Abstract
Recently, mitochondria have been identified as important contributors to the virulence and drug tolerance of human fungal pathogens. In different scenarios, either hypo- or hypervirulence can result from changes in mitochondrial function. Similarly, specific mitochondrial mutations lead to either sensitivity or resistance to antifungal drugs. Here, we provide a synthesis of this emerging field, proposing that mitochondrial function in membrane lipid homeostasis is the common denominator underlying the observed effects of mitochondria in drug tolerance (both sensitivity and resistance). We discuss how the contrasting effects of mitochondrial dysfunction on fungal drug tolerance and virulence could be explained and the potential for targeting mitochondrial factors for future antifungal drug development.
INTRODUCTION
Although it has been studied quite extensively in the model yeast Saccharomyces cerevisiae, mitochondrial function has remained understudied in human fungal pathogens. A possible reason is that several pathogenic fungi, such as Candida albicans and Cryptococcus neoformans, are so-called “petite-negative” yeasts, i.e., they cannot survive mitochondrial genome loss, which is a classic and extensively used tool for studying mitochondrial function in S. cerevisiae. Recent work from several laboratories has revealed that mitochondria have a fundamental role as a control point in the cellular networks impacted by antifungal drugs, as well as a prominent role in fungal virulence. These studies suggest that the functions of mitochondria in these pathways are complex. With antifungal drugs that target cell membranes, such as the azoles and the polyenes, both resistance and sensitivity of mitochondrial mutants have been reported (9, 10, 18, 22, 45, 75, 85, 86). In contrast, only sensitivity has been observed with agents that target the cell wall, which includes the echinocandin class of antifungal drugs (3, 15, 17, 22, 38, 90). A similarly complex picture is observed in regard to virulence of mitochondrial mutants, with both hypo- and hypervirulence of Candida spp. mitochondrial mutants observed in animal infection models (1, 4, 10, 19, 25, 64). These studies pose several questions. What is the biochemical basis for the impact of mitochondria on drug tolerance? When (and why) does a change to mitochondrial function lead to hypo- versus hypervirulence? Would mitochondrial factors be useful targets for antifungal drug development? In this review, we will consider the molecular and cellular mechanisms behind the observed drug sensitivity and virulence phenotypes of mitochondrial mutants, mostly discussing Candida spp., for which the most is known, and mentioning the other pathogens where appropriate. We will also draw on the model fungus S. cerevisiae, because mitochondrial functions have been extensively studied in this yeast and there is reasonable evidence that the mechanisms of mitochondrial involvement in drug resistance are conserved with pathogenic fungi. Finally, we will discuss the potential for targeting mitochondrial factors for the development of future antifungal therapies.
MITOCHONDRIA AND PLASMA MEMBRANE-TARGETING DRUGS
Mitochondrial roles in azole tolerance.
The azole drugs target fungal membrane biogenesis by inhibiting the ergosterol biosynthesis enzyme Erg11, which is a cytochrome P450 enzyme lanosterol 14-α-demethylase. Studies in Candida glabrata, C. albicans, and S. cerevisiae have shown that mitochondrial dysfunction can lead to both resistance and susceptibility to the azoles. We will first consider activation of the drug resistance pathway upon mitochondrial dysfunction and then the examples of azole sensitivity of mitochondrial mutants.
The molecular mechanism of drug resistance induced by mitochondrial dysfunction has been studied in C. glabrata and S. cerevisiae. Compromised mitochondrial function leads to the activation of homologous transcription factors Pdr3 of S. cerevisiae (ScPdr3) and Pdr1 of C. glabrata (CgPdr1), which then results in upregulation of their target genes encoding efflux pumps, such as ScPdr5 or CgCdr1 and CgCdr2 (9, 33, 35, 70, 75, 84, 85, 89; reviewed in references 60 and 79) (Fig. 1). Both C. glabrata and S. cerevisiae can live without mitochondrial DNA (mtDNA), and the drug-resistant mutants most commonly lost their mitochondrial genome (33, 35, 45, 75, 84, 89). In fact, mtDNA loss appears to be a key activating mutation for the drug resistance pathway. Loss of mtDNA and associated drug resistance is relatively common for in vitro cultures of both S. cerevisiae (about 2% of cells) (89) and C. glabrata (frequencies of 2 × 10−4 to 4 × 10−4) (75). However, very few reports of clinical azole resistance in C. glabrata are linked to mitochondrial dysfunction (8, 24), and thus, the relevance of this mechanism in the clinic remains to be studied. Loss of the mitochondrial genome as the key activator of the drug resistance pathway also has to be considered in the context of other major pathogens, such as Cryptococcus spp. or C. albicans, that are unable to survive without mtDNA (16, 83). Consequently, the specific mechanisms behind the role of mitochondria in hyperresistance to azoles might be different in these various pathogens. One study reports that there is a relationship between upregulation of the efflux pump gene MDR1, with its resulting higher azole resistance in C. albicans, and uncoupled mitochondrial oxidative phosphorylation (18). However, the mutant was less susceptible to some azole drugs (fluconazole and voriconazole) but not others (itraconazole and ketoconazole), and the MIC was still in the susceptible range according to CLSI recommendations (18). A study in C. neoformans also reports higher azole resistance related to respiratory deficiency, which was reversible and, thus, not very likely to be due to mtDNA loss (68). How mitochondrial dysfunction leads to azole resistance in the petite-negative yeasts C. albicans and C. neoformans requires further investigation.
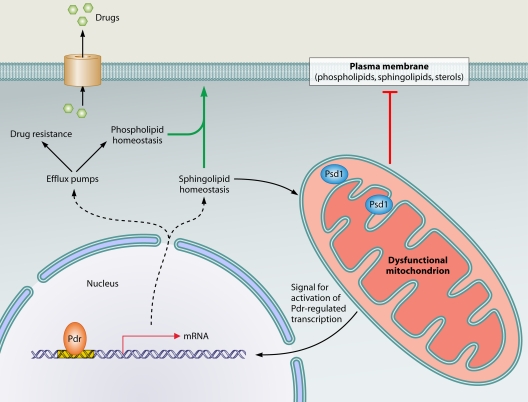
Fig. 1.
