Abstract
Splicing of mRNA is an ancient and evolutionarily conserved process in eukaryotic organisms, but intron-exon structures vary. Plasmodium falciparum has an extreme AT nucleotide bias (>80%), providing a unique opportunity to investigate how evolutionary forces have acted on intron structures. In this study, we developed an in vivo luciferase reporter splicing assay and employed it in combination with lariat isolation and sequencing to characterize 5′ and 3′ splicing requirements and experimentally determine the intron branch point in P. falciparum. This analysis indicates that P. falciparum mRNAs have canonical 5′ and 3′ splice sites. However, the 5′ consensus motif is weakly conserved and tolerates nucleotide substitution, including the fifth nucleotide in the intron, which is more typically a G nucleotide in most eukaryotes. In comparison, the 3′ splice site has a strong eukaryotic consensus sequence and adjacent polypyrimidine tract. In four different P. falciparum pre-mRNAs, multiple branch points per intron were detected, with some at U instead of the typical A residue. A weak branch point consensus was detected among 18 identified branch points. This analysis indicates that P. falciparum retains many consensus eukaryotic splice site features, despite having an extreme codon bias, and possesses flexibility in branch point nucleophilic attack.
INTRODUCTION
Introns are noncoding sequences located inside precursor mRNA (pre-mRNA) transcripts that are excised before nuclear export (39). Splicing of pre-mRNA requires sequence motifs in the intron and is mediated by a ribonucleoprotein complex called the spliceosome (37, 53). While the fundamental mechanisms of splicing are conserved among eukaryotes (4), splice site recognition motifs are short and often only weakly conserved between organisms (20). In addition, most predictive software relies on model organisms, and there has been only limited experimental characterization of intron splicing in deep-branching eukaryotic organisms (50). Introns are predicted to be present in approximately half the genes in the Plasmodium falciparum genome (11). However, inaccuracies in intron prediction have been reported (26, 32, 42). P. falciparum has an extremely AT-rich genome (11), which has implications for both evolutionary adaptations of the spliceosome machinery and accuracy of gene structure predictions.
All spliceosomal introns contain 5′ donor and 3′ acceptor splice sites, usually with GU and AG dinucleotides at the respective intron ends and a branch point located within the intron. Major-class introns often contain a polypyrimidine tract adjacent to the 3′ splice site, which is missing in minor-class introns (29, 39). During splicing, the branch point nucleotide initiates a nucleophilic attack on the 5′ donor splice site. The free end of the upstream intron then initiates a second nucleophilic attack on the 3′ acceptor splice site, releasing the intron as an RNA lariat and covalently combining the two exons (53). Intron size variation is largely due to the distance from the intron's 5′ boundary to the branch site. The vast majority of introns are removed by the major spliceosome, a large molecular machine which contains over 70 proteins and five ribonucleoprotein complexes. Each complex contains one of five small nuclear RNAs (snRNAs), named U1, U2, U4, U5, and U6, plus a set of common proteins and additional proteins that are specific to individual snRNAs (37, 53). Base pairing of the snRNAs to the intron and to each other, plus protein-protein and protein-RNA interactions of splicing factors, position the splice sites for splicing (4, 53).
Five P. falciparum snRNAs have recently been identified (5, 11, 49). They are similar to those of vertebrates and appear to be capable of folding into the same overall conformation (5). The 5′ and 3′ splice site motifs for P. falciparum introns (GU and AG dinucleotides) follow the general eukaryotic pattern (5, 42). However, fewer than 10% of P. falciparum introns contain a sequence that corresponds to the common conserved branch point consensus (20), and no P. falciparum branch point has been experimentally determined thus far. Experimental validation of the P. falciparum intron sequence requirements is therefore of interest.
P. falciparum introns are comparatively short and extremely AT rich, averaging 179 nucleotides (nt) and 86.5% A+T (11). Some are located at the end of an open reading frame, adding a single amino acid (22), or in an untranslated region (UTR) (33, 34). They are implicated in control of var gene expression (8), and instances of stage-specific expression, sex-specific expression, and alternative splicing have been reported (26, 32, 42). These include the 41-3 antigen (23), the surface antigen MAEBL (41), an adenylate cyclase with alternate AUGs due to splicing patterns (30), an aspartyl protease with three variants (52), the SET antigen (for which splicing is sex regulated in gametocytes) (33, 34), and adjacent genes with two shared 5′ exons that are spliced to different downstream exons (51). Recent evaluations of alternative splicing via RNA-Seq analysis have identified numerous additional alternative splicing events in P. falciparum pre-mRNAs (32, 42). In summary, splicing is a key element of gene expression which has been little studied in Plasmodium. In this study, we developed an in vivo luciferase reporter splicing assay and employed it in combination with lariat isolation and sequencing to characterize 5′ and 3′ splicing requirements and experimentally determine the intron branch point.
MATERIALS AND METHODS
Lariat isolation and sequencing.
