Abstract
The original report of sex in fungi dates 2 centuries ago to the species Syzygites megalocarpus (Mucoromycotina). The organism was subsequently used in 1904 to represent self-fertile homothallic species when the concepts of heterothallism and homothallism were developed for the fungal kingdom. In this study, two putative sex/MAT loci were identified in individual strains of S. megalocarpus, accounting for its homothallic behavior. The strains encode both of the high-mobility-group domain-containing proteins, SexM and SexP, flanked by RNA helicase and glutathione oxidoreductase genes that are found adjacent to the mating-type loci in other Mucoromycotina species. The presence of pseudogenes and the arrangement of genes suggest that the origin of homothallism in this species is from a heterothallic relative, obtained via a chromosomal rearrangement to switch two alleles into two separated loci within a single genetic background. Similar events have given rise to homothallic species from heterothallic species in ascomycete fungi, demonstrating that conserved forces shape the evolution of sex determination and speciation in highly diverged fungi.
INTRODUCTION
In research on mating and the mechanisms of sex determination in the fungi, one species stands out for its contributions in this eukaryotic kingdom. Syzygites megalocarpus played two key roles (1). First, it was the species for which sex was first reported, in 1820 (10). Second, it was the main representative of the homothallic (or self-compatible, self-fertile) fungi used in the 1904 research that differentiated fungal species into those with the heterothallic (or self-incompatible, outcrossing) and those with the homothallic mode of reproduction (4).
S. megalocarpus is a Mucoromycotina species (a zygomycete) found in the Northern Hemisphere growing as a parasite on mushrooms. As the first fungus in which sex was reported and probably due in part to the personal interactions between 19th century mycologists, S. megalocarpus was a commonly studied species for investigating sex in fungi. For instance, the name “zygospore” was coined by de Bary for those sexual structures of S. megalocarpus (9). Having been isolated and described on a number of independent occasions, the species and representative strains were also reassessed in the mid-1950s to clarify that this name had priority over an alternative name, Sporodinia grandis, that was in use (18).
The fungi are a kingdom of eukaryotes closely related to the animal kingdom. They are one of the most species-rich groups on earth, with 90,000 described species and an estimated 1.5 million species in total (17). The kingdom is split into multiple lineages, with the ascomycetes and basidiomycetes (collectively, the monophyletic Dikarya) making up about 95% of species and being the best-studied members. The success of the fungi in generating such diversity and inhabiting a wide range of environments can be attributed to many aspects of their physiology, including the production of spores by either sexual or asexual processes. Both spore-forming mechanisms have been extensively investigated because of the direct link between spores as the inocula for plant and animal diseases, because of the commercial propagation of fungi for the production of food, biocontrol agents, and pharmaceuticals, and for insights into the genetics of development. Sexual spore production is controlled by the MAT mating-type loci. These loci are regions of the genomes in fungi that exhibit similarities with sex-determining regions in other eukaryotes, including the presence of transcription factors and dissimilar DNA regions between the alleles of each sex or mating type. Identification, analysis, and comparison of mating-type loci from fungi have established this eukaryotic lineage to be a model for understanding the evolution of sex determination in all eukaryotes (15).
The identification of the sex locus of Phycomyces blakesleeanus represented the initial example of a mating-type locus for a fungus other than a member of the Dikarya (20, 21). P. blakesleeanus is heterothallic. Two strains of different sexes or mating types, designated minus (−) or plus (+) due to the lack of any morphological differences, are needed for successful mating. This leads to formation of a zygospore, in which occurs karyogamy, meiosis, and the mitotic amplification of the progeny in a germsporangium structure to produce haploid germspores (7, 12, 13). The genes responsible were identified by bioinformatics analysis of high-mobility-group (HMG) domain proteins in the genome sequence and examination of their distribution in strains of the (+) and (−) mating types by PCR analysis. Each sex of P. blakesleeanus contains a unique gene, sexM or sexP, at the same position within the genome flanked by the genes tptA and rnhA, which encode a predicted triose phosphate transporter and RNA helicase, respectively (21). HMG domain proteins regulate sex determination in a subset of other fungi, as well as in other organisms. For instance, a well-known animal example is Sry, the HMG domain protein encoded by a gene located on the Y chromosome that regulates male development in humans and other mammals. The (+) and (−) alleles of Phycomyces were defined genetically by Mendelian mapping to within a 38-kb region of the genome, with just seven other genes lying between the closest markers and sex alleles, and none of those seven genes being implicated in mating or meiosis in other organisms.