Pathways linking mitochondrial dysfunction to the activation of multidrug resistance genes. In dysfunctional mitochondria, changes in the membrane association characteristics of the mitochondrial inner membrane protein Psd1 create a signal for activation of the transcription factors ScPdr3 and CgPdr1 (indicated by Pdr in the figure). This results in upregulation of genes encoding efflux pumps (which mediate drug resistance and have additional roles in phospholipid homeostasis), as well as genes required for sphingolipid metabolism. Dysfunctional mitochondria have an effect on membrane lipid homeostasis (depicted by the red “T”). This could be compensated by activation of phospholipid and sphingolipid homeostasis genes via the Pdr pathway (green arrow). Additionally, sphingolipid biosynthesis intermediates can feed into the synthesis of phosphatidylethanolamine by nonmitochondrial pathways, thereby compensating for aberrant lipid composition of mitochondrial membranes in cells with dysfunctional mitochondria.
The relationship between mitochondrial dysfunction and drug resistance is strong, but the mechanistic details, particularly the trigger for activation of drug resistance by mitochondrial dysfunction and why these two pathways are linked, is far from understood. Screening of S. cerevisiae mutants indicates that respiratory deficiency per se is not enough to cause drug resistance (89), and thus, another mechanism of mitochondrial dysfunction related to mtDNA loss must be involved. Other mitochondrial mutations can cause drug resistance in C. glabrata and S. cerevisiae; for example, deletion of the mitochondrial inner membrane translocase gene ScOXA1 (35) that is required for the assembly of mitochondrially encoded subunits of the respiratory complexes into the inner membrane (7, 26, 37, 43). OXA1 is not essential for mtDNA stability, but reports differ as to whether and to what extent the oxa1Δ mutant is affected in mitochondrial genome maintenance, perhaps depending on the yeast strain background (20, 58, 60, 63). In any case, activation of the drug resistance pathway by deletion of OXA1 appears to be distinct from that which occurs in cells that have lost mtDNA (35, 89). Another example is the inactivation of the phosphatidylglycerol synthase gene CgPGS1 in C. glabrata, which is necessary for synthesis of the phospholipids phosphatidylglycerol and cardiolipin in mitochondria (3). pgs1Δ mutants of S. cerevisiae tend to lose mtDNA (90), but activation of the drug resistance genes of the C. glabrata pgs1Δ mutant was complemented by reintroduction of a wild-type copy of PGS1 (3), and it is therefore unlikely to have been due to mtDNA loss (which is expected to be irreversible).
The common denominator in mitochondrial mutations that cause drug resistance is a change to the protein and/or lipid structure of mitochondrial membranes, as demonstrated by the following evidence. Several subunits of the respiratory complexes located in the inner mitochondrial membrane are encoded on the mitochondrial genome. After mtDNA loss, the complexes are not correctly assembled. Moreover, the levels of cardiolipin and phosphatidylglycerol, which are synthesized in mitochondria and are specific to mitochondrial membranes, are altered in the absence of mtDNA, probably because of the interdependency of cardiolipin synthesis and respiration (30). In the S. cerevisiae oxa1Δ mutants, respiratory complexes in the inner membrane are inappropriately assembled (7, 43) and the mutants display lower levels of another important phospholipid in mitochondrial membranes, phosphatidylethanolamine (63). In pgs1Δ mutants of C. glabrata, both phosphatidylglycerol and cardiolipin are absent from mitochondrial membranes (3). Taken together, these data suggest that changes to the structure of mitochondrial membranes could be the trigger for activation of the drug resistance pathway. Recent studies from the Moye-Rowley laboratory suggested that changed mitochondrial membranes in S. cerevisiae cells lacking mtDNA lead to diminished membrane association of Psd1, the phosphatidylserine decarboxylase in the mitochondrial inner membrane (33). This in turn leads to activation of the multidrug resistance pathway (33) (Fig. 1). A similar scenario might be operating in C. glabrata and C. albicans (33, 70). The exact mechanism is not clear, but because the catalytic activity of ScPsd1 was dispensable for activation of drug resistance, it was proposed that the release of a fraction of the protein from the inner membrane into the intermembrane space could permit interactions with another molecule, which then constitutes the signal for activation of the Pdr factors (33, 70) (Fig. 1). Interestingly, in oxa1Δ mutants, ScPsd1 is not properly inserted into the inner membrane (63) and deletion of the mitochondrial protease ScYme1 suppresses both the Psd1 maturation defects and multidrug resistance of oxa1Δ mutants (63, 89), thus supporting a role for changes in ScPsd1 membrane localization in triggering multidrug resistance.
Why do fungi activate the Pdr pathway upon mitochondrial dysfunction? The available evidence points to a link between mitochondrial control over membrane structure and lipid homeostasis and the role of the Pdr pathway in membrane biogenesis (Fig. 1). As has recently been reviewed by Shahi and Moye-Rowley (79), the Pdr transcription factors control the expression of genes required for the homeostasis of two key lipid components of membranes, sphingolipids and phospholipids, and consistently, activation of the Pdr pathway by mitochondrial dysfunction in S. cerevisiae leads to increased expression of genes encoding enzymes of sphingolipid biosynthesis (Fig. 1). Mitochondrial function is important for membrane lipid structure, and thus, activation of lipid homeostasis by the multidrug resistance pathway could serve to compensate for changes to membrane structure/composition upon mitochondrial dysfunction (Fig. 1). This model is supported by several observations. Mitochondria are the predominant cellular site for synthesis of the ubiquitous membrane phospholipid phosphatidylethanolamine in a reaction catalyzed by Psd1 (6, 11, 78, 88) (Fig. 2). Phosphatidylethanolamine also serves as the precursor for synthesis of another major membrane phospholipid, phosphatidylcholine, in the endoplasmic reticulum (ER) (6, 88) (Fig. 2). Sphingolipid homeostasis is disturbed in cells that have lost the mitochondrial genome (34). Additionally, two cytochrome P450 enzymes in the biosynthetic pathway of the membrane lipid ergosterol require heme as a cofactor (21). Heme is synthesized in mitochondria, and it has been shown that mitochondrial mutants have an altered cellular sterol composition (9, 28, 29).