RNA was isolated from saponin-lysed 3D7-infected red blood cells at mixed stages using TRIzol LS (Invitrogen) and chloroform according to the manufacturer's instructions. RNA (100 ng) was incubated with Ambion RNase-free DNase I to remove genomic DNA. Prior to reverse transcription, half of the RNA was treated with 1 μg RNase R in buffer containing 20 mM Tris-HCl (pH 8.2), 100 mM KCl, and 0.25 mM MgCl2 at 37°C for 60 min to digest linear RNAs and enrich lariat RNAs. The other half was mock treated in buffer alone. For reverse transcription, SuperScript III reverse transcriptase (Invitrogen) was employed with a 50:50 mixture of random primers and oligo(dT) primers, following the manufacturer's instructions. cDNA and RNA lariat products were amplified for four different P. falciparum genes (PF08_0019, PF14_0027, PF10_0155, and PFL0190w) using the PCR primers listed in Table S1 in the supplemental material. PCRs were performed using the Expand high-fidelity PCR system (Roche PCR) in a 50-μl reaction volume containing 1× PCR buffer with 1.5 mM MgCl2, 2 μM primers, 0.2 mM deoxynucleoside triphosphates (dNTPs), and 0.05 U/μl Taq polymerase. Thirty cycles were used for cDNA amplification. For lariat amplification, nested primers were designed in divergent orientation so that the PCR product would cross the branch point. A 25-cycle first round of PCR was followed by a 20-cycle second round, with the PCR product purified between rounds using the Qiagen PCR purification kit. The lariat PCR products were extracted from agarose gels, purified with the QIAquick gel extraction kit (Qiagen), cloned into T-easy vector (Promega), and transfected into Escherichia coli DH5 competent cells. The plasmid was extracted with a plasmid minikit (Qiagen) and submitted for DNA sequencing.
Construction of pBtub-fLuc fusion vectors with intron variations for monitoring mRNA splicing.
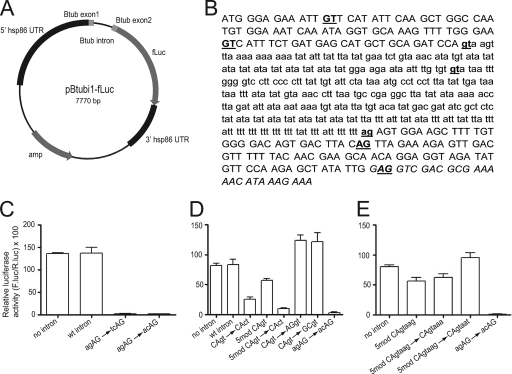
To investigate mRNA splicing requirements in P. falciparum, a fusion vector (pBtubi-fLuc) was designed containing the first intron and flanking exon sequences of β-tubulin (PF10_0084) fused to firefly luciferase in the pPf86 vector backbone (kindly provided by Dyann Wirth and Kevin Militello) (28). A β-tubulin N-terminal fusion vector (pBtub-fLuc) that lacks the β-tubulin intron sequence was constructed as a control for RNA splicing experiments, and clones with intron variations were created from this vector. Construction of the pBtub-fLuc fusion vector and intron variations is detailed in “Supplementary experimental procedures” in the supplemental material. Table S2 in the supplemental material lists all plasmids created during this study as well as the β-tubulin sequence in each plasmid. Plasmids were verified by sequencing prior to transfection. A representative plasmid map for the pBtubi-fLuc vector is shown in Fig. 1, along with the native β-tubulin intron sequence and flanking exon sequence employed in vector construction.
Fig. 1.
Analysis of pre-mRNA splicing in P. falciparum utilizing a β-tubulin-luciferase fusion construct. (A) pBtubi1-fluc reporter construct. The first intron of the P. falciparum β-tubulin gene, with surrounding exonic regions, was fused in frame 5′ to a firefly luciferase reporter gene. (B) Sequence of β-tubulin included in the pBtub-fluc construct. Uppercase, exon sequence; lowercase, intron sequence. Potential alternative in-frame 5′ GT and 3′ AG nucleotides are underlined. (C to E) The relative amount of firefly luciferase activity was normalized by cotransfecting with a Renilla luciferase expressing plasmid. “No intron” is the control fusion plasmid in which the β-tubulin intron has been removed. Plasmid constructs labeled “5mod” are ones in which three alternative in-frame 5′ GT residues were replaced by the nucleotides CT.
P. falciparum culture, transfection, and luciferase assay.
D10 parasites were used for all transfection experiments and were cultured under standard conditions (47) in RPMI 1640 medium supplemented with 0.5% (wt/vol) Albumax II (Invitrogen, Carlsbad, CA) and human red blood cells at 2% hematocrit. Cultures were synchronized using 5% sorbitol (24). Schizonts were isolated using magnetic-activated cell sorting (MACS) purification (48) and allowed to reinvade new red blood cells for 12 h before transfection, resulting in high-parasitemia ring-stage-infected red blood cells at the time of transfection. Infected red blood cells were electroporated using a Bio-Rad Gene Pulser Xcell with 75 μg of experimental plasmid and 50 μg of Renilla control plasmid (kindly provided by Dyann Wirth and Kevin Militello). Electroporations were performed in 0.2-cm cuvettes using low voltage (0.31 kV) and high capacitance (950 μF) (9). Transfectants were maintained on phenol red-free medium and harvested after 16 h by centrifugation. Parasite lysis and luciferase assay were performed according to the manufacturer's protocol with the dual-luciferase reporter assay system (Promega) using a Sirius single-tube luminometer (Berthold).