Similar sex loci have been characterized in other Mucoromycotina species. One potential mating-type allele was found in the completed genome of Rhizopus delemar (32), which is also a heterothallic species. Recent analysis has confirmed a similar gene arrangement in (+) and (−) strains of both R. delemar and Rhizopus oryzae (16). In these two species, the (+) allele contains an additional gene with BTB/POZ, ankyrin, and RCC1 domains. The role of this gene, if any, in controlling sexual reproduction is unknown. Both sex alleles were characterized in strains of Mucor circinelloides (28, 31). Mutation of the sexM gene eliminates the ability to form zygospores, demonstrating the essential role of this gene in sexual development (31). A sexP homolog has yet to be mutated. A remarkable observation was that the Microsporidia, organisms that at one stage were considered basal eukaryotes, also contain loci that are highly similar in gene order to the order seen in the Mucoromycotina sex/MAT loci (28). In the three Microsporidia species analyzed, genes encoding a predicted triose phosphate transporter (tptA) and an RNA helicase (rnhA) usually flank the sex genes. How this conserved synteny evolved is not clear because there are disparate gene phylogenies for the tptA and rnhA genes from the Microsporidia and Mucoromycotina (29). Regardless, this and other conserved gene orders place these enigmatic microbes in a group of organisms related to the Mucoromycotina.
Despite its seminal role in fungal biology and continued cytology until the mid-20th century (reviewed in references 8 and 26), S. megalocarpus has been largely neglected in the last half century. Beyond its distinct place within the history of biology, investigating the mating-type properties of S. megalocarpus presents an opportunity to identify the basis for homothallism in the Mucoromycotina. The underlying genetic basis for homothallism is unknown for any species of fungus outside the Dikarya lineage.
I hypothesized that S. megalocarpus would have mating-type genes and that their identification would explain the homothallism of this Mucoromycotina species. Here, amplification of a piece of the rnhA homolog, adjacent to the sex loci, enabled the subsequent sequencing of two regions of the S. megalocarpus genome that correspond to the sexM and sexP alleles of heterothallic Mucoromycotina species. The genetic arrangement provides an explanation for the homothallic properties of this fungus.
MATERIALS AND METHODS
Strains and cultivation.
S. megalocarpus strains were obtained from the American Type Culture Collection (ATCC; Manassas, VA) or the Centraalbureau voor Schimmelcultures (CBS; Utrecht, Netherlands) and are listed in Table 1. The ATCC strain was revived from a 1963 frozen stock, and the CBS strains were subcultured from slant cultures; strains were grown on potato dextrose agar. Mycelia were produced in liquid yeast extract-peptone-dextrose medium. Genomic DNA was extracted from lyophilized mycelia using a cetyltrimethylammonium bromide extraction buffer protocol (36). Strain ATCC 11807 was selected for initial cloning and analysis because it was the sole strain available from the ATCC and was recommended as typical of the species and suitable for experimentation (3, 18).
Table 1.
Strains, characteristics, and sequence accessions
| Strain name |
Isolationa |
Characteristics | GenBank accession no. |
|||||
|---|---|---|---|---|---|---|---|---|
| From source | From other collection | Yr | Location | Mushroom substrate | EF-1α | sexM | sexP | |
| ATCC 11807 | NRRL 2406 | <1953 | Probably Wisconsin | Unknown | Zygospores form; no germspores produced | JN112238 | JN112239 | JN112240 |
| CBS 372.39 | IMI 122577 | <1939 | Probably Europe | Unknown | No zygospores | JN112234 | JN112226 | JN112230 |
| CBS 715.95 | NAb | 1995 | Flevoland, Netherlands | Agaricus bitorquis | No zygospores | JN112235 | JN112227 | JN112231 |
| CBS 108947 | NA | 2000 | Baarn, Netherlands | Amanita rubescens | Zygospores form; germspores fertile | JN112236 | JN112228 | JN112232 |
| CBS 119041 | NA | 2005 | Merzligen, Switzerland | Ischnoderma benzoinum | Zygospores form; germspores do not germinate | JN112237 | JN112229 | JN112233 |
Isolation information is as provided by ARS, ATCC, or CBS.
NA, not applicable.
Cloning of a fragment of rnhA gene.