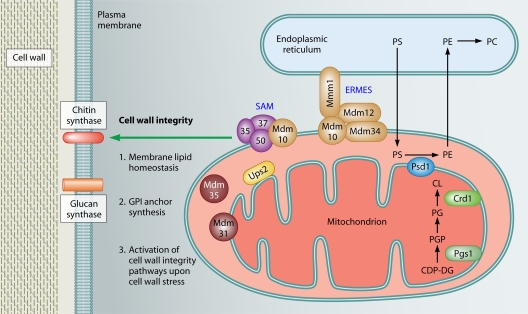
Fig. 2.
Roles of mitochondria in cell wall integrity and resistance to the echinocandin antifungal drugs. Mitochondrial proteins affecting phospholipid homeostasis (Sam37, Mdm10, Mdm35, Mdm31, Ups2, Pgs1, and Psd1) have prominent roles in cell wall integrity. Sam37 and Mdm10 are subunits of the SAM and ERMES complexes, which are necessary for phospholipid trafficking between the ER and the mitochondria. Psd1, Pgs1, and Crd1 are enzymes required for the synthesis of phospholipids PE, PG, and CL. The conversion of PGP to PG is catalyzed by the phosphatidylglycerol phosphatase Gep4, which is not shown in the figure for simplicity's sake. Mitochondrial phospholipid biosynthesis could (1) affect the activity of wall synthesis enzymes in the plasma membrane, (2) impinge on synthesis of the GPI anchor, and (3) be required for the activation of pathways necessary for responding to cell wall stress. PS, phosphatidylserine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PGP, phosphatidylglycerol phosphate; CDP-DG, CDP-diacylglycerol; PG, phosphatidylglycerol; CL, cardiolipin.
It is also tempting to speculate that activation of the multidrug resistance pathway might result from an attempt to compensate not only for problems in general membrane lipid homeostasis but also, more specifically, for defective mitochondrial membrane structure in mitochondrial mutants. Cardiolipin, which is dramatically reduced in mitochondrial membranes of mutants which activate the drug resistance pathway, such as cells lacking mtDNA and pgs1Δ cells, has critical roles in mitochondrial biogenesis. For example, cardiolipin is required for the activity of the respiratory chain complexes in the inner membrane and protein import into mitochondria (44). It has to be stressed here that mitochondria are essential organelles, and even in cells with no mtDNA, mitochondrial function must be maintained. Cardiolipin defects in mitochondrial membranes can be compensated for by phosphatidylethanolamine, with the two phospholipids having overlapping roles (31). Intermediates in sphingolipid biosynthesis can be used as precursors for phosphatidylethanolamine biosynthesis by nonmitochondrial pathways (78), and this could provide a further link between regulation of sphingolipid biosynthesis by the drug resistance pathway and its activation by mitochondrial dysfunction (Fig. 1).
In contrast to the above-described azole resistance, some mitochondrial mutations in C. glabrata and S. cerevisiae lead to increased susceptibility to azoles (22, 38, 86). This further corroborates the point that only very specific mitochondrial mutations and defined changes to mitochondrial physiology lead to activation of the multidrug resistance pathway. In other words, a decrease in mitochondrial function is not sufficient to trigger the changes leading to resistance to antifungal compounds, but rather, specific molecular mechanisms are involved. The reason for decreased azole resistance of some mitochondrial mutants is not fully understood, but it could be related to changes to the sterol content of membranes in the absence of proper mitochondrial function (9, 28, 29). Other mechanisms are also possible, such as fitness defects that are typical for mitochondrial mutants. Determining the molecular mechanism behind increased susceptibility to azoles of cells with dysfunctional mitochondria is key to understanding why some mitochondrial mutants are resistant to azoles while others are sensitive. Data from C. glabrata suggest that the answer to this question will not come down to whether or not the specific mitochondrial mutations activate the Pdr-dependent multidrug resistance pathway, as the azole hypersensitive mitochondrial mutant overexpressed the CgPdr1 target, the CDR1 efflux pump (86).
Mitochondria and polyene drugs.
Polyene drugs (e.g., amphotericin B and nystatin) disrupt membrane integrity by binding to sterols and forming pores. Mitochondrial mutants of C. glabrata are more sensitive to amphotericin B (9, 86). Decreased susceptibility to this drug was also observed for C. albicans treated with tetracycline; this was attributed to the effects of tetracycline on the function of mitochondria (66). Tetracycline also increased the sensitivity to amphotericin B of C. neoformans and Aspergillus fumigatus (66), implying that mitochondrial roles in polyene tolerance are broadly conserved in pathogenic fungi. Moreover, a large study by Hillenmeyer et al. that included 1,144 chemical genomic assays in S. cerevisiae found a significant enrichment in the GO (Gene Ontology) terms “mitochondrial inheritance,” “mitochondrial localization,” and “mitochondrial distribution” among genes required for wild-type fitness in the presence of nystatin (38). However, two reports in C. albicans suggest that mitochondrial mutants display higher resistance to amphotericin B (28, 29), while one report finds no difference (18).
The link between mitochondria and polyene treatment is again lipids. It has been proposed that the increased susceptibility of mitochondrial mutants to polyenes is due to changes in sterol levels (9, 28, 29), which could be due to the requirement for heme as a cofactor for the ergosterol biosynthesis enzymes Erg5 and Erg11.
MITOCHONDRIA AND CELL WALL-TARGETING DRUGS
The echinocandin antifungal drugs inhibit β-1,3 glucan synthase, the enzyme responsible for synthesis of the main glucan component of the yeast cell wall (23). Mitochondrial mutants of C. albicans, Candida parapsilosis, and S. cerevisiae are sensitive to the echinocandins (15, 17, 22, 38, 76). Mitochondrial dysfunction leads to cell wall defects in C. glabrata (3, 10). While sensitivity to the echinocandins has not been tested directly yet, the close relationship between C. glabrata and S. cerevisiae predicts that mitochondria will be required for echinocandin tolerance in C. glabrata also.
