Laboratory in Tucson, AZ. C. hominis DNA was kindly provided by Saul Tzipori. Oocysts were purified using discontinuous sucrose and cesium chloride centrifugation gradients and shipped in an antibiotic solution containing 0.01% Tween 20, 100 U penicillin, and 100 mg of gentamicin per ml. Purified oocysts were stored at 4°C for less than 30 days prior to use. C57BL/6 mice were purchased at Jackson Laboratories (Maine) and maintained in our animal facilities at Virginia Commonwealth University (VCU).
Bioinformatics-based identification of potential vaccine candidates.
The cellular localizations of ∼4,000 potential proteins of C. hominis were predicted by the presence of a signal sequence or peptide, the number of potential transmembrane domains, and the presence of GPI anchor signal using SignalP, Big-HHMTOP, TMHMM, PI predictor, and GPI SOM software available at http://www.expasy.ch/tools/.
Cloning of C. hominis genes.
Primers for PCR amplification were based on the sequence for the selected genes from the Cryptosporidium hominis genome (GenBank accession number 3415519; CryptoDB Chro.60368, Chro.60194, and Chro.30189). The primers selected were as follows (underlining indicates sequences that allowed the directional cloning of the amplified products into the EK/LIC cloning site of the pTriEx-4/EKLIC vector [Novagen] using the ligation-independent cloning method): cp15F, 5′GACGACGACAAGATGGCAGATACTGAACAAAAG3′; cp15R, 5′GAGGAGAAGCCCGGTTTACTTTAGAGGAATGAATCTGGA3′; profilinF, 5′GACGACGACAAGATGTCTGAATGGGATGAT3′; profilinR, 5′GAGGAGAAGCCCGGTTTAGTATCCCTGAGATACGAG3′; capyF, 5′GACGACGACAAGATACAGGAAAGGAGGGTTTGCACTG3′; and capyR, 5′GAGGAGAAGCCCGGTTTATATAAATTCTATCCCCTCGTA3′.
We used the Taq recombinant polymerase kit (Invitrogen) to amplify C. hominis genomic DNA (1 μg) with 120 pmol of each primer essentially as recommended by the manufacturer. The recombinant vectors were transformed into NovaBlue cells (Novagen) for plasmid propagation and maintenance. The resulting constructs were checked for correct insertion of the selected genes by sequencing using the T7 promoter and T7 terminator primers.
Construction of the live vector vaccine expressing Cryptosporidium antigens.
The selected Cryptosporidium hominis genes were amplified using the pTriEX-4 constructs as templates together with the nucleotide sequence for the His6 tag using a common forward primer and a gene-specific reverse primer. The primers used are as follows: CommonF, 5′CTAGCTAGCCACCATCACCACCATCACTCT3′; cp15salR, 5′GGCCCCTAGGATTACTTTAGAGGAATGAA3′; profilinsalR, 5′GGCCCCTAGGATTAGTATCCCTGAGATACGA3′; capysalR, 5′GGCCCCTAGGATTATATAAATTCTATCCCCT3′.
The amplified products were cut with NheI and XbaI and ligated with the pSEC10 plasmid which had been linearized with NheI and AvrII (36). The resulting plasmid was transformed into Salmonella serovar Typhi CVD 908-htrA. Bacteria were grown in 2× LB medium with 25 μg/ml kanamycin alone or supplemented with 0.0001% 2,3-dihydroxybenzoic acid (Sigma, St. Louis, MO) as described previously (12). The ClyA fusion proteins were visualized in sodium dodecyl sulfate-polyacrylamide gels and designated ClyA-Cp15, ClyA-profilin, and ClyA-CApy (CApy is the Cryptosporidium apyrase).
Expression in Escherichia coli and purification of recombinant Cryptosporidium antigens.
Cells of E. coli BL21(DE3) were transformed with pTriEx-4/Cp15, pTriEx-4/profilin, or pTriEx-4/CApy. All Cryptosporidium-derived genes were fused C terminally to a His6/S peptide (S-tag) (Novagen). For expression of the recombinant Cryptosporidium genes, whose products were designated rCp15, r-profilin, and rCApy, TB medium (Express overnight autoinduction system; Novagen) supplemented with ampicillin (100 μg/ml) was inoculated with E. coli cultures carrying the respective plasmid and incubated at 37°C overnight (16 h) under agitation. The bacteria were harvested by centrifugation at 4,193 × g for 10 min at 4°C. The cell pellets were resuspended in BugBuster protein extraction reagent with Lysonase (Novagen) and lysed for 30 min at room temperature. After centrifugation at 15,339 × g for 30 min at 4°C, rCApy and rCp15 were found largely in inclusion bodies in the pellet, whereas r-profilin was largely soluble in the supernatant (unpublished observation).
