Abstract
We used comparative proteomics to analyze eosinophils from a patient with hypereosinophilia due to fascioliasis. Using 2-dimensional electrophoresis and mass spectrometry, we demonstrated that the eosinophil proteome was significantly altered compared to those of healthy controls.
CASE REPORT
A 48-year-old female presented to the Instituto de Medicina Tropical Alexander von Humboldt, Universidad Peruana Cayetano Heredia, Lima, Peru, with complaints of colicky abdominal pain in the right upper quadrant. She had experienced symptoms for several years and had undergone a cholecystectomy 6 years before. She had previously worked in an area where fascioliasis is hyperendemic. One month prior to hospitalization, her pain increased in intensity, and symptomatic treatment did not alleviate the pain. On presentation, physical examination revealed a palpable liver 4 cm below the right costal margin and right upper quadrant abdominal tenderness. A complete blood count revealed an absolute eosinophil count of 3,060/μl. Subsequent blood counts confirmed hypereosinophilia. Bilirubin, alkaline phosphatase, aspartate aminotransferase (AST), and alanine aminotransferase (ALT) were within normal limits. A computed tomography (CT) scan revealed small liver lesions. Fasciola hepatica infection (fascioliasis) was diagnosed by enzyme-linked immunosorbent assay (ELISA) (3), and the patient responded to triclabendazole with resolution of symptoms and improvement in eosinophilia over the next few weeks.
Prior to treatment, she consented to enroll in a study approved by the Institutional Review Board for research with human subjects at Universidad Peruana Cayetano Heredia. Granulocytes were isolated by gradient centrifugation and erythrocytes lysed with hypotonic saline as described previously (10). Granulocytes were incubated with anti-CD16 immunomagnetic beads and loaded onto a VarioMACS (Miltenyi Biotec, Auburn CA) column, and a magnetic field was used to separate neutrophils from eosinophils. The isolated eosinophils (>97% pure by Giemsa staining) were lysed in DeStreak rehydration buffer, frozen (−70 C), and transported on dry ice to the University of Texas Medical Branch, Galveston, TX, where the whole-cell lysate was resolved via 2-dimensional electrophoresis (2-DE) as described previously (10). Gels were stained with Sypro Ruby protein stain (Bio-Rad, Hercules CA) and imaged, and the normalized volumes of protein spots compared to a baseline eosinophil proteome map from control donors using Nonlinear SameSpots software (Nonlinear Dynamics, Durham NC). Proteins differentially expressed by ≥2-fold were excised and analyzed by matrix-assisted laser desorption ionization-tandem time of flight mass spectrometry (MALDI-TOF-TOF MS) (Applied Biosystems 4800) (10). Applied Biosystems GPS ExplorerTM (version 3.6) software was used along with MASCOT (Matrix Science, London, United Kingdom) to search protein databases using both MS and tandem MS (MS-MS) spectral data for protein identification.
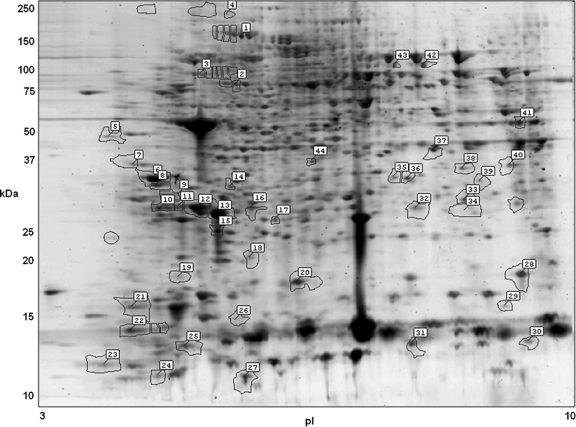
Of over 3,000 protein spots identified by 2-DE, 60 were differentially expressed by ≥2-fold as measured by SameSpots analysis when compared to a healthy control profile averaged from 8 donors (Fig. 1) (10). Of the 60 protein spots that were differentially expressed, some 44 were subjected to MALDI-TOF-TOF MS, and their identifications are given in Table 1. Thirty-eight of the 44 had statistically significant expectation values of <10−3 (10). Of the 38 significant proteins, 23 were unique within the data set after discounting redundant proteins. Redundant proteins typically represent a variety of posttranslational modifications that are frequently observed in eosinophils (10). They are usually represented by protein spots whose observed pIs and Mrs are among those that differ due to posttranslational modification.