Degenerate oligonucleotides were designed to conserved regions in the tptA and rnhA genes. Those that amplified the rnhA homolog were 5′-AA(C/T)GA(A/G)CA(C/T)GA(A/G)GC(A/C/G/T)AA (A/G)(A/T)T(C/G/T)GC-3′ and 5′-TC(C/T)TC(A/C/G/T)CC(C/T)TG(A/G)TA (A/C/G/T)CC(A/G)TC(A/C/G/T)AC-3′ for amino acid residues NEHEAK(F/M) and VDGYQGEE. Those that amplified the tptA homolog were 5′-AA(T/C)TG(T/C)TG(T/C)ATGTGGTA(T/C)(A/G)-3′ and 5′-(T/C)TG(A/G)TACATCCA(A/C/G/T)AG(A/C/G/T)CC-3′ for residues NCCMWY(V/I) and GLWMYQ. PCRs were performed with Takara ExTaq in an Eppendorf Mastercycler thermal cycler. Amplicons were cloned into the pCR2.1 TOPO vector (Invitrogen, Carlsbad, CA), and the resulting plasmids were sequenced.
Nucleic acid manipulations.
For inverse PCR, ∼2 μg of genomic DNA was digested with a restriction enzyme and self-ligated with T4 DNA ligase (New England BioLabs, Ipswich, MA). Twelve enzymes were used: BamHI, BglII, ClaI, EcoRI, HindIII, KpnI, NcoI, PstI, SalI, SpeI, XbaI, and XhoI. The 12 ligations served as the templates for PCRs. While the majority of DNA sequencing was directly from inverse or conventional PCR products, for areas with stretches of nucleotide repeats that reduced sequence quality due to polymerase slippage, PCR products were cloned into the pCR2.1 TOPO plasmid and multiple independent plasmids were sequenced to provide a consensus. 5′ and 3′ rapid amplifications of cDNA ends (RACEs) to define transcript ends were conducted with a GeneRacer kit (Invitrogen). Dot plot comparison of the two regions utilized the YASS (33) and PipMaker (11) programs.
Part of the elongation factor 1 alpha (EF-1α) gene was amplified and sequenced to confirm that all strains were S. megalocarpus by comparisons with the sequences in the GenBank database. The primers used to amplify the EF-1α gene were MEF-1 (5′-ATGGGTAAAGA(A/G)AAGACTCACG-3′) and MEF-4 (5′-ATGACACC(A/G)ACAGCGACGGTTTG-3′) (34). These two primers and the internal primers ALID1247 (5′-AAGCTGGTGCCAAGTCTG-3′) and ALID1248 (5′-ACATGTTATCACCGTGCC-3′) were used to sequence the PCR products.
Phylogenetics analyses.
Predicted amino acid sequences were downloaded from GenBank or organism-specific genome databases. Protein sequences were aligned with the ClustalW program, and the alignment was inspected by eye. Neighbor-joining (1,000 bootstraps) and maximum-likelihood (100 bootstraps) methods were used in MEGA5 software (24), producing similar results.
RESULTS AND DISCUSSION
The strategy to find the candidate regions that control mating in Syzygites megalocarpus was first to identify the conserved genes flanking the sex loci in the Mucoromycotina lineage rather than directly amplify the HMG domain proteins encoded by sexM or sexP. Analysis of the genomes of Mucoromycotina species reveals a number of possible HMG domain proteins that could be implicated in mating type (21). This makes cloning based on sequence similarity of the few reported sexM and sexP homologs a challenge. The MAT loci of both basidiomycetes and ascomycetes have conserved genes on either side of the idiomorphic regions, and those flanks can be used as the primary targets for identifying full mating-type loci (6, 22, 23). Alignments of the predicted amino acids encoded by the tptA and rnhA genes of the three available Mucoromycotina species were made, and degenerate oligonucleotides were designed. A 200-bp fragment of the rnhA gene was amplified from DNA extracted from strain ATCC 11807. The tptA gene could not be amplified, whereas the primers worked to amplify this gene from another Mucoromycotina species (strain NRRL A-10032). The sequence of the fragment of the RNA helicase, which included the 46 bp of primer sequences and 154 bp of unique sequence, was used as the starting point for a sequential series of inverse PCRs that enabled sequencing in either direction. Two different sequences were obtaining starting with the RNA helicase, yielding 20,845 and 25,552 bp with both DNA strands sequenced (Fig. 1).
Fig. 1.
Two sex loci are present in S. megalocarpus. The color coding indicates HMG-domain-encoding genes (red, sexM and sexP), those conserved in Mucoromycotina species (green, blue, or brown), and those not associated with sex loci (gray). ψ, a pseudogene or fragment of a degenerated transposable element (Tn). The dark blue section of ψ rnhA indicates the inverted region within this pseudogene. Details about each of the genetic elements are provided in Table S1 in the supplemental material. Scale marks, 1 kb.