Sequencing of β-tubulin reporter construct mRNA.
RNA was harvested by TRIzol LS (Invitrogen)–1-bromo-3-chloropropane extraction from D10-infected red blood cells transiently transfected with the pβtubi1-fLuc plasmid containing the wild-type intron. Plasmid DNA contamination of this RNA was reduced by treatment with the Turbo DNA-free kit (Ambion) and a cocktail of restriction enzymes, including AluI, MnlI, and NlaIII (NEB), and by phenol-chloroform and 1-bromo-3-chloropropane extractions. Reverse transcription-PCR (RT-PCR) was performed using TaqMan reverse transcription reagents (Applied Biosystems) and the Expand high-fidelity PCR System (Roche). RT-PCR was performed using a forward primer in β-tubulin exon 1 (5′ CATATTCAAGCTGGCCAATGTGGAAATC) and a reverse primer in firefly luciferase (5′ AGTTGCTCTCCAGCGGTTCCATC). The product was isolated from an agarose gel using the Wizard SV gel and PCR clean-up system (Promega) and then cloned into pGEM-T Easy (Promega). Plasmids were prepared with the FastPlasmid miniprep kit (5 PRIME) and sequenced using SP6 and T7 primers.
Sequence analysis of P. falciparum and human introns.
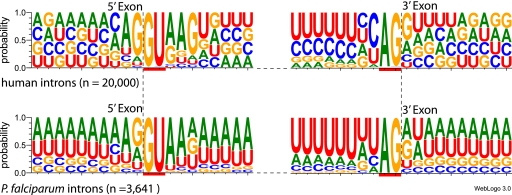
Sequences surrounding the 5′ donor and 3′ acceptor splice sites were obtained by using the Program to Assemble Spliced Alignments (PASA) (15) to assemble P. falciparum expressed sequence tags (ESTs) to the 3D7 genome sequence. ESTs were obtained from two sources: 14,362 P. falciparum asexual ESTs and 5,814 P. falciparum gametocyte ESTs were obtained from GenBank, and 7,683 P. falciparum 3D7 ESTs were generated in the Gardner lab from mixed asexual stages (Z. Hang and M. Gardner, unpublished data). The PASA alignments generated 3,641 P. falciparum EST-confirmed intron sequences and the exon sequences flanking each intron. Data for 20,000 introns were obtained from a set of 1,533 human genes in the NCBI RefSeq database (36). Twenty-nucleotide-long sequences surrounding the splice sites were used to generate sequence logos with WebLog 3 (http://weblogo.threeplusone.com/) (6) using the default parameters, except that the classic color scheme was used and the units were plotted as “probability.”
RESULTS AND DISCUSSION
Bioinformatic analysis of P. falciparum introns.
The P. falciparum genome annotation relies upon a mixture of sequence-confirmed and computationally predicted exon-intron structures (11). Because some predictions may be inaccurate, PASA was used to align and assemble 23,817 P. falciparum ESTs against the 3D7 genome in order to produce a validated set of P. falciparum introns for analysis. A total of 3,641 EST-confirmed introns were identified (Table 1), along with exon sequences flanking the 5′ and 3′ splice sites. The close fit of size distributions of our intron data set compared with the original genome annotation (11) indicates that this is a representative sample of P. falciparum introns (Table 1). Intron length ranged from 38 nucleotides to almost 4 kb, but the average size is small, with only 20 introns exceeding 1 kb. Analysis of the positions flanking each junction shows a good match to the eukaryotic consensus for splice sites. The canonical 5′GU-AG 3′ sequence was almost 100% conserved, and the immediately flanking exon sequences had a consensus sequence of WR for the 5′ and RW for the 3′ splice site (Table 2). On average, 87% of P. falciparum intron residues are A or T (11). This bias is also generally observed in the intron sequences immediately adjacent to the GT-AG splice sites, except that the fifth nucleotide position at the 5′ splice site is an equal distribution of A, T, or G nucleotides (Fig. 2). In other organisms, a G nucleotide is more common in this position (Fig. 2), so this difference was investigated in an RNA splicing assay described below. Many U2 spliceosomal introns have a polypyrimidine tract immediately adjacent to the 3′ splice site, which is recognized by the splicing factor U2AF65. Interactions of this protein with U2AF35, which recognizes the 3′ splice site, and proteins involved in branch point recognition are important elements for positioning the components needed for splicing (29). P. falciparum has putative orthologs for U2AF35 (PF11_0200) and U2AF65 (PF14_0656), as well as other key splicing factors, and, as expected, most introns contain a polypyrimidine tract adjacent to the 3′ splice site consensus sequence (YAGRW). However, the P. falciparum polypyrimidine tract contains fewer C nucleotides than human introns and has more A nucleotides (Fig. 2).
Table 1.
Intron characteristics
| Intron characteristic | Value from: |
|
|---|---|---|
| Annotated sequence (11) | PASA assemblies | |
| No. | 7,406 | 3,641 |
| Largest (nt) | 3,040 | 3,998 |
| Smallest (nt) | 3 | 38 |
| Avg (nt) | 179 | 179 |
Table 2.