Purification and refolding of rCApy and rCp15.
The pellets were suspended in BugBuster protein extraction reagent, and rLysozyme solution (Novagen) was added. Following incubation for 5 min at room temperature, six volumes of 1:10-diluted BugBuster protein extraction reagent was added, and the solution was mixed by vortexing for 1 min. The suspension was centrifuged for 15 min at 4,193 × g at 4°C, and the supernatant was removed. Inclusion bodies carrying Cryptosporidium proteins were resuspended in 0.5 volume of 1:10-diluted BugBuster protein extraction reagent and centrifuged at 15,339 × g for 15 min at 4°C. Following removal of the supernatant, the final pellet was resuspended in 20 mM sodium phosphate (pH 7.4), 500 mM NaCl, 4 M guanidine hydrochloride, 10 mM imidazole (buffer 1). The solubilized inclusion bodies were loaded onto a Ni2+ column (GE Healthcare) equilibrated with buffer 1. Column-bound proteins were washed with buffer 2 (buffer 1 but with 50 mM [instead of 10 mM] imidazole) and eluted with buffer 3 (buffer 1 but with 500 mM [instead of 10 mM] imidazole). The eluted material was renatured by immediate dialysis against 100 volumes of 100 mM Tris (pH 8.0), 1 M arginine, 2 mM EDTA, 1 mM glutathione (GSH), 0.1 mM oxidized glutathione (GSSG), 5% glycerol followed by a final dialysis against a minimum of 500 volumes of phosphate-buffered saline (PBS), pH 7.4. All dialysis was performed at 4°C.
Purification of r-profilin.
After bacterial lysis, the resultant supernatant was diluted 1:1 with 20 mM sodium phosphate (pH 7.4), 500 mM NaCl, 10 mM imidazole (buffer 4) and loaded onto a Ni2+ column (GE Healthcare) equilibrated with buffer 4. Column bound proteins were washed with buffer 5 (buffer 4 but with 50 mM imidazole) and eluted with buffer 6 (buffer 4 but with 500 mM imidazole). The eluted material was dialyzed immediately against a minimum of 500 volumes of PBS, pH 7.4. All dialysis was performed at 4°C. The purity of the protein preparations was examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
Immunization of mice and collection of serum and intestinal contents.
In standard experiments, groups of five female, 6- to 8-week-old C57BL/6 mice (Jackson Lab, Bar Harbor, ME) were inoculated intranasally on day 0 with 10 μl of PBS, pH 7.4, containing ∼1 × 109 CFU Salmonella serovar Typhi organisms expressing ClyA-Cp15, ClyA-profilin, ClyA-CApy, or ClyA (vector alone), essentially as previously described (12). At day 14, a protein booster of 20 μg recombinant protein (rCp15, r-profilin, rCApy, or bovine serum albumin [BSA]) in Freund's complete adjuvant was administered intraperitoneally, followed by a second intraperitoneal booster at day 28 of protein in Freund's incomplete adjuvant. Additional immunization schemes were tested, including an extra protein booster in Freund's incomplete adjuvant at day 42 or 56. Mice were sacrificed 14 days after the last immunization. Blood was collected via intracardiac puncture and allowed to clot, and serum was separated by centrifugation. Stool samples (500 mg) were collected, placed in 1 ml of a 3:1 mixture of PBS (pH 7.4) with 0.1 M EDTA and 0.1 mg/ml trypsin inhibitor (Sigma), and homogenized by vortexing until completely suspended. The mixture was centrifuged at 270 × g for 10 min. The supernatant was recovered, and 20 μl of 100 mM phenylmethylsulfonyl fluoride (PMSF) (Sigma) was added to each 1 ml of recovered supernatant. Serum and stool samples were aliquoted and stored at −20°C and −70°C, respectively, until subsequent analysis.
Cytokine profile.