The gene content in these regions was predicted using FGENESH software and BLAST searches against the sequences in the GenBank and the R. delemar, M. circinelloides, and P. blakesleeanus genome databases. For the two regions analyzed, the rnhA homolog has to the 5′ side a gene named either sexM or sexP, both encoding an HMG domain-containing protein (Fig. 1; see Table S1 in the supplemental material). The ends of the sexM and sexP genes were defined by 5′ and 3′ rapid amplification of cDNA ends (RACE), also confirming that both genes are expressed. BLAST analysis with the predicted protein sequences and phylogenetic analysis showed that the closest matches are the SexM and SexP proteins of the Mucoromycotina (Fig. 2; see Fig. S1 in the supplemental material).
Fig. 2.
Phylogeny of the predicted SexM and SexP HMG domains. Sequences of the SexM and SexP proteins of Mucoromycotina species (16, 21, 28, 31, 39) were downloaded from GenBank, and 85 amino acids centered on the HMG domains were aligned and compared by neighbor-joining analysis. The numbers in gray adjacent to nodes are percent bootstrap support from 1,000 replicates, with values less than 65% omitted.
Dot plot comparisons revealed the extent of DNA conservation for the two putative sex loci in S. megalocarpus (see Fig. S2 in the supplemental material; Fig. 3). Similarity is across a region that includes the RNA helicase rnhA and glutathione oxidoreductase glrA homologs. Sequence comparison indicates that the RNA helicase adjacent to sexP and the glutathione oxidoreductase near sexM are pseudogenes. While the remnant RNA helicase shares a high degree of DNA sequence similarity, it has an ∼600-bp inversion within the middle of the gene and a ∼1.4-kb deletion (Fig. 3). Also, a predicted 4-bp deletion in the first exon and 1-bp deletion in the second would cause frameshift mutations. A 676-bp deletion removes part of the first exon of the sexM-associated glutathione oxidoreductase. Four other pseudogenes are the remnants of transposable or repetitive elements adjacent to the sexP gene. BLAST matches of these four elements in the R. delemar genome are all represented by multiple DNA sequences.
Fig. 3.
Dot plot comparison of the two sex loci in S. megalocarpus, covering sexM and sexP (red) and their 5′ regions. DNA common to both loci includes the RNA helicase (blue) and glutathione oxidoreductase (brown) genes. However, there are an inversion and a deletion within the RNA helicase adjacent to sexP, and there is a deletion that includes part of the first exon of the glutathione oxidoreductase associated with sexM. Scale marks, 1 kb. The plot was generated with YASS software using an entropy filter set at 4.4 and all other parameters set at default. A comparison of the entire sequenced regions is presented as Fig. S2 in the supplemental material.
The closest relative to S. megalocarpus thus far analyzed in phylogenetic studies is Rhizopus stolonifer (34, 40), a heterothallic species. This species or its MAT locus has not been sequenced. However, two alleles were described for both sexes of R. delemar and R. oryzae (16). Adjacent to the S. megalocarpus sexM gene is a gene (named arbA) encoding a multiple-domain (ankyrin-3× RCC1-BTB/POZ) protein, with this being the homolog of a gene within the sexP allele of the two Rhizopus species. In addition, a glutathione oxidoreductase found adjacent to the locus in R. delemar and M. circinelloides lies to the 3′ side of the S. megalocarpus RNA helicases. Examination of the flanks for the sex loci in the sequenced Mucoromycotina species revealed an additional conserved gene adjacent to the cluster for P. blakesleeanus and M. circinelloides, although not for S. megalocarpus (Fig. 4). This gene (named sagA) is of unknown function and may be a transcription factor since it contains a pfam04082 domain found in other transcriptional regulators. The conserved gene order surrounding Mucoromycotina sex loci further implicates the two regions sequenced in S. megalocarpus as being involved in mating.
Fig. 4.
Arrangement of sex loci for the sexM (M) or sexP (P) alleles or loci with their adjacent genes in Mucoromycotina species. Conserved genes do not extend beyond the regions illustrated. Fragments surrounding the sex loci have been sequenced from M. circinelloides (+), R. delemar (−), and P. blakesleeanus (+), hence the truncated alignments for these alleles. For clarity, the idiomorphic regions of the heterothallic species have been omitted; these lie between the tptA and rnhA genes. Enterocytozoon bieneusi is a member of the Microsporidia.
A curious aspect of the S. megalocarpus loci is the close proximity between the sexM and sexP genes and the RNA helicases. The remnant idiomorphic regions encompass only 43 and 40 bp between the stop codon of the sex genes and the start codon of the RNA helicases (Fig. 5). Analysis of transcript ends by RACE revealed that the sexM and rnhA genes produce overlapping transcripts, with the sexM transcript reading fully across the first exon of rnhA. This finding suggests that the RNA helicase gene may have been recruited into the mating-type locus. A similar observation has been made regarding the idiomorphic region and the promoters of the flanking genes in different clades of M. circinelloides, in which the promoter region of the flanking gene lies within the mating-type locus (28, 31). The longest transcript for rnhA identified by 5′ RACE starts 12 bp from the sexM stop codon (Fig. 5). It is thus also unclear what DNA acts as the promoter for the S. megalocarpus rnhA gene.