Splice junction sequences
| Splice site and characteristic | Value at nt position (relative to splice junction): |
||||||
|---|---|---|---|---|---|---|---|
| −3 | −2 | −1 | 1 | 2 | 3 | ||
| 5′ | |||||||
| % of base: | |||||||
| G | 8.9 | 53.7 | 100 | —a | 2.8 | ||
| A | 64.2 | 26.7 | 0 | — | 85.5 | ||
| U | 19.4 | 14.4 | 0 | 99.7 | 10.2 | ||
| C | 7.2 | 5.1 | 0 | — | — | ||
| Consensusb | |||||||
| P. falciparum | W | R | g | u | a | ||
| Eukaryote | A | G | g | u | r | ||
| 3′ | |||||||
| % of base: | |||||||
| G | <1 | — | 99.5 | 36 | 12.3 | ||
| A | 12.1 | 96.8 | — | 39.6 | 34.7 | ||
| U | 68 | — | — | 15.7 | 41.7 | ||
| C | 19.3 | — | — | 5.5 | 10.8 | ||
| Consensus | |||||||
| P. falciparum | y | a | g | R | W | ||
| Eukaryote | y | a | g | G | U | ||
—, less than 0.5%.
Exon and intron sequences are in upper- and lowercase, respectively.
Fig. 2.
Comparison of 5′ and 3′ splice site WebLogos for humans and P. falciparum. The height of each letter indicates the preference strength for that nucleotide at each position.
Effect of 5′ and 3′ splice site mutations on mRNA splicing efficiency in P. falciparum.
To investigate mRNA splicing requirements in P. falciparum, we developed a transient-transfection assay in which firefly luciferase expression is dependent on proper intron splicing (Fig. 1A). P. falciparum β-tubulin has two short introns, 350 and 168 nt long (11). The first intron of the P. falciparum β-tubulin gene, with short flanking exon regions, was fused in frame 5′ to a firefly luciferase reporter gene (Fig. 1). The intron contains stop codons in all three reading frames to stop translation if splicing does not occur. The reporter plasmid was cotransfected with a Renilla luciferase-expressing plasmid into P. falciparum-infected red blood cells to normalize activity between experiments. A firefly construct containing only the β-tubulin exon sequences, without an intron, was included as a positive control. The intron-containing and intronless constructs produced similar amounts of luciferase activity (Fig. 1C and D, first two bars). The validity of the splicing assay was checked by RT-PCR and sequencing of the spliced mRNA fusion product, which was found to be spliced at the expected sites of the native β-tubulin gene (data not shown).
To explore sequence requirements for efficient splice site recognition, a number of plasmids containing modified β-tubulin sequences were prepared and tested in the splicing assay. At the 3′ splice site, the native sequence, agAG, was mutated to tcAG and acAG (intron sequences are in lowercase). Both changes from the consensus completely abolished firefly luciferase activity (Fig. 1C). In comparison, mutation of the 5′ splice site from CAgt to CAct significantly reduced but did not abolish luciferase activity (Fig. 1D, bar 3), possibly due to the presence of potential alternate 5′ splice sites both upstream and downstream of the exon-intron boundary (Fig. 1B, underlined GTs).
The first intron in P. falciparum β-tubulin has a rare 5′ splice acceptor site, CAgt (Table 3). One of the potential alternate in-frame splice sites would be the much more common AAgt 5′ splice acceptor site (Table 3). To investigate sequence features that influence 5′ splice recognition in P. falciparum, we mutated the AAgt and two other potential alternative splice sites from GT to CT (Fig. 1B, underlined nucleotides) while retaining the native 5′ splice site (construct 5mod CAgt). This combination of mutations only partially reduced luciferase activity (Fig. 1D, bar 4), indicating that the native site was the preferred recognition site in the reporter construct despite being a rare type. Mutating the native 5′ splice site in addition to the possible alternate sites reduced luciferase activity almost to background levels (Fig. 1D, bar 5). Of interest, there are also alternative in-frame AG sequences downstream of the 3′ splice site (Fig. 1B, underlined AGs). These do not seem to be able to compensate for loss of the 3′ splice site, probably because they are not adjacent to a polypyrimidine tract (Fig. 1B) and are therefore not recognized by U2AF65. Taken together, these data demonstrate that P. falciparum has conventional 5′GU-AG3′ splice site motifs.
Table 3.
5′ Splice junction frequencies
| Sequencea | No. (3,641 total introns) |
|---|---|
| AGgt | 1,207 |
| AAgt | 662 |
| ATgt | 350 |
| ACgt | 119 |
| GAgt | 93 |
| GGgt | 169 |
| GTgt | 46 |
| GCgt | 17 |
| TAgt | 125 |
| TGgt | 455 |
| TTgt | 96 |
| TCgt | 30 |
| CGgt | 116 |
| CAgt | 92 |
| CTgt | 32 |
| CCgt | 21 |
Upper- and lowercase indicate exon and intron sequences, respectively.