Which mitochondrial function is the most critical for cell wall integrity and tolerance of the echinocandin drugs? A recent screen of a collection of S. cerevisiae mitochondrial mutants in our laboratory suggests that respiratory deficiency does play a role, as mutants that are respiratory deficient are hypersensitive to the echinocandin drug caspofungin (22). A similar conclusion can be drawn from the large study by Hillenmeyer et al. (38). Moreover, in C. parapsilosis, treatment with inhibitors of the mitochondrial respiratory pathways resulted in lowering of the MIC of caspofungin (15), also suggesting a role for mitochondrial respiration in cell wall integrity and echinocandin tolerance. However, the extent of caspofungin hypersensitivity did not correlate with respiratory deficiency, but again, the strongest link appeared to be the function of mitochondria in lipid homeostasis (22). The mitochondrial mutants most sensitive to caspofungin were those with impaired mitochondrial biosynthesis of the phospholipids phosphatidylethanolamine and cardiolipin (22, 49, 67, 82) (Fig. 2). Consistently, deletion of the phosphatidylglycerol synthase gene PGS1 leads to cell wall defects in S. cerevisiae and C. glabrata (3, 90, 91) and to caspofungin sensitivity in S. cerevisiae (76). Moreover, the C. albicans mutants with deletions of the genes for the phospholipid-biosynthetic enzymes phosphatidylserine synthase (CHO1) and phosphatidylserine decarboxylase (PSD1 and PSD2) have defective cell walls and dysfunctional mitochondria and are sensitive to caspofungin (17). Additionally, the C. albicans mutant lacking the mitochondrial outer membrane SAM (Sorting and Assembly Machinery) complex subunit Sam37 is sensitive to caspofungin (22). The SAM complex functions in the assembly of outer membrane proteins (14), and at the cellular level, it is required for both mitochondrial phospholipid biosynthesis and cell wall integrity (22).
What aspect of cell wall integrity is affected by mitochondrial dysfunction? Three possibilities can be envisaged and are depicted in Fig. 2. Impairment of mitochondrial functions could lead to changes in the lipid composition of the plasma membrane, which could in turn have an impact on the activity of biosynthesis enzymes located in the plasma membrane, such as glucan synthase and chitin synthase. In support of this mechanism, the S. cerevisiae pgs1Δ mutant, cells lacking mtDNA, and the C. albicans mutant with a mutation in the posttranscriptional regulator Ccr4 that coordinates the network linking cell wall integrity to mitochondria and phospholipid homeostasis all have lower levels of cell wall β-glucans (22, 90). Also, in the S. cerevisiae pgs1Δ mutant, the steady-state protein level of the glucan synthase catalytic subunit Fks1 decreases and its activity is lower (91). Another mechanism linking mitochondrial phospholipid biosynthesis and cell wall integrity is the requirement for phosphatidylethanolamine in synthesis of the glycosylphosphatidylinositol (GPI) anchor (5, 41), with a large number of cell wall proteins and cell wall biosynthesis enzymes being GPI anchored (47, 71) (Fig. 2). Finally, studies with the S. cerevisiae pgs1Δ mutant suggest that mitochondrial phospholipid biosynthesis is required for activation of the PKC-dependent cell wall integrity pathway (91), although the exact mechanism is not clear. Of note, links have been reported between cell wall integrity signaling and lipid metabolism. For example, the kinases Pkh1 and Pkh2, which are activated by the long-chain base phytosphingosine (an intermediate in sphingolipid biosynthesis), have roles in cell wall integrity by acting on the central PKC-dependent cell wall integrity pathway, as well as via a parallel pathway involving the Ypk1 and Ypk2 kinases (13, 27, 42, 52, 73, 77; reviewed in reference 51). In this context, it is interesting that work from the Moye-Rowley laboratory in S. cerevisiae linked mitochondrial dysfunction and consequent activation of the Pdr pathway to regulation of the expression of genes encoding enzymes of sphingolipid biosynthesis, as well as the membrane transporter gene RSB1 that is required for tolerance of long-chain bases (34, 48, 69, 79) (Fig. 1). Whether and how mitochondrial dysfunction, sphingolipid metabolism and signaling, and cell wall integrity are functionally interconnected is unclear at this stage, but it is certainly worth exploring. Moreover, an important goal for the future is to use directed biochemical experiments to determine exactly how membrane lipids and mitochondria affect the activity of the wall biosynthesis enzymes in the plasma membrane. In addition, we need a large-scale study of the effects of mitochondrial dysfunction on cell wall carbohydrate and protein composition, particularly in pathogens such as C. albicans and C. glabrata, using diverse mitochondrial mutants and the “-omics” approaches of systems biology.
MITOCHONDRIA AND VIRULENCE OF HUMAN FUNGAL PATHOGENS
In addition to being required for antifungal drug tolerance, mitochondria have also been associated with the ability of fungal pathogens to cause disease. This link is supported by direct assays of mitochondrial mutants in animal infection models, and it can also be inferred indirectly from the requirement for mitochondrial function in cellular pathways associated with virulence. We will first consider the direct assays and then the indirect evidence.
The majority of the direct assays support a positive correlation between mitochondrial function and virulence of human fungal pathogens. Loss of mitochondrial function in C. glabrata cripples virulence (10). Moreover, in C. albicans, inactivation of the mitochondrial protein Goa1, putative subunits of the respiratory complex I, or factors required for mitochondrial biogenesis and morphology all result in attenuated virulence in the mouse model of systemic candidiasis (1, 4, 64). Moreover, deletion of the mitochondrial superoxide dismutase gene SOD2 in C. neoformans reduced virulence in the mouse model (62). The attenuation of virulence of strains with dysfunctional mitochondria is probably due to a combination of reduced fitness, metabolic changes, and sensitivity to oxidative stress, which is linked to higher production of reactive oxygen species (ROS) because of defective respiration. In this context, it has been shown that inactivation of the mitochondrial alternative oxidase in A. fumigatus causes an increase in ROS and enhanced killing of the fungus by macrophages (54). Additional support for a positive role for mitochondrial function in virulence comes from studies of the Vancouver Island and North American outbreaks of hypervirulent Cryptococcus gattii. It was proposed that the hypervirulence was associated with more efficient mitochondrial function, which occurred via a change in mitochondrial morphology toward more tubular organelles (12, 53). The effects of tubular mitochondria on fungal survival in macrophages were proposed as the mechanism, but the molecular basis of this remains to be determined (53).