Splenocytes were isolated from immunized C57BL/6 mice 2 weeks after the third immunization and suspended in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES, 2 mM l-glutamine, 1 mM sodium pyruvate, 1% (vol/vol) nonessential amino acid solution, 1% (vol/vol) minimal essential medium vitamin solution (all from Life Technologies), and 100 U of penicillin and streptomycin (both from Sigma) per ml. Cell viability was determined by trypan blue exclusion. The number of cells and viability did not vary substantially among the experimental conditions. Cells were adjusted to 5 × 106 viable cells per well in 96-well flat-bottom plates (Costar), suspended in 0.2 ml culture medium containing the respective antigens (r-profilin, rCp15, and rCApy) at concentrations of 5 to 20 μg/ml. Concanavalin A (ConA) (Sigma) was used as a proliferation control at a concentration of 10 μg/ml. After incubation for 3 days at 37°C in an atmosphere containing 5% CO2, the cell supernatants were collected. Concentrations of IL-2, IL-6, IL-12, and IFN-γ in supernatants were determined by capture enzyme-linked immunosorbent assay (ELISA) using commercially available kits (BD OptEIA) following the manufacturer's recommendations. All assays were done in triplicate; the results are reported as means ± standard deviations.
Measurement of antigen-specific antibody isotype response.
Microtiter plates (96 well) were coated with recombinant protein (1 μg/ml of r-profilin, 2 μg/ml rCp15, or 2 μg/ml rCApy) in 50 mM sodium carbonate buffer (pH 9.6) by overnight incubation with 0.1 ml at 4°C. Plates were washed twice with TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) before incubation for 1 h at room temperature with blocking buffer (TBST containing 1% milk powder), followed by three washes with TBST. Sera or supernatants of intestinal contents were diluted serially in blocking buffer and incubated with antigen-coated plates for 2 h at room temperature. After four washes with TBST, goat anti-mouse IgG1, IgG2a, IgG2b, IgG3, or IgA conjugated with horseradish peroxidase (Southern Biotech) diluted 1:5,000 in blocking buffer was added to plates, which were then incubated for 1 h at room temperature. After six washes with TBST, o-phenylenediamine dihydrochloride (SigmaFAST OPD) dissolved in water was added as a substrate. Endpoint dilutions were determined as the dilutions at which the optical density at 450 nm was 1.0 after 30 min of incubation.
Immunofluorescence assay and microscopy.
Cryptosporidium parvum oocysts were excysted as previously described (6). Excysted parasites were fixed with 3,5% formaldehyde for 30 min, washed three times in phosphate-buffered saline (PBS), blocked with 5% bovine serum albumin (BSA) in PBS, and incubated with antiserum derived from previously immunized animals (see above) diluted in 1% BSA. After three washes with PBS, the parasites were incubated with anti-mouse IgG conjugated with fluorescein isothiocyanate. The coverslips were examined on a Zeiss LSM 510 metaconfocal scanning microscope.
RESULTS
Prediction of vaccine candidates from the Cryptosporidium genome. (i) In silico selection of surface associated proteins.
A bioinformatics/genomics-based filtering strategy for identification of surface-linked Cryptosporidium antigens was implemented. Thus, all of the approximately 4,000 C. hominis and C. parvum genes were carefully functionally annotated and screened using a series of public available software (see Materials and Methods) for the presence of signal peptide sequence, transmembrane domains, GPI anchors, hydrophobicity and hydrophilicity, similarity to known virulence factors, and other features that suggest that the protein might be a promising vaccine target (see Table S1 in the supplemental material).
(ii) Sporozoite-expressed protein candidates.
To further focus our selection, we investigated the expression patterns of these genes in sporozoites, the infective form of Cryptosporidium parasites, by microarray and proteomics analysis (M. Serrano et al., unpublished data; P. Manque et al., unpublished data). This combined strategy identified hundreds of genes that encode proteins that exhibit high levels of both mRNA and protein expression in C. parvum sporozoites and that were identified as candidate immunogens by the bioinformatics screening described above.
(iii) Characterization of CApy, Cp15, and profilin.
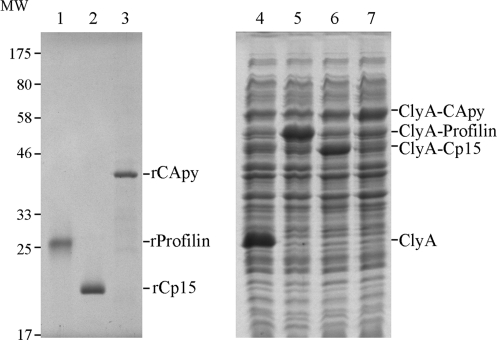
Nine of the selected proteins described above were selected for initial analysis (see Table S2 in the supplemental material) based primarily on the likelihood of their being expressed on the surface of the parasite and their ability to be produced as recombinant proteins; here, we report on the results of the first three, i.e., CApy, Cp15, and profilin. These candidate genes were amplified by PCR from C. hominis genomic DNA, cloned into pTriEx-4 and pSEC10 expression vectors, transformed, and expressed in E. coli and Salmonella typhi, respectively. The apparent molecular masses of the three recombinant His6/S-tagged candidates were estimated by SDS-PAGE at 42 kDa for rCApy, 26 kDa for r-profilin, and 20 kDa for rCp15 (Fig. 1). After purification using Ni2+ columns, the purity of our recombinant protein preparations exceeded 95% when evaluated by SDS-PAGE (Fig. 1).