To further investigate how rare 5′ splice recognition sequences are recognized by the P. falciparum spliceosomal machinery, we also explored the role of exon sequence in splicing efficiency. Unexpectedly, mutation of the exon side of the 5′ splice site from the native CAgt to the more common AGgt or the extremely rare GCgt (Table 3) both led to a significant increase in luciferase activity (Fig. 1D, bars 6 and 7). Thus, the major factor determining splice site preference for this intron is not the immediately preceding exon dinucleotide but presumably other features in the flanking exon or intron sequence. These data also suggest that the exon sequences immediately preceding an intron may influence splicing efficiency or otherwise affect protein levels. All 16 dinucleotide combinations occur at least occasionally prior to P. falciparum introns (Table 3). The potential that they may have a role in regulating expression is intriguing.
In humans and Saccharomyces cerevisiae (5), the fifth position of the intron is a highly conserved G. In contrast, A, G, and T are equally common at this position in P. falciparum (Fig. 2), despite the fact that the U1 snRNA and U6 snRNA residues proposed to interact with this position (25, 40) are unchanged between S. cerevisiae and P. falciparum (5). Mutation of G5 in S. cerevisiae has been shown to reduce splicing efficiency (10) and is the second leading cause of activation of new, aberrant 5′ splice sites in human genetic diseases, including beta-thalassemia, McLeod neuroacanthocytosis syndrome, and Duchenne muscular dystrophy (1, 3, 7, 46). To test whether the fifth nucleotide also influences splicing activity in P. falciparum, we mutated this residue from G (the native residue in the first intron of ß-tubulin) to A or T. Both are common at this position in P. falciparum and are much rarer in humans (Fig. 2) or S. cerevisiae (39). These mutations were made in the β-tubulin plasmid construct in which possible alternate 5′ splice sites were mutated to ensure that the native 5′ splice site was used. Unexpectedly, the constructs with an A substitution had splicing activity comparable to that of the native G construct, while substituting a T produced an increase (Fig. 1E). Therefore, P. falciparum is relatively tolerant of A or T substitutions at this location, despite the fact that U1 and U6 residues predicted to interact with this nucleotide should preferentially base pair with a G at position 5 (5).
P. falciparum branch point identification and branch site consensus motif.
In the first step of pre-mRNA splicing, spliceosome assembly involves base pairing between a sequence in U2 snRNA and the branch point (4, 53). In most eukaryotes, the branch point is typically located near the 5′ end of a polypyrimidine tract and base pairs with a 6-nt motif in U2 snRNA (4, 53). For instance, S. cerevisiae introns each contain one branch point at an A 10 to 155 nucleotides upstream of the 3′ splice site. This branch point (underlined A) is located within a highly conserved UACUAAC sequence and base pairs with an 8-nt U2 snRNA sequence (43). The mammalian branch point is found in a less-conserved YURAY sequence, usually 18 to 37 nucleotides upstream of the 3′ splice site (39, 53). Despite the fact that the P. falciparum U2 snRNA sequence is identical to those of S. cerevisiae and human U2 snRNAs at the branch point interaction region (5), fewer than 10% of P. falciparum introns have an apparent consensus branch point sequence similar to those of S. cerevisiae or humans (39). Therefore, we experimentally determined branch points for four P. falciparum introns.
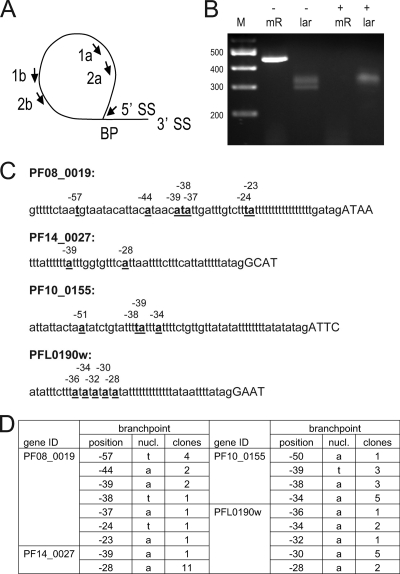
During splicing, two sequential transesterifications produce an RNA lariat. Lariats are formed from pre-mRNA by the nucleophilic attack of a bulged A at the branch point on the 5′ donor splice site. This creates a 2′-5′ linkage rather than the typical 3′-5′ bond, producing a circular RNA with a short linear tail (53). RNA lariats are short-lived but can be detected at a low concentration in total RNA (44). Using nested primers in divergent orientation, RT-PCR products that cross the branch point can be obtained (Fig. 3). We focused on genes that are moderately to highly expressed to increase the chance of finding intron lariats. The choice of intron within each gene was guided by intron size and sequence. We obtained RT-PCR products for four P. falciparum introns. These were cloned and sequenced, and the branch point was identified by comparison to the genomic DNA sequence. Altogether, 18 branch sites were identified (Fig. 3). Unexpectedly, there were multiple branch points for all four introns, and both A and U branch points were found, although A branch points predominated (14 of 18). As a control, the branch points of the human β-globin intron and of an S. cerevisiae ribosomal protein intron were analyzed. For both, all clones had the same branch point, located at an A (data not shown). This approach, i.e., PCR amplification across the branch point, was also recently used to identify branch points for trans-spliced Trypanosoma brucei RNAs. Here too, multiple branch points per intron were detected (27). Most were A branch points, but C branch points were observed. Although they are less common, U or C branch points have been reported for cis splicing (12, 13, 16).