Fig. 5.
Expansion of the sex loci to acquire the adjacent RNA helicase gene. (A) Alignment of the 3′ ends of sexM and sexP with the start of sequence similarity, just prior to the start codon of the RNA helicase genes. The stop codons are in gray boxes, and the start codons are in black boxes. *, the position corresponding to where the poly(A) tail is attached to the sexP transcript, determined on the basis of 3′ RACE. Lowercase nucleotides represent the first intron in the rnhA homologs. (B) Overlapping transcripts for sexM and rnhA genes. Ends were amplified by 5′ or 3′ RACE, cloned, and sequenced. The poly(A) tail sequences for the sexM transcripts have been removed for clarity in alignment. Four different transcripts for each gene are illustrated.
One mechanism predicted to lead to homothallism in the Mucoromycotina is the generation of aneuploids, diploids, or heterokaryons containing the chromosomes encoding both alleles of the MAT loci. The earliest investigations into the underlying basis for homothallism and heterothallism in fungi were performed by Blakeslee on S. megalocarpus, Mucor mucedo, and P. blakesleeanus (5). Germination of S. megalocarpus zygospores produced progeny that were always homothallic. The situation in P. blakesleeanus was more complicated: Blakeslee described (+) or (−) progeny arising from zygospores but also, occasionally, as he described them, homothallic strains. Those strains produce mating-like pseudophore structures and in very rare cases zygospores. However, the trait is mitotically unstable, with reversion to strains showing either mating type and containing single sex alleles as assessed by PCR analysis (5, 21). The findings from P. blakesleeanus suggest that containing two nuclei or an aneuploid content may represent a mechanism to generate homokaryotic species in this subphylum. Other processes can lead to homothallism in fungi, with evidence that homothallism is derived from heterothallic states (30, 42), but, conversely, the possibility for the evolution of heterothallism from homothallic species also exists (2, 27). Thus, a number of options were possible to account for the homothallic behavior of S. megalocarpus.
Homothallism in S. megalocarpus can be explained by the presence of both sexM and sexP genes expressed within the same cell, with the two loci predicted to be contained within the same haploid nucleus. A model to explain the current arrangement of genes in S. megalocarpus is highly consistent with evolution from a heterothallic ancestor. Either a series of translocation events or a single segmental translocation occurred between two chromosomes (or at a distance within the same chromosome) to give rise to the current arrangement seen in this species (Fig. 6). In contrast, in many other homothallic fungi, the MAT gene organization can be explained by a single break in the MAT locus or its associated chromosome and a subsequent chromosomal rearrangement (Fig. 6). The flanks of sexM and sexP in S. megalocarpus are unusual because they have common regions, i.e., those including the RNA helicase and glutathione oxidoreductase genes, with the duplicated DNA supporting evolution of homothallism from a Rhizopus-like heterothallic ancestor being the most parsimonious option. The presence of remnant transposable elements adjacent to sexP may reflect a role for these elements in illegitimate recombination events that could drive chromosomal rearrangements.
Fig. 6.
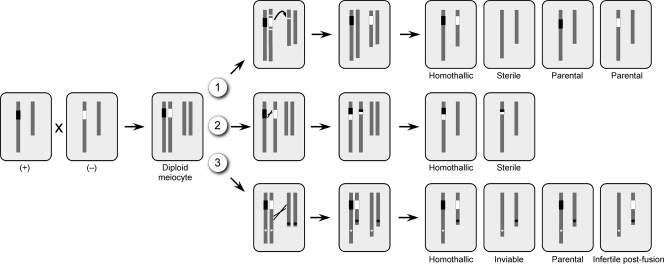
Model for the evolution of homothallic species from heterothallic ancestors through chromosomal rearrangements. Two chromosomes are illustrated, with the sex locus alleles colored black or white. The rearrangements are illustrated in a diploid state after two heterothallic parents of (+) and (−) sex undergo cell fusion, although situations 1 and 3 could also occur in a haploid cell. Reduction of the diploid to a haploid state would occur by meiosis or a parasexual loss of chromosomes. In scenario 1, a segmental translocation moves one allele to a new chromosome. Examples include S. megalocarpus, C. cymbopogonis, N. fischeri, and E. crustaceum. In scenario 2, asymmetric recombination between MAT alleles could generate strains with both mating type-determining genes. Examples include Gibberella zeae, Cochliobolus homomorphus, C. kusanoi, and C. luttrellii. In scenario 3, a reciprocal translocation could occur between the chromosome bearing MAT and another chromosome. Centromere positions are marked as circles. An example of such an event may have occurred in A. nidulans.