The idea that mitochondrial function is necessary for virulence is challenged by a recent report of more virulent mitochondrial mutants of C. glabrata (25). These were clinical isolates that lost mtDNA in the host. Surprisingly, inducing mtDNA loss in the parental clinical isolate in vitro by ethidium bromide mutagenesis gave the opposite result: the mutant was not more virulent than the wild type and even showed slightly reduced fungal burdens in the kidneys (25). This raises the question of whether the clinical mitochondrial mutants acquired other mutations unrelated to mitochondrial dysfunction that rendered them more virulent. It is also possible that pressures operating on respiration-deficient mutants in the host are different from those in an in vitro environment, activating compensatory pathways specifically in the host upon loss of mitochondrial function which then contribute to virulence.
Indirect evidence for a role of mitochondria in the survival of fungal pathogens within the host includes a requirement for mitochondrial function for metabolic pathways necessary for virulence (such as the glyoxylate cycle and gluconeogenesis) (2, 72), the activation of genes required for mitochondrial respiration during colonization of the central nervous system by C. neoformans (80), and the involvement of mitochondria in survival under oxidative stress in C. albicans, C. neoformans, A. fumigatus, and Paracoccidioides brasiliensis (1, 54, 56, 62), as well as the role of mitochondria in fungal morphogenesis (1, 46, 56, 57, 87). With respect to morphogenesis, C. albicans mutants lacking the mitochondrial protein Goa1, as well as those with inactivation of the NADH dehydrogenase Ndh51 or subunits of the mitochondrial pyruvate dehydrogenase complex, all display a defect in switching from an ovoid yeast to a filamentous cell morphology, which is a key aspect of virulence (1, 46, 57, 87). There is, however, a report that inhibiting respiration by using antimycin A induces filamentous growth in C. albicans (61). Whether this discrepancy is due to differences in strain backgrounds of C. albicans or the nature of the mitochondrial mutations remains to be determined. In the dimorphic pathogen P. brasiliensis, inhibition of mitochondrial respiration blocks the switch from filamentous to yeast morphology that is important for virulence (56). In conclusion, the available data from both direct and indirect studies support the notion that wild-type mitochondrial function is required for virulence.
TARGETING MITOCHONDRIAL FACTORS FOR DRUG DISCOVERY
Would mitochondrial functions/factors represent promising targets for antifungal therapy? We would argue that they would, as inactivating mitochondrial factors is likely to substantially cripple virulence for multiple reasons, such as effects on fitness, metabolism, and oxidative stress responses. While the resistance to azole drugs that is associated with some mitochondrial mutations is a concern, it has to be noted that resistance is only seen with specific mutations, and several mitochondrial mutants are known which are sensitive to the azoles (22, 38, 86). Also, azole resistance due to mitochondrial mutations is rarely observed in the clinic (8, 24). Moreover, even the drug-resistant mitochondrial mutants of C. glabrata and C. albicans were crippled for virulence in animal models, probably due to a substantial fitness defect (10, 18, 19).
Which mitochondrial factors could be promising drug targets? Some predictions can be made based on studies in the model yeast S. cerevisiae and in the pathogen C. albicans. In S. cerevisiae, a deletion collection covering all nonessential open reading frames has been screened for genes required for mitochondrial respiration (so-called “pet genes,” because they give rise to mitochondrial or “petite” mutants) (58). Based on three independent screens, a core set of 129 genes encoding mitochondrial proteins have been identified that are likely to be required for mitochondrial respiration regardless of the strain background (58). Petite mutants of fungal pathogens can be predicted to have reduced virulence in animals due their reduced fitness, as well as other defects discussed above. Therefore, the identified pet genes from S. cerevisiae can be the starting point for creating mitochondrial mutants in human fungal pathogens and testing their requirement for virulence. The functions of the pet genes should be tested directly in several human fungal pathogens, as inferring the function of a gene based on knowledge from the model yeast system can be complicated by profound changes to the organization and regulation of cellular pathways (50, 55). In the context of mitochondrial function, S. cerevisiae has a strikingly different respiratory chain than the Candida species: it does not have the respiratory complex I or alternative respiratory pathways (36, 40, 59, 74, 81). Also, not all mitochondrial mutants will have the same effect on virulence, as exemplified by the study of Becker et al. (4). Among 11 mitochondrial mutants tested, 10 of which were not viable in vitro, some were essential for virulence in mice while some were only attenuated (4).
One group of S. cerevisiae pet genes that we would like to discuss in greater detail encompasses a set of factors required for maintenance of the mitochondrial genome. In the most recent screen of the S. cerevisiae deletion collection, 162 mutants were identified which tend to lose mtDNA (58). While these mtDNA maintenance genes are not essential for viability in S. cerevisiae, they are likely to be essential for viability in petite-negative pathogens, such as C. albicans or C. neoformans (4, 58, 83), and could thus represent potentially attractive drug targets. This idea is supported by a recently published screen of C. albicans strains (4): seven mitochondrial genes were identified which are essential in C. albicans but not in S. cerevisiae. For all of them, the likely reason for the essential function in C. albicans is an effect on mtDNA stability: four genes encode mitochondrial ribosomal proteins, with mitochondrial translation being required for mtDNA maintenance (20); the other three are MGM101, the mitochondrial RNA polymerase gene RPO41, and the ERMES (ER-mitochondria-encounter structure) subunit gene MMM1, all of which affect mtDNA stability in S. cerevisiae (32, 39, 58).
CONCLUDING REMARKS
While recent studies have highlighted the importance of mitochondrial function for tolerance of antifungal drugs and for virulence, our understanding of the roles of this organelle in human fungal pathogens is still limited. Functions in lipid homeostasis are likely to be at the heart of the involvement of mitochondria in tolerance of antifungal drugs, but the exact molecular mechanisms are not fully understood. Biochemical experiments in mitochondrial mutants addressing lipid composition, activities of plasma membrane enzymes, and the structure of the cell wall should answer some of the outstanding questions. Also important is to test the virulence of many more mutants affected in diverse mitochondrial functions. This could produce a list of potentially attractive targets for future antifungal drug development. It is of interest that a large number of mitochondrial proteins are likely to be essential for viability in petite-negative pathogens, such as C. albicans and C. neoformans. An important point is that several of these mitochondrial factors do not have close orthologs in humans; for example, see reference 65. Collectively, these observations indicate that studies into how mitochondria contribute to the fitness, pathogenicity, and drug tolerance of human fungal pathogens are worthwhile.