Fig. 1.
Two-dimensional gel image of the eosinophils from a patient with Fasciola hepatica infection and hypereosinophilia, stained with Sypro Ruby fluorescent stain. Highlighted protein spots indicate proteins whose levels differed by at least 2-fold between the Fasciola patient and healthy controls. The spot numbers accompanying the highlighted proteins are the pick numbers in Table 1.
Table 1.
Proteins with altered expression in eosinophils from a patient with hypereosinophilia from fascioliasis
| Protein name | SwissProt accession no. | Fold changea | Theoretical/observed |
Peptide count | Expectation valueb | Function | Pick no.c | |
|---|---|---|---|---|---|---|---|---|
| pI | Mr | |||||||
| 14-3-3 protein zeta/delta | P63104 | 2.1 | 4.73/5.17 | 27.9/21.31 | 12 | 1.58E-29 | Cell signaling | 10 |
| 60S ribosomal protein L4 | P36578 | 5.6 | 11.07/7.6 | 47.95/73.91 | 12 | 0.16 | Protein synthesis | 42 |
| Actin, cytoplasmic 2 | P63261 | 4.9 | 5.31/4.91 | 42.11/11.06 | 4 | 1.26E-08 | Actin-related protein | 22 |
| Actin, cytoplasmic 2 | P63261 | 5.1 | 5.31/6.23 | 42.11/19.43 | 6 | 7.92E-16 | Actin-related protein | 17 |
| Actin, cytoplasmic 2 | P63261 | 2.8 | 5.31/5.7 | 42.11/20.2 | 9 | 3.97E-22 | Actin-related protein | 13 |
| Actin, cytoplasmic 2 | P63261 | 2.2 | 5.31/5.51 | 42.11/21.43 | 13 | 3.97E-42 | Actin-related protein | 12 |
| Actin, cytoplasmic 2 | P63261 | 2.5 | 5.31/5.35 | 42.11/21.89 | 12 | 1.26E-37 | Actin-related protein | 11 |
| Actin, cytoplasmic 2 | P63261 | 2.9 | 5.31/5.34 | 42.11/24.02 | 8 | 2.51E-29 | Actin-related protein | 9 |
| Actin, cytoplasmic 2 | P63261 | 4.0 | 5.31/5.82 | 42.11/24.93 | 10 | 6.29E-23 | Actin-related protein | 14 |
| Actin, cytoplasmic 2 | P63261 | 4.2 | 5.31/5.15 | 42.11/25 | 9 | 6.29E-28 | Actin-related protein | 8 |
| Actin, cytoplasmic 2 | P63261 | 2.0 | 5.31/5.1 | 42.11/25.09 | 12 | 1.58E-42 | Actin-related protein | 6 |
| Actin-like protein 3 | P61158 | 4.4 | 5.61/7.33 | 47.8/25.03 | 16 | 3.97E-25 | Actin-related protein | 35 |
| Actin-like protein 3 | P61158 | 3.1 | 5.61/7.46 | 47.8/25.07 | 15 | 5.00E-34 | Actin-related protein | 36 |
| Alpha-enolase | P06733 | 2.7 | 7.01/6.58 | 47.48/24.16 | 15 | 9.98E-22 | Carbohydrate metabolism | 44 |
| Annexin A1 | P04083 | 3.2 | 6.57/7.98 | 38.92/23.24 | 14 | 9.98E-36 | Ca2+/phospholipid binding/exocytosis | 33 |
| Annexin A1 | P04083 | 3.2 | 6.57/7.69 | 38.92/24.22 | 20 | 1.99E-43 | Ca2+/phospholipid binding/exocytosis | 37 |
| Annexin A1 | P04083 | 6.1 | 6.57/8.12 | 38.92/25.1 | 6 | 7.92E-03 | Ca2+/phospholipid binding/exocytosis | 39 |
| Calreticulin precursor | P27797 | 2.4 | 4.29/4.7 | 48.28/27.01 | 5 | 1.99E-11 | Ca2+/phospholipid binding/exocytosis | 5 |
| Cofilin-1 | P23528 | 2.1 | 8.22/6.44 | 18.72/15.89 | 12 | 6.29E-25 | Actin-related protein | 20 |