Two unlinked MAT loci are also observed in the homothallic ascomycetes Neosartorya fischeri and Eupenicillium crustaceum, both species presumed to be derived from heterothallic ancestors (37, 38). In N. fischeri, both flanks of the MAT locus are duplicated, and for one locus the conserved APN2 and SLA2 genes are pseudogenes. This contrasts to the related homothallic species Aspergillus nidulans, in which homothallism likely arose from a single chromosomal reciprocal translocation (35). In E. crustaceum, the MAT1-2-1 gene and two adjacent genes moved with one duplicated copy of SLA2, now a pseudogene. A third example of a segmental translocation event may also be found in the homothallic species Cochliobolus cymbopogonis (42). The ascomycete examples parallel what is seen in S. megalocarpus, which also has duplications and subsequent formation of pseudogenes. Thus, as a representative of a subphylum distant from the ascomycetes, the nature of the loci in S. megalocarpus indicates that similar genomic forces can shape the evolution of mating-type loci in the fungi and lead to the evolution of new species with homothallic properties from heterothallic ancestors.
A caveat to this research is the experimental evidence that the two putative sex loci are required for sexual reproduction. There are a limited number of tools for studying Mucoromycotina species. Attempts to isolate uracil auxotrophs of S. megalocarpus by plating on 5-fluoroorotic acid, as a step toward isolating homokaryotic strains or transformation for gene disruption, were unsuccessful. Staining of both asexual spores and sexual germspores with the nuclear dye 4′,6-diamidino-2-phenylindole revealed a large number (20+) of nuclei, accounting for this result and highly limiting the ability to isolate homokaryotic strains or creating gene disruption strains. A homokaryon could provide evidence that both loci are present in a single nucleus. Demonstrating that sexM and sexP are essential for zygospore production requires a DNA transformation system and functional RNA interference to silence the genes.
Different researchers have reached conflicting conclusions about S. megalocarpus biology. There is disagreement about the environmental conditions that trigger asexual sporulation, zygospore formation, and zygospore germination and the nuclear behavior within the zygospores (e.g., see discussions in references 8, 18, 25, and 41). Falck (14) and Blakeslee (5) reported that S. megalocarpus zygospores germinated and gave rise to homothallic progeny, while other researchers were unable to induce germination (26). In reviewing research on S. megalocarpus and with additional cytology, Cutter even suggested that the species may not undergo a meiotic cycle but rather undergoes apomixis and that the zygospore functioned as an asexual structure (8). In this study, strain ATCC 11807 was used for sequencing; however, while ATCC 11807 produces zygospores and they germinate, they do not make germspores. As a consequence, additional strains were sought to ensure that the DNA sequences of ATCC 11807 reflected the species in general and fertile strains.
S. megalocarpus is rare in culture collections. The World Federation for Culture Collections lists strains available from only five locations, one being a herbarium, with likely no more than seven strains still viable. This paucity in collections is not due to the rarity of the species but, rather, to the inability of the species to survive the lyophilization process that was and still is commonly used to preserve fungal species (19). For instance, lyophils of the strains used by Hesseltine (18), dating from the 1950s and 1960s and provided from the NRRL collection of the Agricultural Research Service (ARS), USDA, were tested but were not viable. Four strains were acquired from the CBS and examined for strain-specific differences. The five strains used in this study were isolated from different countries and over more than 65 years (Table 1). Zygospores formed for three of the five strains. The zygospores were placed on wet filter paper, and they germinated in 2 to 3 weeks to form the germsporangium structures. However, the three zygospore-producing strains behaved differently. As mentioned previously, the zygospores of strain ATCC 11807 germinated but did not make germspores. Those of CBS 119041 germinated and produced germspores, but these spores were inviable when plated. CBS 108947 zygospores germinated and made fertile germspores. Of 24 progeny tested, each derived from a separate zygospore of strain CBS 108947, all 24 were self-fertile and produced zygospores.
The sexM, sexP, and part of the EF-1α genes were amplified from the four CBS strains and sequenced. All four strains had the same sequences. These four strains are from Europe. Alignment of the sequences with those from strain ATCC 11807, which was isolated in the United States, reveal 32/4,567 (0.7%) nucleotide polymorphisms. The sequence that covered sexP also extended into the downstream RNA helicase gene that is a pseudogene in ATCC 11807. The same inversion event was observed in the four CBS strains, indicating that this is not a unique feature of ATCC 11807. Neither sexM nor sexP bore any mutation that could account for the observed differences in fertility, suggesting that other genes are responsible for the variation in fertility among strains. The phenotypic variation highlights the need to study a selection of strains to understand the reproductive biology of any fungus and is consistent with the previous conflicting reports on S. megalocarpus.