Fig. 1.
Electrophoretic analysis of purified recombinant vaccinogens and their expression in the Salmonella live vaccine vector. Results of Coomassie brilliant blue-stained SDS-PAGE analysis of purified His6/S-tagged r-profilin, rCp15, and rCApy proteins expressed in E. coli (A, lanes 1 to 3) and whole bacterial lysates of live vaccine S. Typhi 908 transformed with pSEC10 plasmid vector expressing ClyA alone, ClyA-profilin, ClyA-Cp15, and ClyA-CApy (B, lanes 4 to 7) are shown. The molecular weight standard (MW, in thousands) applies to both gels.
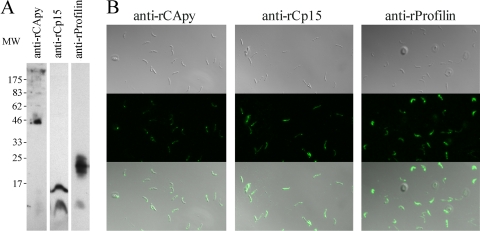
Western blot and immunofluorescence assay (IFA) experiments provided further confirmation of the expression of the selected antigens on the surfaces of the infective forms of the parasite. Thus, the proteins were recognized by antibodies from mice immunized with purified recombinant antigens in Western blots of sporozoite extracts and in IFAs of intact, nonpermeabilized sporozoites (Fig. 2A and B, respectively). Recognition was specific, since serum from immunized animals with vector alone or BSA failed to recognize sporozoites (data not shown).
Fig. 2.
Interaction of antibodies raised against rCApy, rCp15, and r-profilin with C. parvum sporozoites. (A) Detection of native CApy, Cp15, and profilin. C. parvum sporozoites were separated by SDS-PAGE on a 12% polyacrylamide gel, transferred to nitrocellulose, and probed with anti-rCApy, anti-rCp15, and anti-r-profilin. MW, molecular weight (in thousands). (B) Localization of CApy, Cp15, and profilin on the surfaces of C. parvum sporozoites.
Immune response generated by immunization with recombinant Cryptosporidium proteins.
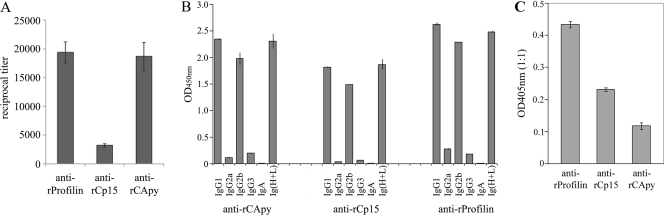
To characterize the immune response induced by the selected vaccine candidates, we immunized C57BL/6 mice intranasally with Salmonella live vector expressing Cryptosporidium antigens followed by one intraperitoneal booster with recombinant protein in Freund's complete adjuvant and two with Freund's incomplete adjuvant, as described in Materials and Methods. Serum from mice immunized with the respective antigen exhibited significant titers of antibodies specific for the antigens used for immunization (Fig. 3). However, each of the three antigens displayed a unique antibody response. Immunization with profilin and CApy produced the highest absolute titers, whereas Cp15 generated the lowest (Fig. 3A). Interestingly, no major differences in the overall response were observed when we reduced the number of protein boosters to two in our immunization protocol, as described in Materials and Methods (data not shown). In addition, as shown in Fig. 3B, antigen-specific isotyping of immunoglobulins in the sera of immunized animals revealed the highest levels of IgG1 and IgG2b for all antigens, suggesting that all antigens induced a Th1-like immune response. Moreover, all selected vaccinogens induced variable titers of mucosal IgA (Fig. 3C), indicating that our vaccination protocol raised mucosal immunity.
Fig. 3.