| Cofilin-1 | P23528 | 6.6 | 8.22/8.5 | 18.72/16.15 | 11 | 3.97E-20 | Actin-related protein | 28 |
| Corticotropin-lipotropin precursor | P01189 | 2.3 | 7.56/5.34 | 29.8/16.05 | 4 | 6.29 | Precursor for ACTH, MSH, beta-endorphin, and met-enkephalin | 19 |
| Ezrin | P15311 | 4.4* | 5.94/5.87 | 69.48/12.49 | 6 | 0.16 | Cytoskeletal protein | 26 |
| Fructose-bisphosphate aldolase A | P04075 | 5.2 | 8.3/8.49 | 39.85/34.54 | 14 | 1.58E-26 | Carbohydrate metabolism | 41 |
| Gelsolin precursor | P06396 | 2.9* | 5.9/5.92 | 86.04/85.18 | 26 | 3.97E-56 | Actin-related protein | 1 |
| Guanine nucleotide-binding protein subunit beta 2-like 1 | P63244 | 2.8 | 7.6/7.96 | 35.51/24.55 | 6 | 5.00E-06 | Cell signaling | 38 |
| Heat shock protein HSP 90-alpha | P07900 | 2.2 | 4.94/6 | 85.01/16.56 | 7 | 1.58E-14 | Molecular chaperone | 18 |
| Hemoglobin subunit beta | P68871 | 3.9 | 6.75/7.5 | 16.1/9.33 | 4 | 1.26E-07 | Oxygen transport | 31 |
| Histone-lysine N-methyltransferase SETDB1 | Q15047 | 5.0 | 5.74/5.7 | 145.12/18.12 | 15 | 0.20 | Regulation of gene expression | 15 |
| Keratin, type I cytoskeletal 10 | P13645 | 6.6 | 5.13/5.96 | 59.7/5 | 8 | 0.01 | Intermediate filaments | 27 |
| Keratin, type I cytoskeletal 10 | P13645 | 3.3 | 5.13/4.66 | 59.7/6.57 | 20 | 5.00E-28 | Intermediate filaments | 23 |
| Keratin, type I cytoskeletal 9 | P35527 | 4.3 | 5.19/8.35 | 62.32/14.29 | 10 | 5.00E-07 | Intermediate filaments | 29 |
| Leukocyte elastase inhibitor | P30740 | 3.0 | 5.9/4.92 | 42.83/14.17 | 8 | 5.00E-26 | Protease inhibition | 21 |
| Macrophage migration inhibitory factor | P14174 | 6.5 | 7.74/8.57 | 12.64/9.55 | 3 | 9.98E-05 | Proinflammatory cytokine | 30 |
| Neuroepithelial cell-transforming gene 1 protein | Q7Z628 | 7.3 | 9.31/8.4 | 68.15/24.37 | 8 | 0.25 | Cell signaling | 40 |
| Neutral alpha-glucosidase AB precursor | Q14697 | 2.2* | 5.74/5.79 | 107.26/101.61 | 10 | 1.26E-08 | Glycoprotein alteration | 4 |
| Plastin-2 | P13796 | 2.4 | 5.2/4.93 | 70.82/24.33 | 9 | 1.99E-08 | Actin-related protein | 7 |
| Protein disulfide-isomerase A3 precursor | P30101 | 2.1* | 5.98/5.88 | 57.15/65.95 | 19 | 9.98E-42 | Protein rearrangement/alteration | 2 |
| Protein disulfide-isomerase A3 precursor | P30101 | 2.0* | 5.98/5.55 | 57.15/70.5 | 15 | 3.15E-14 | Protein rearrangement/alteration | 3 |
| Protein S100-A9 | P06702 | 2.4 | 5.71/5.16 | 13.29/5.22 | 4 | 6.29E-03 | Ca2+ binding | 24 |
| Protein S100-A9 | P06702 | 4.7 | 5.71/5.41 | 13.29/8.96 | 5 | 9.98E-06 | Ca2+ binding | 25 |
| Synaptic vesicle membrane protein VAT-1 homolog | Q99536 | 3.9* | 5.88/7.98 | 42.12/20.97 | 4 | 9.98E-03 | Unknown function | 34 |
| Transketolase | P29401 | 5.2 | 7.58/7.34 | 68.52/73.91 | 17 | 9.98E-20 | Carbohydrate alteration | 43 |