ACKNOWLEDGMENTS
We thank Kip Gabriel and Trevor Lithgow for comments on the manuscript.
Footnotes
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Bambach A., et al. 2009. Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot. Cell 8:1706–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barelle C. J., et al. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell. Microbiol. 8:961–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Batova M., et al. 2008. Functional characterization of the CgPGS1 gene reveals a link between mitochondrial phospholipid homeostasis and drug resistance in Candida glabrata. Curr. Genet. 53:313–322 [DOI] [PubMed] [Google Scholar]
- 4. Becker J. M., et al. 2010. Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model. Proc. Natl. Acad. Sci. U. S. A. 107:22044–22049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Birner R., Burgermeister M., Schneiter R., Daum G. 2001. Roles of phosphatidylethanolamine and of its several biosynthetic pathways in Saccharomyces cerevisiae. Mol. Biol. Cell 12:997–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Birner R., Daum G. 2003. Biogenesis and cellular dynamics of aminoglycerophospholipids. Int. Rev. Cytol. 225:273–323 [DOI] [PubMed] [Google Scholar]
- 7. Bonnefoy N., Chalvet F., Hamel P., Slonimski P. P., Dujardin G. 1994. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 239:201–212 [DOI] [PubMed] [Google Scholar]
- 8. Bouchara J. P., et al. 2000. In-vivo selection of an azole-resistant petite mutant of Candida glabrata. J. Med. Microbiol. 49:977–984 [DOI] [PubMed] [Google Scholar]
- 9. Brun S., et al. 2004. Mechanisms of azole resistance in petite mutants of Candida glabrata. Antimicrob. Agents Chemother. 48:1788–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brun S., et al. 2005. Biological consequences of petite mutations in Candida glabrata. J. Antimicrob. Chemother. 56:307–314 [DOI] [PubMed] [Google Scholar]
- 11. Burgermeister M., Birner-Grunberger R., Nebauer R., Daum G. 2004. Contribution of different pathways to the supply of phosphatidylethanolamine and phosphatidylcholine to mitochondrial membranes of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1686:161–168 [DOI] [PubMed] [Google Scholar]
- 12. Byrnes E. J., III, et al. 2010. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 6:e1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Casamayor A., Torrance P. D., Kobayashi T., Thorner J., Alessi D. R. 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9:186–197 [DOI] [PubMed] [Google Scholar]
- 14. Chacinska A., Koehler C. M., Milenkovic D., Lithgow T., Pfanner N. 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138:628–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chamilos G., Lewis R. E., Kontoyiannis D. P. 2006. Inhibition of Candida parapsilosis mitochondrial respiratory pathways enhances susceptibility to caspofungin. Antimicrob. Agents Chemother. 50:744–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen X. J., Clark-Walker G. D. 2000. The petite mutation in yeasts: 50 years on. Int. Rev. Cytol. 194:197–238 [DOI] [PubMed] [Google Scholar]
- 17. Chen Y. L., et al. 2010. Phosphatidylserine synthase and phosphatidylserine decarboxylase are essential for cell wall integrity and virulence in Candida albicans. Mol. Microbiol. 75:1112–1132 [DOI] [PubMed] [Google Scholar]
- 18. Cheng S., Clancy C. J., Nguyen K. T., Clapp W., Nguyen M. H. 2007. A Candida albicans petite mutant strain with uncoupled oxidative phosphorylation overexpresses MDR1 and has diminished susceptibility to fluconazole and voriconazole. Antimicrob. Agents Chemother. 51:1855–1858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng S., et al. 2007. Uncoupling of oxidative phosphorylation enables Candida albicans to resist killing by phagocytes and persist in tissue. Cell. Microbiol. 9:492–501 [DOI] [PubMed] [Google Scholar]
- 20. Contamine V., Picard M. 2000. Maintenance and integrity of the mitochondrial genome: a plethora of nuclear genes in the budding yeast. Microbiol. Mol. Biol. Rev. 64:281–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cresnar B., Petric S. 2011. Cytochrome P450 enzymes in the fungal kingdom. Biochim. Biophys. Acta 1814:29–35 [DOI] [PubMed] [Google Scholar]
- 22. Dagley M. J., et al. 2011. Cell wall integrity is linked to mitochondria and phospholipid homeostasis in Candida albicans through the activity of the post-transcriptional regulator Ccr4-Pop2. Mol. Microbiol. 79:968–989 [DOI] [PubMed] [Google Scholar]
- 23. Douglas C. M., et al. 1997. Identification of the FKS1 gene of Candida albicans as the essential target of 1,3-beta-d-glucan synthase inhibitors. Antimicrob. Agents Chemother. 41:2471–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferrari S., et al. 2009. Gain of function mutations in CgPDR1 of Candida glabrata not only mediate antifungal resistance but also enhance virulence. PLoS Pathog. 5:e1000268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ferrari S., et al. 2011. Loss of mitochondrial functions associated with azole resistance in Candida glabrata results in enhanced virulence in mice. Antimicrob. Agents Chemother. 55:1852–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fiumera H. L., et al. 2009. Translocation and assembly of mitochondrially coded Saccharomyces cerevisiae cytochrome c oxidase subunit Cox2 by OxaI and Yme1 in the absence of Cox18. Genetics 182:519–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Friant S., Lombardi R., Schmelzle T., Hall M. N., Riezman H. 2001. Sphingoid base signaling via Pkh kinases is required for endocytosis in yeast. EMBO J. 20:6783–6792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Geraghty P., Kavanagh K. 2003. Disruption of mitochondrial function in Candida albicans leads to reduced cellular ergosterol levels and elevated growth in the presence of amphotericin B. Arch. Microbiol. 179:295–300 [DOI] [PubMed] [Google Scholar]
- 29. Geraghty P., Kavanagh K. 2003. Erythromycin, an inhibitor of mitoribosomal protein biosynthesis, alters the amphotericin B susceptibility of Candida albicans. J. Pharm. Pharmacol. 55:179–184 [DOI] [PubMed] [Google Scholar]
- 30. Gohil V. M., et al. 2004. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J. Biol. Chem. 279:42612–42618 [DOI] [PubMed] [Google Scholar]