Heterologous vaccination with CApy, Cp15, and profilin induces a strong antibody response in mice. C57BL/6 mice were immunized with the antigen-expressing Salmonella live vector, followed by three boosters with purified recombinant antigens in Freund's adjuvant. (A) Reciprocal titers. (B) Antigen-specific immunoglobulin isotype responses measured 2 weeks after the final immunization. Data are representative of two independent experiments for all panels. (C) IgA titers in intestinal contents.
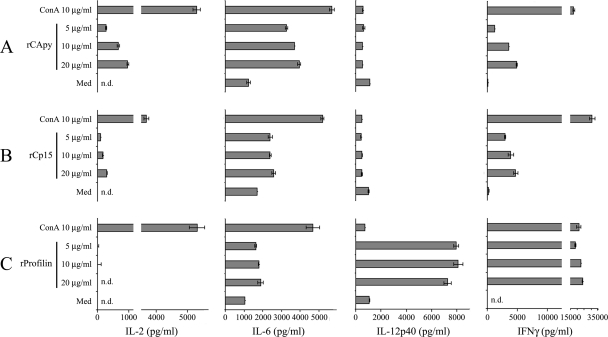
To assess the induction of Cryptosporidium-specific T-cell responses after immunization with the selected vaccinogens, we measured production of IL-2, IL-6, IL-12, and IFN-γ by spleen cells from mice immunized using our immunization protocol. Upon in vitro restimulation with recombinant proteins derived from Cryptosporidium, we observed antigen-specific proliferation (data not shown) and cytokine production in cultured spleen cells from immunized mice (Fig. 4). All antigens induced production of IFN-γ and IL-6, consistent with a Th1 response. Interestingly, profilin very strongly induced IFN-γ at levels comparable to those obtained with ConA. In addition, only profilin induced significant levels of IL-12, whereas IL-2 was produced only by splenocytes stimulated with Cp15 or CApy.
Fig. 4.
Cytokine profile after stimulation with specific antigens. Production of cytokines of splenocytes isolated from vaccinated mice. Groups of five C56BL/6 mice were vaccinated with one dose of Salmonella live vectors expressing the Cryptosporidium antigen CApy, Cp15, or profilin as a ClyA fusion followed by three doses of the respective purified recombinant antigen as described in Materials and Methods. A total of 107 splenocytes from vaccinated mice were restimulated in vitro with 5, 10, or 20 μg of rCApy (A), rCp15 (B), or r-profilin (C) per ml of medium, with 10 μg/ml ConA as a positive control, or with medium as a background control. After 96 h of stimulation, IL-2, IL-6, IL-12, and IFN-γ levels were measured by ELISA in cell-free supernatants. Data are representative of two independent experiments.
Thus, our data suggest that both humoral and cellular responses are induced upon vaccination of mice with each of these antigens and that the reaction to each of these three antigens is unique.
DISCUSSION
In the present study, we examined the potential of three candidate Cryptosporidium vaccinogens to induce immune responses that could lead to protection against Cryptosporidium infection. These antigens were identified using a reverse vaccinology approach combined with expression profile analysis using microarrays and proteomics of the infective sporozoite forms of the parasite. Initially, we selected these antigens based on four main criteria: (i) a bioinformatics analysis indicating that these genes could be localized to the surface of the parasite and may play a role in pathogenesis; (ii) mRNA expression in the infective sporozoite; (iii) protein expression in the infective sporozoite; and (iv) the ability to be expressed in E. coli and to be obtained in soluble form. Our analysis revealed that each of the selected antigens displayed a series of interesting features that were explored independently, leading to the observation that each could be associated with the invasion process as well as the activation of the innate immunity. Thus, CApy is an apyrase that is involved in the invasion process of Cryptosporidium (Manque et al., unpublished data). Profilin has been characterized as a potent agonist of the innate immune system through its recognition by Toll-like receptor 11 and is described as essential for the invasion of Toxoplasma gondii (27). Cp15 has been identified as an immunodominant antigen (19, 29, 30, 33, 34, 39), and we are exploring its apparent role in the invasion of mammalian cells by C. parvum sporozoites (Manque et al. unpublished). Bioinformatics analysis of the selected vaccine candidates did not reveal any consequential polymorphism between these genes in C. hominis, which is essentially an exclusively human pathogen, and C. parvum, which infects humans but is primarily responsible for veterinary cryptosporidiosis (37). Thus, these antigens may have utility in both human and veterinary vaccines.