| Vimentin | P08670 | 4.7 | 5.06/7.53 | 53.68/21.54 | 23 | 1.99E-35 | Mesenchymal intermediate filament | 32 |
| Vimentin | P08670 | 2.1 | 5.06/6.02 | 53.68/21.66 | 27 | 6.29E-29 | Mesenchymal intermediate filament | 16 |
Eosinophils are important mediators of allergies, asthma, and adverse drug reactions. In contrast, the adaptive role of eosinophils in human biology is thought to relate to host defenses against helminth infections (1, 6), which, even today, affect billions of people. Invasive infections with helminths are typically characterized by eosinophilia. However, eosinophil function in host defenses is incompletely understood.
In this preliminary analysis, we studied a single sample from a single time point and conducted comparative proteomic analysis of eosinophils using available data (∼3,000 protein spots) from a healthy control (10). We noted that the level of expression of the vast majority of proteins was similar to that previously reported for quiescent peripheral blood eosinophils (control) (10). However, using a stringent statistical cutoff (P < 0.001), four proteins were statistically significantly upregulated in eosinophils from the Fasciola patient. These proteins included gelsolin, protein disulfide isomerase, neutral alpha-glucosidase, and the synaptic vesicle membrane protein vesicle amine transport 1 (VAT-1). Protein disulfide isomerase has been shown to play a role in L-selectin (CD62L) shedding by activated eosinophils (2). Gelsolin is thought to play a role in degranulation (4) and adherence to target cells. The synaptic vesicle protein VAT-1 may play a role in antiapoptotic and prosurvival mechanisms, typical of several cytokines (11). The spot for ezrin was also increased, but this was not statistically significant. Ezrin is known to be expressed and phosphorylated in activated eosinophils (9). Among the downregulated proteins, annexin-1, S100-A9, and macrophage migration inhibitory factor are released during degranulation (7, 8). The downregulation of the observed protein spots could also have been due to mobility changes during 2-DE as a result of posttranslational modification, and thus, they may not represent downregulated proteins. We and others have observed that activated eosinophils show considerable phosphorylation (9, 13). Further studies using more patients are planned to evaluate phosphorylation of eosinophil proteins as a result of Fasciola infection. Proteomic studies of circulating eosinophils from patients with atopic dermatitis or pollen allergy did not correlate well with each other or the current study (12, 13). This suggests that the eosinophil proteome may differ depending on the route of sensitization or the allergen, even in patients with eosinophilia.