Fig. 3.
Summary of branch point mapping in P. falciparum. (A) Schematic of the RT-PCR approach to map the branch points in four P. falciparum introns. (B) Agarose gel of PCR products amplified from RNA lariats of receptor for activated C kinase homolog (PfRACK, PF08_0019). + or − indicates whether the RNA sample was treated with RNase R prior to reverse transcription. M, DNA marker; mR, mRNA; lar, lariat. DNA marker sizes in base pairs are on the left. (C) The experimentally determined branch points are indicated by underlined letters. The distance of each branch point from the 3′ splice site is indicated. (D) Summary of branch point information.
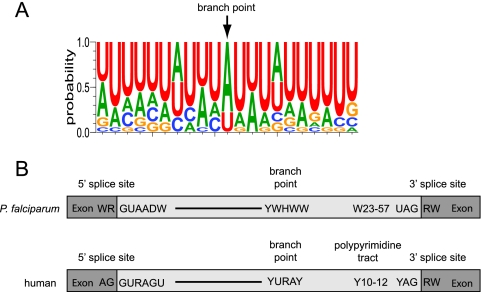
In P. falciparum, branch points were located 23 to 57 nt upstream of the 3′ splice site (Fig. 3). Comparison of the 18 branch points identified a YWHWW consensus motif (Fig. 4), which is related to the human consensus sequence (YURAY) but has more A/T bias and is less conserved. Although the branch point interaction site in P. falciparum U2 snRNA is conventional (5), we failed to find strong base pairing potential between U2 and the observed P. falciparum branch points.
Fig. 4.
Consensus sequences found in P. falciparum introns. (A) WebLogo for the 18 branch points identified in four P. falciparum sequences. (B) The consensus sequences at the 5′ and 3′ splice sites and branch point sequences are indicated. The cutoffs for the 5′ and 3′ splice site consensus sequences were set at 20%, and that for the branch point was set at 10% because there were fewer sequences. The distance between the 3′ splice site and branch point is shown. W = A or T, R = A or G, D = T or A or G, Y = C or T, and H = C or A or T.
Base pairing between snRNAs and intron sequences is a key element for bringing the 5′ and 3′ splice sites into proximity, allowing transesterification to occur. The snRNAs are complexed with proteins, some of which also recognize and bind intron motifs and other proteins, contributing to proper positioning (14, 38, 45, 53). These elements cooperate and may be able to compensate when another is mutated or lost. Branch point consensus motifs are generally stronger in intron-poor organisms and weaker in intron-rich organisms (21). Branch points in some organisms, notably Caenorhabditis elegans, lack strong U2 complementarity. Instead, the final eight nucleotides of C. elegans introns are highly conserved and are recognized and bound by protein components of the U2 snRNP, U2AF35 and U2AF65 (17, 55, 56), properly positioning the intron and U2 snRNA. S. cerevisiae has no U2AF35 ortholog, and its introns lack a polypyrimidine tract, the binding site for its U2AF65 ortholog. However, the tightly conserved UACUAAC branch point sequence in S. cerevisiae (35, 54) provides a longer region of complementarity with U2 than is common for intron-rich organisms. Maintaining accurate positioning with these differences may foster strong conservation of motifs, to provide an “anchor” for splicing machinery. These points suggest the possibility of compensatory changes in the P. falciparum spliceosome to accommodate the weak branch point motif.
Conclusion.
Comparative genomics has revealed that the strength of intron consensus sequences varies across eukaryotic organisms (39). P. falciparum has one of the highest AT contents (81%) of any sequenced genome (21) and therefore presents a unique opportunity to investigate spliceosome adaptation in a deep-branching eukaryotic organism with extreme AT sequence bias. With rare exceptions (42, 50), U2 spliceosomal introns are characterized by 5′GU-AG3′ intron boundaries. These canonical features are also maintained in P. falciparum but with strong conservation of the 5′ (WR/GUAADW) and 3′ (UAGRW) intron boundary motifs. The adjacent 3′ polypyrimidine tract has fewer C nucleotides in P. falciparum than in human introns (Fig. 4). Predicted conventional branch point motifs are rare in P. falciparum (42), and our data suggest that the branch point consensus motif is relatively weak, with considerable tolerance for A/U substitutions. Notably, multiple branch points per intron were detected, with some at U instead of A residues. Non-A branch points and multiple branch points in an intron are rare but have been previously reported (16, 27), and the use of alternate branch points is a factor in some alternative splicing (18, 19) and human genetic diseases (2, 31). Alternative splicing has been reported in P. falciparum (32, 42), but whether alternative branch points are factors in any of these instances is unknown.
Sequencing of the P. falciparum genome and analyses of its transcriptome have generated multiple papers assessing predicted introns and conserved sequences. In this study, we have moved from predictions to experimental evaluation of intron sequence features that influence splicing efficiency in P. falciparum. Our data confirm some expected features of eukaryotic splicing and identify unexpected flexibility in the branch point motif and branch site selection.
Supplementary Material
ACKNOWLEDGMENTS
We thank Gowthaman Ramasamy for bioinformatics assistance.