In summary, nearly 200 years ago Christian Ehrenberg observed zygospores of S. megalocarpus and for the first time in fungi proposed that these were sexual structures. The organism was instrumental in defining the processes of sexual reproduction in the kingdom. Here, two loci that are implicated in the production of zygospores in S. megalocarpus and the homothallic properties of this fungus are identified. The presence of a pair of sex loci supports the original conjecture that the S. megalocarpus zygospores are formed through a sexual process, although it is unclear whether or not meiosis occurs within the zygospore. Further understanding of the evolution and function of the sex loci may be achieved through genome sequencing of S. megalocarpus or analysis of closely related homothallic species, such as Rhizopus homothallicus or R. sexualis, as well as characterization of these loci in more distant relatives.
Supplementary Material
ACKNOWLEDGMENTS
I thank James Swezey and Kerry O'Donnell (ARS, USDA) for providing strains and advice.
This research was supported by funds from the UMKC School of Biological Sciences and the National Science Foundation (grant MCB-0920581).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Ainsworth G. C. 1976. Introduction to the history of mycology. Cambridge University Press, Cambridge, United Kingdom [Google Scholar]
- 2. Amselem J., et al. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Benny G. L., O'Donnell K. L. 1978. Syzygites megalocarpus, p. 127–128In Fuller M. S.(ed.), Lower fungi in the laboratory. University of Georgia, Athens, GA [Google Scholar]
- 4. Blakeslee A. F. 1904. Sexual reproduction in the Mucorineae. Proc. Am. Acad. Arts Sci. 40:205–319 [Google Scholar]
- 5. Blakeslee A. F. 1906. Zygospore germinations in the Mucorineae. Annal. Mycolog. 4:1–28 [Google Scholar]
- 6. Butler G., et al. 2004. Evolution of the MAT locus and its Ho endonuclease in yeast species. Proc. Natl. Acad. Sci. U. S. A. 101:1632–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cerdá-Olmedo E. 1975. The genetics of Phycomyces blakesleeanus. Genet. Res. 25:285–296 [DOI] [PubMed] [Google Scholar]
- 8. Cutter V. M., Jr 1942. Nuclear behavior in the Mucorales. II. The Rhizopus, Phycomyces and Sporodinia patterns. Bull. Torrey Club 69:480–508 [Google Scholar]
- 9. de Bary A. 1864. I. Syzygites megalocarpus, p. 74–88In Beiträge zur Morphologie und Physiologie der Pilze. Heinrich Ludwig Brönner's Verlag, Frankfurt, Germany [Google Scholar]
- 10. Ehrenberg C. G. 1820. Syzygites, eine neue Schimmelgattung, nebst Beobachtungen über sichtbare Bewegung in Schimmeln. Verhandl. Gesamte Naturf. Freunde, Berlin I:98–109 [Google Scholar]
- 11. Elnitski L., Riemer C., Schwartz S., Hardison R., Miller W. 2003. PipMaker: a World Wide Web server for genomic sequence alignments. Curr. Protoc. Bioinformatics Chapter 10:Unit 10.12. [DOI] [PubMed] [Google Scholar]
- 12. Eslava A. P., Alvarez M. I. 1996. Genetics of Phycomyces, p. 385–406In Bos C. J.(ed.), Fungal genetics: principles and practice. Marcel Dekker, Inc., New York, NY [Google Scholar]
- 13. Eslava A. P., Alvarez M. I., Delbrück M. 1975. Meiosis in Phycomyces. Proc. Natl. Acad. Sci. U. S. A. 72:4076–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Falck R. 1901. Die Bedingungen und die Bedeutung der Zygotenbildung bei Sporodinia grandis. Beiträge Biol. Pflanzen VIII:213–306 [Google Scholar]
- 15. Fraser J. A., et al. 2004. Convergent evolution of chromosomal sex-determining regions in the animal and fungal kingdoms. PLoS Biol. 2:e384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gryganskyi A. P., et al. 2010. Structure, function, and phylogeny of the mating locus in the Rhizopus oryzae complex. PLoS One 5:e15273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hawksworth D. L. 2001. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 105:1422–1432 [Google Scholar]
- 18. Hesseltine C. W. 1957. The genus Syzygites (Mucoraceae). Lloydia 20:228–237 [Google Scholar]
- 19. Hwang S.-W. 1966. Long-term preservation of fungus cultures with liquid cultures with liquid nitrogen refrigeration. Appl. Microbiol. 14:784–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Idnurm A., James T. Y., Vilgalys R. 2007. Sex in the rest: mysterious mating in the Chytridiomycota and Zygomycota, p. 407–418In Heitman J., Kronstad J. W., Taylor J. W., Casselton L. A.(ed.), Sex in fungi: molecular determination and evolutionary implications. ASM Press, Washington, DC [Google Scholar]