Using data from healthy controls, this proteomic analysis demonstrated that not only were eosinophil numbers increased but there was also a set of proteins that were differentially expressed during parasitic infection. These results suggest significant activation or priming of the eosinophil by the Fasciola infection. Similarly, Klion et al. report that compared to eosinophils from healthy donors, those isolated from individuals that suffer from parasitic infections appear to degranulate in the periphery and show upregulation of the cell surface markers CD69, CD25, and others (5).
This preliminary report points toward the usefulness of comparative proteomics in studying the effects of parasitic infections on eosinophils or other cells within the body. This approach revealed a set of novel proteins that were differentially expressed during Fasciola infection, which suggests the potential of this approach for proteomic investigations into eosinophil function. We propose that future unbiased characterization of a comprehensive set of proteins from different patients, modulated in the course of parasitic infection, will provide novel insights into the molecular circuitry, signaling pathways, and cytokines that may play a role in the pathogenesis of parasite-induced eosinophilic inflammation.
Acknowledgments
This study was funded by the National Institutes of Health (grants NO1-HV-00245 to A.K., R01TW007642 to M.M., and R25TW008129 to A.C.W.) and a UTMB James W. McLaughlin Predoctoral Fellowship to C.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print on 28 September 2011.
REFERENCES
- 1. Abraham D., et al. 2004. Immunoglobulin E and eosinophil-dependent protective immunity to larval Onchocerca volvulus in mice immunized with irradiated larvae. Infect. Immun. 72:810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bennett T. A., Edwards B. S., Sklar L. A., Rogelj S. 2000. Sulfhydryl regulation of L-selectin shedding: phenylarsine oxide promotes activation-independent L-selectin shedding from leukocytes. J. Immunol. 164:4120–4129 [DOI] [PubMed] [Google Scholar]
- 3. Espinoza J. R., et al. 2007. Evaluation of FAS2-ELISA for the serological detection of fasciola hepatica infection in humans. Am. J. Trop. Med. Hyg. 76:977–982 [PubMed] [Google Scholar]
- 4. Fukuda T., et al. 1985. Calcium ionophore-A23187 calcium-dependent cytolytic de-granulation in human eosinophils. J. Immunol. 135:1349–1356 [PubMed] [Google Scholar]
- 5. Klion A. D., et al. 2004. Familial eosinophilia: a benign disorder? Blood 103:4050–4553 [DOI] [PubMed] [Google Scholar]
- 6. Meeusen E. N., Balic A. 2000. Do eosinophils have a role in the killing of helminth parasites? Parasitol. Today 16:95–101 [DOI] [PubMed] [Google Scholar]
- 7. Perretti M., D'Acquisto F. 2009. Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nat. Rev. Immunol. 9:62–70 [DOI] [PubMed] [Google Scholar]
- 8. Rossi A. G., et al. 1998. Human circulating eosinophils secrete macrophage migration inhibitory factor (MIF). Potential role in asthma. J. Clin. Invest. 101:2869–2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Starosta V., Pazdrak K., Boldogh I., Svider T., Kurosky A. 2008. Lipoxin A4 counterregulates GM-CSF signaling in eosinophilic granulocytes. J. Immunol. 181:8688–8699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Straub C., et al. 2009. Toward the proteome of the human peripheral blood eosinophil. Proteomics Clin. Appl. 3:1151–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ueki S., et al. 2008. Retinoic acids are potent inhibitors of spontaneous human eosinophil apoptosis. J. Immunol. 81:7689–7698 [DOI] [PubMed] [Google Scholar]
- 12. Woschnagg C., et al. 2009. The human eosinophil proteome. Changes induced by birch pollen allergy. J. Proteome Res. 8:2720–2732 [DOI] [PubMed] [Google Scholar]
- 13. Yoon S. W., Kim T. Y., Sung M. H., Kim C. J., Poo H. 2005. Comparative proteomic analysis of peripheral blood eosinophils from healthy donors and atopic dermatitis patients with eosinophilia. Proteomics 5:1987–1995 [DOI] [PubMed] [Google Scholar]