- 31. Gohil V. M., Thompson M. N., Greenberg M. L. 2005. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae. J. Biol. Chem. 280:35410–35416 [DOI] [PubMed] [Google Scholar]
- 32. Greenleaf A. L., Kelly J. L., Lehman I. R. 1986. Yeast RPO41 gene product is required for transcription and maintenance of the mitochondrial genome. Proc. Natl. Acad. Sci. U. S. A. 83:3391–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gulshan K., Schmidt J. A., Shahi P., Moye-Rowley W. S. 2008. Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase: role in Pdr3-dependent retrograde regulation of PDR5 expression. Mol. Cell. Biol. 28:5851–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hallstrom T. C., et al. 2001. Coordinate control of sphingolipid biosynthesis and multidrug resistance in Saccharomyces cerevisiae. J. Biol. Chem. 276:23674–23680 [DOI] [PubMed] [Google Scholar]
- 35. Hallstrom T. C., Moye-Rowley W. S. 2000. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J. Biol. Chem. 275:37347–37356 [DOI] [PubMed] [Google Scholar]
- 36. Helmerhorst E. J., Stan M., Murphy M. P., Sherman F., Oppenheim F. G. 2005. The concomitant expression and availability of conventional and alternative, cyanide-insensitive, respiratory pathways in Candida albicans. Mitochondrion 5:200–211 [DOI] [PubMed] [Google Scholar]
- 37. Herrmann J. M., Bonnefoy N. 2004. Protein export across the inner membrane of mitochondria: the nature of translocated domains determines the dependence on the OxaI translocase. J. Biol. Chem. 279:2507–2512 [DOI] [PubMed] [Google Scholar]
- 38. Hillenmeyer M. E., et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hobbs A. E., Srinivasan M., McCaffery J. M., Jensen R. E. 2001. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 152:401–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huh W. K., Kang S. O. 2001. Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem. J. 356:595–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Imhof I., Canivenc-Gansel E., Meyer U., Conzelmann A. 2000. Phosphatidylethanolamine is the donor of the phosphorylethanolamine linked to the alpha1,4-linked mannose of yeast GPI structures. Glycobiology 10:1271–1275 [DOI] [PubMed] [Google Scholar]
- 42. Inagaki M., et al. 1999. PDK1 homologs activate the Pkc1-mitogen-activated protein kinase pathway in yeast. Mol. Cell. Biol. 19:8344–8352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jia L., Dienhart M. K., Stuart R. A. 2007. OxaI directly interacts with Atp9 and mediates its assembly into the mitochondrial F1Fo-ATP synthase complex. Mol. Biol. Cell 18:1897–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Joshi A. S., Zhou J., Gohil V. M., Chen S., Greenberg M. L. 2009. Cellular functions of cardiolipin in yeast. Biochim. Biophys. Acta 1793:212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kaur R., Castano I., Cormack B. P. 2004. Functional genomic analysis of fluconazole susceptibility in the pathogenic yeast Candida glabrata: roles of calcium signaling and mitochondria. Antimicrob. Agents Chemother. 48:1600–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim S. Y., Kim J. 2010. Roles of dihydrolipoamide dehydrogenase Lpd1 in Candida albicans filamentation. Fungal Genet. Biol. 47:782–788 [DOI] [PubMed] [Google Scholar]
- 47. Klis F. M., Brul S., De Groot P. W. 2010. Covalently linked wall proteins in ascomycetous fungi. Yeast 27:489–493 [DOI] [PubMed] [Google Scholar]
- 48. Kolaczkowski M., Kolaczkowska A., Gaigg B., Schneiter R., Moye-Rowley W. S. 2004. Differential regulation of ceramide synthase components LAC1 and LAG1 in Saccharomyces cerevisiae. Eukaryot. Cell 3:880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kornmann B., et al. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 325:477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lavoie H., et al. 2010. Evolutionary tinkering with conserved components of a transcriptional regulatory network. PLoS Biol. 8:e1000329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Levin D. E. 2005. Cell wall integrity signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 69:262–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu K., Zhang X., Lester R. L., Dickson R. C. 2005. The sphingoid long chain base phytosphingosine activates AGC-type protein kinases in Saccharomyces cerevisiae including Ypk1, Ypk2, and Sch9. J. Biol. Chem. 280:22679–22687 [DOI] [PubMed] [Google Scholar]
- 53. Ma H., et al. 2009. The fatal fungal outbreak on Vancouver Island is characterized by enhanced intracellular parasitism driven by mitochondrial regulation. Proc. Natl. Acad. Sci. U. S. A. 106:12980–12985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Magnani T., et al. 2008. Silencing of mitochondrial alternative oxidase gene of Aspergillus fumigatus enhances reactive oxygen species production and killing of the fungus by macrophages. J. Bioenerg. Biomembr. 40:631–636 [DOI] [PubMed] [Google Scholar]
- 55. Martchenko M., Levitin A., Hogues H., Nantel A., Whiteway M. 2007. Transcriptional rewiring of fungal galactose-metabolism circuitry. Curr. Biol. 17:1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Martins V. P., et al. 2011. Involvement of an alternative oxidase in oxidative stress and mycelium-to-yeast differentiation in Paracoccidioides brasiliensis. Eukaryot. Cell 10:237–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. McDonough J. A., Bhattacherjee V., Sadlon T., Hostetter M. K. 2002. Involvement of Candida albicans NADH dehydrogenase complex I in filamentation. Fungal Genet. Biol. 36:117–127 [DOI] [PubMed] [Google Scholar]
- 58. Merz S., Westermann B. 2009. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 10:R95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Milani G., et al. 2001. Respiratory chain network in mitochondria of Candida parapsilosis: ADP/O appraisal of the multiple electron pathways. FEBS Lett. 508:231–235 [DOI] [PubMed] [Google Scholar]
- 60. Moye-Rowley W. S. 2005. Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene 354:15–21 [DOI] [PubMed] [Google Scholar]
- 61. Mulhern S. M., Logue M. E., Butler G. 2006. Candida albicans transcription factor Ace2 regulates metabolism and is required for filamentation in hypoxic conditions. Eukaryot. Cell 5:2001–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Narasipura S. D., Chaturvedi V., Chaturvedi S. 2005. Characterization of Cryptococcus neoformans variety gattii SOD2 reveals distinct roles of the two superoxide dismutases in fungal biology and virulence. Mol. Microbiol. 55:1782–1800 [DOI] [PubMed] [Google Scholar]