- 21. Idnurm A., Walton F. J., Floyd A., Heitman J. 2008. Identification of the sex genes in an early diverged fungus. Nature 451:193–196 [DOI] [PubMed] [Google Scholar]
- 22. James T. Y., Kües U., Rehner S. A., Vilgalys R. 2004. Evolution of the gene encoding mitochondrial intermediate peptidase and its cosegregation with the A mating-type locus of mushroom fungi. Fungal Genet. Biol. 41:381–390 [DOI] [PubMed] [Google Scholar]
- 23. James T. Y., Srivilai P., Kües U., Vilgalys R. 2006. Evolution of the bipolar mating system of the mushroom Coprinellus disseminatus from its tetrapolar ancestors involves loss of mating-type-specific pheromone receptor function. Genetics 172:1877–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamura K., et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaplan J. D., Goos R. D. 1982. The effect of water potential on zygospore formation in Syzygites megalocarpus. Mycologia 74:684–686 [Google Scholar]
- 26. Keene M. L. 1914. Cytological studies of the zygospores of Sporodinia grandis. Ann. Bot. 28:455–470 [Google Scholar]
- 27. Lee J., Lee T., Lee Y.-W., Yun S.-H., Turgeon B. G. 2003. Shifting fungal reproductive mode by manipulation of mating type genes: obligatory heterothallism of Gibberella zeae. Mol. Microbiol. 50:145–152 [DOI] [PubMed] [Google Scholar]
- 28. Lee S. C., et al. 2008. Microsporidia evolved from ancestral sexual fungi. Curr. Biol. 18:1675–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lee S. C., et al. 2010. Evolution of the sex-related locus and genomic features shared in Microsporidia and fungi. PLoS One 5:e10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee S. C., Ni M., Li W., Shertz C., Heitman J. 2010. The evolution of sex: a perspective from the fungal kingdom. Microbiol. Mol. Biol. Rev. 74:298–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li C. H., et al. 2011. Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 7:e1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma L.-J., et al. 2009. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 5:e1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Noe L., Kucherov G. 2005. YASS: enhancing the sensitivity of DNA similarity search. Nucleic Acids Res. 33:W540–W543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. O'Donnell K., Lutzoni F., Ward T. J., Benny G. L. 2001. Evolutionary relationships among the mucoralean fungi (Zygomycota): evidence for family polyphyly on a large scale. Mycologia 93:286–296 [Google Scholar]
- 35. Paoletti M., et al. 2007. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr. Biol. 17:1384–1389 [DOI] [PubMed] [Google Scholar]
- 36. Pitkin J. W., Panaccione D. G., Walton J. D. 1996. A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant-pathogenic fungus Cochliobolus carbonum. Microbiology 142:1557–1565 [DOI] [PubMed] [Google Scholar]
- 37. Pöggeler S., O'Gorman C. M., Hoff B., Kück U. 2011. Molecular organization of the mating-type loci in the homothallic ascomycete Eupenicillium crustaceum. Fungal Biol. 115:615–624 [DOI] [PubMed] [Google Scholar]
- 38. Rydholm C., Dyer P. S., Lutzoni F. 2007. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot. Cell 6:868–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sugui J. A., Christensen J. A., Bennett J. E., Zelazny A. M., Kwon-Chung K. J. 2011. Hematogenously disseminated skin disease caused by Mucor velutinosus in a patient with acute myeloid leukemia. J. Clin. Microbiol. 49:2728–2732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Voigt K., Wöstemeyer J. 2001. Phylogeny and origin of 82 zygomycetes from all 54 genera of the Mucorales and Mortierellales based on combined analysis of actin and translation elongation factor EF-1α genes. Gene 270:113–120 [DOI] [PubMed] [Google Scholar]
- 41. Wenger C. J., Lilly V. G. 1966. The effects of light on carotenogenesis, growth, and sporulation of Syzygites megalocarpus. Mycologia 58:671–680 [PubMed] [Google Scholar]
- 42. Yun S.-H., Berbee M. L., Yoder O. C., Turgeon B. G. 1999. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc. Natl. Acad. Sci. U. S. A. 96:5592–5597 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.