The project described was supported by grants R21AI072681 and R01 AI47953 from NIH.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of NIH.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Atweh G. F., et al. 1987. A new mutation in IVS-1 of the human beta globin gene causing beta thalassemia due to abnormal splicing. Blood 70:147–151 [PubMed] [Google Scholar]
- 2. Blencowe B. J. 2000. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem. Sci. 25:106–110 [DOI] [PubMed] [Google Scholar]
- 3. Buratti E., et al. 2007. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 35:4250–4263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Burge C. B., Tuschi T., Sharp P. A. 2011. Splicing of precursors to mRNAs by the spliceosomes, p. 525–560.In: Gesteland R. F., Cech T. R., Atkins J. F. (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 5. Chakrabarti K., et al. 2007. Structural RNAs of known and unknown function identified in malaria parasites by comparative genomics and RNA analysis. RNA 13:1923–1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daniels G. L., et al. 1996. A combination of the effects of rare genotypes at the XK and KEL blood group loci results in absence of Kell system antigens from the red blood cells. Blood 88:4045–4050 [PubMed] [Google Scholar]
- 8. Deitsch K. W., Calderwood M. S., Wellems T. E. 2001. Malaria. Cooperative silencing elements in var genes. Nature 412:875–876 [DOI] [PubMed] [Google Scholar]
- 9. Fidock D. A., Wellems T. E. 1997. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. U. S. A. 94:10931–10936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fouser L. A., Friesen J. D. 1986. Mutations in a yeast intron demonstrate the importance of specific conserved nucleotides for the two stages of nuclear mRNA splicing. Cell 45:81–93 [DOI] [PubMed] [Google Scholar]
- 11. Gardner M. J., et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goux-Pelletan M., et al. 1990. In vitro splicing of mutually exclusive exons from the chicken beta-tropomyosin gene: role of the branch point location and very long pyrimidine stretch. EMBO J. 9:241–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Green M. R. 1986. Pre-mRNA splicing. Annu. Rev. Genet. 20:671–708 [DOI] [PubMed] [Google Scholar]
- 14. Gupta A., Jenkins J. L., Kielkopf C. L. 2011. RNA induces conformational changes in the SF1/U2AF65 splicing factor complex. J. Mol. Biol. 405:1128–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haas B. J., et al. 2003. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31:5654–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hartmuth K., Barta A. 1988. Unusual branch point selection in processing of human growth hormone pre-mRNA. Mol. Cell. Biol. 8:2011–2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hollins C., Zorio D. A., MacMorris M., Blumenthal T. 2005. U2AF binding selects for the high conservation of the C. elegans 3′ splice site. RNA 11:248–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hovhannisyan R. H., Carstens R. P. 2005. A novel intronic cis element, ISE/ISS-3, regulates rat fibroblast growth factor receptor 2 splicing through activation of an upstream exon and repression of a downstream exon containing a noncanonical branch point sequence. Mol. Cell. Biol. 25:250–263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hovhannisyan R. H., Warzecha C. C., Carstens R. P. 2006. Characterization of sequences and mechanisms through which ISE/ISS-3 regulates FGFR2 splicing. Nucleic Acids Res. 34:373–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Irimia M., Penny D., Roy S. W. 2007. Coevolution of genomic intron number and splice sites. Trends Genet. 23:321–325 [DOI] [PubMed] [Google Scholar]
- 21. Irimia M., Roy S. W. 2008. Evolutionary convergence on highly-conserved 3′ intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome. PLoS Genet. 4:e1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knapp B., Gunther K., Lingelbach K. 1991. In vitro translation of Plasmodium falciparum aldolase is not initiated at an unusual site. EMBO J. 10:3095–3097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knapp B., Nau U., Hundt E., Kupper H. A. 1991. Demonstration of alternative splicing of a pre-mRNA expressed in the blood stage form of Plasmodium falciparum. J. Biol. Chem. 266:7148–7154 [PubMed] [Google Scholar]
- 24. Lambros C., Vanderberg J. P. 1979. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol. 65:418–420 [PubMed] [Google Scholar]
- 25. Lesser C. F., Guthrie C. 1993. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science 262:1982–1988 [DOI] [PubMed] [Google Scholar]
- 26. Lu F., et al. 2007. cDNA sequences reveal considerable gene prediction inaccuracy in the Plasmodium falciparum genome. BMC Genomics 8:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lucke S., Jurchott K., Hung L. H., Bindereif A. 2005. mRNA splicing in Trypanosoma brucei: branch-point mapping reveals differences from the canonical U2 snRNA-mediated recognition. Mol. Biochem. Parasitol. 142:248–251 [DOI] [PubMed] [Google Scholar]
- 28. Militello K. T., Wirth D. F. 2003. A new reporter gene for transient transfection of Plasmodium falciparum. Parasitol. Res. 89:154–157 [DOI] [PubMed] [Google Scholar]