A Cryptosporidium infection is initiated when the host ingests oocysts, from which invasive sporozoites emerge and infect the intestinal mucosa (35). It is likely that establishment of the infection and severity of the disease depends upon early events associated with the interaction of the parasite with the intestinal mucosa, suggesting that an effective vaccine should combine mucosal and systemic immunity. Thus, we decided to use a heterologous prime-boost strategy consisting of mucosal priming with Salmonella serovar Typhi expressing a fragment of the cytolysin ClyA fused to our selected vaccinogens, followed by intramuscular boosting with their corresponding recombinant proteins. This strategy and variations of it that include heterologous boosts with DNA and protein have proven successful in the generation of strong immune responses against other mucosal pathogens (5, 10). Furthermore, heterologous prime-boost regimens have been found to be effective raising immune responses against poorly immunogenic antigens (7, 23). Supporting these findings, our immunization protocol was able to generate a strong immune response to each of the three vaccinogens. We observed high specific-antibody titers after the second protein booster. Recombinant profilin was found to induce the highest titers. Isotype analysis also revealed differences among the selected antigens; IgG1 was the predominant isotype, whereas IgG2B was higher in Cp15- and profilin-immunized animals than in those that were immunized with CApy. Since the isotype profiles reflect the Th1/Th2 pathways that are activated, our results suggest that each antigen induces a particular pattern of T-cell response. Further studies are under way to characterize the particular T-cell subpopulations that are involved in each response. In addition, our heterologous prime-boost strategy yielded variable levels of mucosal immunity, as observed by the IgA titers detected (Fig. 3C). We believe that this variation primarily reflects technical difficulties associated with the detection of IgA in intestinal contents due to its high susceptibility to degradation. Previous studies using a Salmonella vector achieved the induction of a mucosal response with detectable titers of IgA (32), consistent with our observation that our vaccination protocol led to measurable mucosal immune responses in the GI tract.
The cellular response elicited by our heterologous prime-boost strategy was robust, and similarly to the observed humoral response, the pattern of cytokine production was antigen dependent and antigen specific. Significantly, IFN-γ, a critical cytokine associated with a protective Th1 memory response and protection against Cryptosporidium infection (20), was produced by spleen cells from animals immunized with each of the selected antigens after restimulation with their corresponding protein. Interestingly, profilin induced extremely high levels of IFN-γ, similar to the levels induced by ConA. As discussed above, profilin has been reported to be an agonist of TLR11 (27), suggesting that these high levels of IFN-γ may be at least partly due to macrophage activation during the cytokine production assay.
Both humoral and cellular responses were elicited using a Salmonella strain-and-vector combination which delivered Cp23 and Cp40 fused to the C-terminal fragment of tetanus toxins (3). However, the delivery of our antigens as ClyA fusion proteins that are expressed on the surface of the bacteria or secreted into the periplasmic space has been reported to result in more efficient antigen processing and presentation (11).
We are now examining the ability of our antigens to confer protection against Cryptosporidium infection in two recently established animal models, the neonatal mouse and the malnourished mouse models. Thus, our results suggest that our selected vaccinogens will lead to some level of protection via an orchestrated activation of mucosal, humoral, and cellular immune responses.
In summary, our results underscore the value of a reverse vaccinology strategy to identify new vaccine candidates in protozoan parasites. Further studies are necessary to verify the protection induced by these antigens as well as the associated immune mechanisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jennifer Fettweis for the assistance with the confocal microscopy, James Galen for generously providing the Salmonella vector system, and the rest of the members of G. Buck's laboratory at VCU for helpful discussions.
This project was supported by funding from NIH-NIAID grant U54 AI57168. Services and products used in this research project were generated in VCU's Nucleic Acids Research Facilities, Flow Cytometry Core Resource, and Imaging Shared Resource, which are all supported, in part, by funding from NIH-NCI Cancer Center Support Grant 5 P30 CA016059.
Footnotes
Supplemental material for this article may be found at http://cvi.asm.org/.