- 63. Nebauer R., Schuiki I., Kulterer B., Trajanoski Z., Daum G. 2007. The phosphatidylethanolamine level of yeast mitochondria is affected by the mitochondrial components Oxa1p and Yme1p. FEBS J. 274:6180–6190 [DOI] [PubMed] [Google Scholar]
- 64. Noble S. M., French S., Kohn L. A., Chen V., Johnson A. D. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Okamoto K., Shaw J. M. 2005. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 39:503–536 [DOI] [PubMed] [Google Scholar]
- 66. Oliver B. G., et al. 2008. Tetracycline alters drug susceptibility in Candida albicans and other pathogenic fungi. Microbiology 154:960–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Osman C., et al. 2009. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. J. Cell Biol. 184:583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Panepinto J. C., et al. Overexpression of TUF1 restores respiratory growth and fluconazole sensitivity to a Cryptococcus neoformans vad1Delta mutant. Microbiology 156:2558–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Panwar S. L., Moye-Rowley W. S. 2006. Long chain base tolerance in Saccharomyces cerevisiae is induced by retrograde signals from the mitochondria. J. Biol. Chem. 281:6376–6384 [DOI] [PubMed] [Google Scholar]
- 70. Paul S., Schmidt J. A., Moye-Rowley W. S. 2011. Regulation of the CgPdr1 transcription factor from the pathogen Candida glabrata. Eukaryot. Cell 10:187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Plaine A., et al. 2008. Functional analysis of Candida albicans GPI-anchored proteins: roles in cell wall integrity and caspofungin sensitivity. Fungal Genet. Biol. 45:1404–1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Price M. S., et al. 2011. Cryptococcus neoformans requires a functional glycolytic pathway for disease but not persistence in the host. mBio 2:e00103–e00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Roelants F. M., Torrance P. D., Bezman N., Thorner J. 2002. Pkh1 and Pkh2 differentially phosphorylate and activate Ypk1 and Ykr2 and define protein kinase modules required for maintenance of cell wall integrity. Mol. Biol. Cell 13:3005–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ruy F., Vercesi A. E., Kowaltowski A. J. 2006. Inhibition of specific electron transport pathways leads to oxidative stress and decreased Candida albicans proliferation. J. Bioenerg. Biomembr. 38:129–135 [DOI] [PubMed] [Google Scholar]
- 75. Sanglard D., Ischer F., Bille J. 2001. Role of ATP-binding-cassette transporter genes in high-frequency acquisition of resistance to azole antifungals in Candida glabrata. Antimicrob. Agents Chemother. 45:1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sarinova M., Ticha E., Obernauerova M., Gbelska Y. 2007. Impact of mitochondrial function on yeast susceptibility to antifungal compounds. Folia Microbiol. (Praha) 52:223–229 [DOI] [PubMed] [Google Scholar]
- 77. Schmelzle T., Helliwell S. B., Hall M. N. 2002. Yeast protein kinases and the RHO1 exchange factor TUS1 are novel components of the cell integrity pathway in yeast. Mol. Cell. Biol. 22:1329–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schuiki I., Schnabl M., Czabany T., Hrastnik C., Daum G. 2010. Phosphatidylethanolamine synthesized by four different pathways is supplied to the plasma membrane of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801:480–486 [DOI] [PubMed] [Google Scholar]
- 79. Shahi P., Moye-Rowley W. S. 2009. Coordinate control of lipid composition and drug transport activities is required for normal multidrug resistance in fungi. Biochim. Biophys. Acta 1794:852–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Steen B. R., et al. 2003. Cryptococcus neoformans gene expression during experimental cryptococcal meningitis. Eukaryot. Cell 2:1336–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Stuart R. A. 2008. Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J. Bioenerg. Biomembr. 40:411–417 [DOI] [PubMed] [Google Scholar]
- 82. Tamura Y., Endo T., Iijima M., Sesaki H. 2009. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J. Cell Biol. 185:1029–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Toffaletti D. L., Nielsen K., Dietrich F., Heitman J., Perfect J. R. 2004. Cryptococcus neoformans mitochondrial genomes from serotype A and D strains do not influence virulence. Curr. Genet. 46:193–204 [DOI] [PubMed] [Google Scholar]
- 84. Traven A., Wong J. M., Xu D., Sopta M., Ingles C. J. 2001. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J. Biol. Chem. 276:4020–4027 [DOI] [PubMed] [Google Scholar]
- 85. Tsai H. F., Krol A. A., Sarti K. E., Bennett J. E. 2006. Candida glabrata PDR1, a transcriptional regulator of a pleiotropic drug resistance network, mediates azole resistance in clinical isolates and petite mutants. Antimicrob. Agents Chemother. 50:1384–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vandeputte P., et al. 2009. Hypersusceptibility to azole antifungals in a clinical isolate of Candida glabrata with reduced aerobic growth. Antimicrob. Agents Chemother. 53:3034–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Vellucci V. F., Gygax S. E., Hostetter M. K. 2007. Involvement of Candida albicans pyruvate dehydrogenase complex protein X (Pdx1) in filamentation. Fungal Genet. Biol. 44:979–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Voelker D. R. 2000. Interorganelle transport of aminoglycerophospholipids. Biochim. Biophys. Acta 1486:97–107 [DOI] [PubMed] [Google Scholar]
- 89. Zhang X., Moye-Rowley W. S. 2001. Saccharomyces cerevisiae multidrug resistance gene expression inversely correlates with the status of the F(0) component of the mitochondrial ATPase. J. Biol. Chem. 276:47844–47852 [DOI] [PubMed] [Google Scholar]
- 90. Zhong Q., Gvozdenovic-Jeremic J., Webster P., Zhou J., Greenberg M. L. 2005. Loss of function of KRE5 suppresses temperature sensitivity of mutants lacking mitochondrial anionic lipids. Mol. Biol. Cell 16:665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhong Q., Li G., Gvozdenovic-Jeremic J., Greenberg M. L. 2007. Up-regulation of the cell integrity pathway in saccharomyces cerevisiae suppresses temperature sensitivity of the pgs1Delta mutant. J. Biol. Chem. 282:15946–15953 [DOI] [PubMed] [Google Scholar]