- 29. Moore M. J. 2000. Intron recognition comes of AGe. Nat. Struct. Biol. 7:14–16 [DOI] [PubMed] [Google Scholar]
- 30. Muhia D. K., et al. 2003. Multiple splice variants encode a novel adenylyl cyclase of possible plastid origin expressed in the sexual stage of the malaria parasite Plasmodium falciparum. J. Biol. Chem. 278:22014–22022 [DOI] [PubMed] [Google Scholar]
- 31. Nissim-Rafinia M., Kerem B. 2005. The splicing machinery is a genetic modifier of disease severity. Trends Genet. 21:480–483 [DOI] [PubMed] [Google Scholar]
- 32. Otto T. D., et al. 2010. New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 76:12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pace T., Birago C., Janse C. J., Picci L., Ponzi M. 1998. Developmental regulation of a Plasmodium gene involves the generation of stage-specific 5′ untranslated sequences. Mol. Biochem. Parasitol. 97:45–53 [DOI] [PubMed] [Google Scholar]
- 34. Pace T., et al. 2006. Set regulation in asexual and sexual Plasmodium parasites reveals a novel mechanism of stage-specific expression. Mol. Microbiol. 60:870–882 [DOI] [PubMed] [Google Scholar]
- 35. Pel H. J., Grivell L. A. 1993. The biology of yeast mitochondrial introns. Mol. Biol. Rep. 18:1–13 [DOI] [PubMed] [Google Scholar]
- 36. Pruitt K. D., Tatusova T., Klimke W., Maglott D. R. 2009. NCBI reference sequences: current status, policy and new initiatives. Nucleic Acids Res. 37:D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Reed R. 2000. Mechanisms of fidelity in pre-mRNA splicing. Curr. Opin. Cell Biol. 12:340–345 [DOI] [PubMed] [Google Scholar]
- 38. Rino J., Desterro J. M., Pacheco T. R., Gadella T. W., Jr., Carmo-Fonseca M. 2008. Splicing factors SF1 and U2AF associate in extraspliceosomal complexes. Mol. Cell. Biol. 28:3045–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roy S. W., Irimia M. 2009. Splicing in the eukaryotic ancestor: form, function and dysfunction. Trends Ecol. Evol. 24:447–455 [DOI] [PubMed] [Google Scholar]
- 40. Seraphin B., Kretzner L., Rosbash M. 1988. A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J. 7:2533–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Singh N., et al. 2004. Conservation and developmental control of alternative splicing in maebl among malaria parasites. J. Mol. Biol. 343:589–599 [DOI] [PubMed] [Google Scholar]
- 42. Sorber K., Dimon M. T., DeRisi J. L. 2011. RNA-Seq analysis of splicing in Plasmodium falciparum uncovers new splice junctions, alternative splicing and splicing of antisense transcripts. Nucleic Acids Res. 39:3820–3835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Spingola M., Grate L., Haussler D., Ares M., Jr 1999. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA 5:221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suzuki H., et al. 2006. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thickman K. R., Swenson M. C., Kabogo J. M., Gryczynski Z., Kielkopf C. L. 2006. Multiple U2AF65 binding sites within SF3b155: thermodynamic and spectroscopic characterization of protein-protein interactions among pre-mRNA splicing factors. J. Mol. Biol. 356:664–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Thi Tran H. T., et al. 2005. A G-to-A transition at the fifth position of intron-32 of the dystrophin gene inactivates a splice-donor site both in vivo and in vitro. Mol. Genet. Metab. 85:213–219 [DOI] [PubMed] [Google Scholar]
- 47. Trager W., Jensen J. B. 1976. Human malaria parasites in continuous culture. Science 193:673–675 [DOI] [PubMed] [Google Scholar]
- 48. Trang D. T., Huy N. T., Kariu T., Tajima K., Kamei K. 2004. One-step concentration of malarial parasite-infected red blood cells and removal of contaminating white blood cells. Malar. J. 3:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Upadhyay R., Bawankar P., Malhotra D., Patankar S. 2005. A screen for conserved sequences with biased base composition identifies noncoding RNAs in the A-T rich genome of Plasmodium falciparum. Mol. Biochem. Parasitol. 144:149–158 [DOI] [PubMed] [Google Scholar]
- 50. Vanacova S., Yan W., Carlton J. M., Johnson P. J. 2005. Spliceosomal introns in the deep-branching eukaryote Trichomonas vaginalis. Proc. Natl. Acad. Sci. U. S. A. 102:4430–4435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. van Dooren G. G., Su V., D'Ombrain M. C., McFadden G. I. 2002. Processing of an apicoplast leader sequence in Plasmodium falciparum and the identification of a putative leader cleavage enzyme. J. Biol. Chem. 277:23612–23619 [DOI] [PubMed] [Google Scholar]
- 52. Volkman S. K., et al. 2001. Recent origin of Plasmodium falciparum from a single progenitor. Science 293:482–484 [DOI] [PubMed] [Google Scholar]
- 53. Wahl M. C., Will C. L., Luhrmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136:701–718 [DOI] [PubMed] [Google Scholar]
- 54. Wentz-Hunter K., Potashkin J. 1995. The evolutionary conservation of the splicing apparatus between fission yeast and man. Nucleic Acids Symp. Ser. 226–228 [PubMed] [Google Scholar]
- 55. Zhang H., Blumenthal T. 1996. Functional analysis of an intron 3′ splice site in Caenorhabditis elegans. RNA 2:380–388 [PMC free article] [PubMed] [Google Scholar]
- 56. Zorio D. A., Blumenthal T. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402:835–838 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.