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Abrahamsen M. S., et al. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–445 [DOI] [PubMed] [Google Scholar]
- 2. Barrier M., et al. 2006. Oral and intraperitoneal administration of phosphorothioate oligodeoxynucleotides leads to control of Cryptosporidium parvum infection in neonatal mice. J. Infect. Dis. 193:1400–1407 [DOI] [PubMed] [Google Scholar]
- 3. Benitez A. J., McNair N., Mead J. R. 2009. Oral immunization with attenuated Salmonella enterica serovar Typhimurium encoding Cryptosporidium parvum Cp23 and Cp40 antigens induces a specific immune response in mice. Clin. Vaccine Immunol. 16:1272–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brantley R. K., et al. 2003. AIDS-associated diarrhea and wasting in Northeast Brazil is associated with subtherapeutic plasma levels of antiretroviral medications and with both bovine and human subtypes of Cryptosporidium parvum. Braz. J. Infect. Dis. 7:16–22 [DOI] [PubMed] [Google Scholar]
- 5. Chinchilla M., et al. 2007. Enhanced immunity to Plasmodium falciparum circumsporozoite protein (PfCSP) by using Salmonella enterica serovar Typhi expressing PfCSP and a PfCSP-encoding DNA vaccine in a heterologous prime-boost strategy. Infect. Immun. 75:3769–3779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cohn B., et al. 2010. Putative cis-regulatory elements associated with heat shock genes activated during excystation of Cryptosporidium parvum. PLoS One 5:e9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dunachie S. J., Hill A. V. 2003. Prime-boost strategies for malaria vaccine development. J. Exp. Biol. 206:3771–3779 [DOI] [PubMed] [Google Scholar]
- 8. Ehigiator H. N., McNair N., Mead J. R. 2007. Cryptosporidium parvum: the contribution of Th1-inducing pathways to the resolution of infection in mice. Exp. Parasitol. 115:107–113 [DOI] [PubMed] [Google Scholar]
- 9. Ehigiator H. N., Romagnoli P., Priest J. W., Secor W. E., Mead J. R. 2007. Induction of murine immune responses by DNA encoding a 23-kDa antigen of Cryptosporidium parvum. Parasitol. Res. 101:943–950 [DOI] [PubMed] [Google Scholar]
- 10. Galen J. E., et al. 2009. Mucosal immunization with attenuated Salmonella enterica serovar Typhi expressing protective antigen of anthrax toxin (PA83) primes monkeys for accelerated serum antibody responses to parenteral PA83 vaccine. J. Infect. Dis. 199:326–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galen J. E., et al. 2009. Salmonella enterica serovar Typhi live vector vaccines finally come of age. Immunol. Cell Biol. 87:400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Galen J. E., et al. 2004. Adaptation of the endogenous Salmonella enterica serovar Typhi clyA-encoded hemolysin for antigen export enhances the immunogenicity of anthrax protective antigen domain 4 expressed by the attenuated live-vector vaccine strain CVD 908-htrA. Infect. Immun. 72:7096–7106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gookin J. L., et al. 2006. NF-κB-mediated expression of iNOS promotes epithelial defense against infection by Cryptosporidium parvum in neonatal piglets. Am. J. Physiol. Gastrointest. Liver Physiol. 290:G164–174 [DOI] [PubMed] [Google Scholar]
- 14. Guerrant R. L., et al. 2002. Magnitude and impact of diarrheal diseases. Arch. Med. Res. 33:351–355 [DOI] [PubMed] [Google Scholar]
- 15. Guillen S., et al. 2010. Opportunistic infections and organ-specific diseases in HIV-1-infected children: a cohort study (1990-2006). HIV Med. 11:245–252 [DOI] [PubMed] [Google Scholar]
- 16. He H., et al. 2004. The humoral and cellular immune responses in mice induced by DNA vaccine expressing the sporozoite surface protein of Cryptosporidium parvum. DNA Cell Biol. 23:335–339 [DOI] [PubMed] [Google Scholar]
- 17. Hlavsa M. C., Watson J. C., Beach M. J. 2005. Cryptosporidiosis surveillance—United States 1999-2002. MMWR Surveill. Summ. 54:1–8 [PubMed] [Google Scholar]
- 18. Hong-Xuan H., et al. 2005. Expression of the recombinant fusion protein CP15-23 of Cryptosporidium parvum and its protective test. J. Nanosci. Nanotechnol. 5:1292–1296 [DOI] [PubMed] [Google Scholar]
- 19. Jenkins M. C., Fayer R. 1995. Cloning and expression of cDNA encoding an antigenic Cryptosporidium parvum protein. Mol. Biochem. Parasitol. 71:149–152 [DOI] [PubMed] [Google Scholar]
- 20. Lacroix S., Mancassola R., Naciri M., Laurent F. 2001. Cryptosporidium parvum-specific mucosal immune response in C57BL/6 neonatal and gamma interferon-deficient mice: role of tumor necrosis factor alpha in protection. Infect. Immun. 69:1635–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leav B. A., Mackay M., Ward H. D. 2003. Cryptosporidium species: new insights and old challenges. Clin. Infect. Dis. 36:903–908 [DOI] [PubMed] [Google Scholar]
- 22. Mariam Z. T., Abebe G., Mulu A. 2008. Opportunistic and other intestinal parasitic infections in AIDS patients, HIV seropositive healthy carriers and HIV seronegative individuals in southwest Ethiopia. East Afr. J. Public Health 5:169–173 [PubMed] [Google Scholar]
- 23. Moorthy V. S., et al. 2004. Phase 1 evaluation of 3 highly immunogenic prime-boost regimens, including a 12-month reboosting vaccination, for malaria vaccination in Gambian men. J. Infect. Dis. 189:2213–2219 [DOI] [PubMed] [Google Scholar]
- 24. O'Hara S., Tietz Bogert P. S., Trussoni C. E., Chen X., Larusso N. F. 2011. Tlr4 promotes Cryptosporidium parvum clearance in a mouse model of biliary cryptosporidiosis. J. Parasitol. 97:813–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. O'Hara S. P., et al. 2010. NF-κB p50-CCAAT/enhancer-binding protein beta (C/EBPβ)-mediated transcriptional repression of microRNA let-7i following microbial infection. J. Biol. Chem. 285:216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pantenburg B., et al. 2008. Intestinal immune response to human Cryptosporidium sp. infection. Infect. Immun. 76:23–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Plattner F., et al. 2008. Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe 3:77–87 [DOI] [PubMed] [Google Scholar]
- 28. Preidis G. A., et al. 2007. Seropositive human subjects produce interferon gamma after stimulation with recombinant Cryptosporidium hominis gp15. Am. J. Trop. Med. Hyg. 77:583–585 [PubMed] [Google Scholar]
- 29. Reperant J. M., Naciri M., Chardes T., Bout D. T. 1992. Immunological characterization of a 17-kDa antigen from Cryptosporidium parvum recognized early by mucosal IgA antibodies. FEMS Microbiol. Lett. 78:7–14 [DOI] [PubMed] [Google Scholar]
- 30. Reperant J. M., Naciri M., Iochmann S., Tilley M., Bout D. T. 1994. Major antigens of Cryptosporidium parvum recognised by serum antibodies from different infected animal species and man. Vet. Parasitol. 55:1–13 [DOI] [PubMed] [Google Scholar]
- 31. Robinson P., et al. 2001. Expression of tumor necrosis factor alpha and interleukin 1 beta in jejuna of volunteers after experimental challenge with Cryptosporidium parvum correlates with exposure but not with symptoms. Infect. Immun. 69:1172–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rojas R. L., et al. 2010. Salmonella enterica serovar Typhimurium vaccine strains expressing a nontoxic Shiga-like toxin 2 derivative induce partial protective immunity to the toxin expressed by enterohemorrhagic Escherichia coli. Clin. Vaccine Immunol. 17:529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sagodira S., Buzoni-Gatel D., Iochmann S., Naciri M., Bout D. 1999. Protection of kids against Cryptosporidium parvum infection after immunization of dams with CP15-DNA. Vaccine 17:2346–2355 [DOI] [PubMed] [Google Scholar]
- 34. Sagodira S., Iochmann S., Mevelec M. N., Dimier-Poisson I., Bout D. 1999. Nasal immunization of mice with Cryptosporidium parvum DNA induces systemic and intestinal immune responses. Parasite Immunol. 21:507–516 [DOI] [PubMed] [Google Scholar]
- 35. Smith H. V., Nichols R. A., Grimason A. M. 2005. Cryptosporidium excystation and invasion: getting to the guts of the matter. Trends Parasitol. 21:133–142 [DOI] [PubMed] [Google Scholar]
- 36. Stokes M. G., et al. 2007. Oral administration of a Salmonella enterica-based vaccine expressing Bacillus anthracis protective antigen confers protection against aerosolized B. anthracis. Infect. Immun. 75:1827–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tanriverdi S., et al. 2008. Inferences about the global population structures of Cryptosporidium parvum and Cryptosporidium hominis. Appl. Environ. Microbiol. 74:7227–7234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tzipori S., Ward H. 2002. Cryptosporidiosis: biology, pathogenesis and disease. Microbes Infect. 4:1047–1058 [DOI] [PubMed] [Google Scholar]
- 39. Ungar B. L., Nash T. E. 1986. Quantification of specific antibody response to Cryptosporidium antigens by laser densitometry. Infect. Immun. 53:124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu P., et al. 2004. The genome of Cryptosporidium hominis. Nature 431:1107–1112 [DOI] [PubMed] [Google Scholar]
- 41. Yarovinsky F., et al. 2005. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science 308:1626–1629 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.