Abstract
Curcumin, a polyphenol derived from turmeric, is an ancient therapeutic used in India for centuries to treat a wide array of ailments. Interest in curcumin has increased recently, with ongoing clinical trials exploring curcumin as an anticancer therapy and as a protectant against neurodegenerative diseases. In vitro, curcumin chelates metal ions. However, although diverse physiological effects have been documented for this compound, curcumin's mechanism of action on mammalian cells remains unclear. This study uses yeast as a model eukaryotic system to dissect the biological activity of curcumin. We found that yeast mutants lacking genes required for iron and copper homeostasis are hypersensitive to curcumin and that iron supplementation rescues this sensitivity. Curcumin penetrates yeast cells, concentrates in the endoplasmic reticulum (ER) membranes, and reduces the intracellular iron pool. Curcumin-treated, iron-starved cultures are enriched in unbudded cells, suggesting that the G1 phase of the cell cycle is lengthened. A delay in cell cycle progression could, in part, explain the antitumorigenic properties associated with curcumin. We also demonstrate that curcumin causes a growth lag in cultured human cells that is remediated by the addition of exogenous iron. These findings suggest that curcumin-induced iron starvation is conserved from yeast to humans and underlies curcumin's medicinal properties.
INTRODUCTION
Curcumin is the major chemical component of turmeric, a dietary spice made from the root of the Curcuma longa Linn plant and used extensively in traditional Indian medicine (38). Curcumin is a potent bioactive compound that is used to treat cancer (5, 35), atherosclerosis (33), and neurodegenerative diseases, such as Alzheimer's (26, 45) and Parkinson's (44) disease, as well as to promote wound healing (15, 36). Curcumin is particularly appealing as a therapeutic agent because of its extremely low toxicity. Many biological activities have been ascribed to curcumin. For example, curcumin suppresses inflammatory responses in cultured cells and in animals and also exhibits antioxidant properties. Furthermore, curcumin's ability to inhibit tumorigenesis and proliferation of a wide variety of cancerous cells has been well documented. Curcumin is a polyphenol and complexes readily with a number of different metal ions. In aqueous solutions of neutral pH, curcumin is an effective chelator of Fe(III) (2). Curcumin is also lipophilic and readily crosses membranes (19), so therefore it may also chelate metal ions intracellularly. How these chemical properties contribute to curcumin's biological activities, however, is not understood.
Identifying relevant in vivo targets of small molecules is technically challenging. Recently, several genetic and genomic approaches have been developed that use the simple eukaryote Saccharomyces cerevisiae, or budding yeast, to study the mechanism of drug action (17, 27, 31). One such method, termed homozygous profiling, uses a comprehensive collection of 4,700 homozygous diploid deletion yeast strains, each bearing a deletion of a single nonessential gene (39), to examine growth in the presence of a bioactive compound (12). Mutant strains that display increased sensitivity to the compound are identified, and the identity of the genes deleted in these hypersensitive strains is used to infer the biological effects of the compound.
We carried out such a screen to identify yeast mutants whose growth is strongly inhibited by curcumin. The results of this study indicate that curcumin antagonizes yeast growth by chelating iron. Furthermore, iron supplementation alleviates the growth-inhibiting effect of curcumin on both yeast and cultured human cells, suggesting a common mechanism. Previous studies established that curcumin treatment causes mouse cells and tissues to display iron depletion characteristics (20). The findings presented here also indicate that curcumin chelates iron in vivo and suggest that iron chelation may underlie many of curcumin's therapeutic activities.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The yeast growth medium and basic methods were as described in reference 34. Curcumin, bathophenanthroline disulfonic acid (BPS), bathocuproine disulphonate (BCS), ferrous sulfate (Fe2SO4), copper chloride (CuCl2), and ferric chloride (FeCl3) were from Sigma-Aldrich, Inc. (St. Louis, MO). BY4743 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 LYS2/lys2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0) or BY4741 (MATa his3Δ1 leu2Δ0 LYS2 met15Δ0 ura3Δ0) was used as the wild-type (WT) strain. The homozygous diploid deletion collection, based on BY4743, was purchased from Open Biosystems (Huntsville, AL). Cells used in growth analysis on plates were grown to saturation and then plated in 5-fold serial dilutions, with a starting concentration of approximately 1.0 × 107 cells.
Pooled competitive growth assays.
The experimental design for pooled competitive growth assays is detailed in reference 11. In brief, yeast strains from the homozygous deletion collection were pooled as described previously (12, 32) and stored at −80°C. The pooled cells were then thawed and diluted in yeast extract-peptone-dextrose (YPD) to an optical density at 600 nm (OD600) of 0.0625. This dilution ensures that 300 individual cells from each deletion strain are represented at the onset of the experiment. A 150 μM concentration of curcumin was added to the pooled cells, and the cells were grown for five generations (to an OD600 of 2.0) in a Tecan GENios microtiter plate reader (Tecan, Durham, NC). Cells were harvested, and genomic DNA was extracted and used as a template in PCR amplification of “Uptag” and “Downtag” sequences that mark each gene deletion. The fluorescently tagged PCR products were hybridized to TAG3 microarrays from Affymetrix (Santa Clara, CA) (11), and the fluorescence intensity of each pair of tags was determined. Fitness defect scores were calculated for each strain in the pool. These scores are based on a tag-specific algorithm that takes into account the intensities of each tag on the experimental array and the corresponding intensities on a set of control arrays, performed using pooled cells grown without the compound of interest (control set) (12). Tag intensities were log transformed and mean normalized, and the intensities for replicates were averaged into a single value. Means and standard deviations were calculated for both the Uptag and Downtag hybridizations. Z scores for the Uptag and Downtag for each strain were calculated by subtracting the control average intensity from the experimental (curcumin-treated) average intensity and dividing by the standard deviation of the control set intensities. This generates z score values for both the Uptag and Downtag sequences, which are then averaged into a single fitness defect score for the strain.
β-Galactosidase assays.
BY4741 cells were transformed by the lithium acetate method with a plasmid bearing the PFET3-lacZ reporter (provided by Caroline Philpott, NIDDK, National Institutes of Health) or a plasmid containing the PFRE2-lacZ reporter (provided by Dennis Winge, University of Utah) (28). Cells were then grown to an OD600 of 1.0 in synthetic complete medium (lacking the required amino acid for plasmid selection) made with yeast nitrogen base (YNB), lacking iron and copper, from Bio101 (Solon, OH). Cultures were diluted to an OD600 of 0.01 in fresh, low-iron/copper synthetic complete medium with additions of curcumin or BPS, in the presence or absence of Fe2SO4, and grown an additional 12 h. Cells were harvested, and proteins were extracted for use in β-galactosidase activities as described in reference 4. The β-galactosidase activity reported is the maximum rate of OD415 change/minute/amount of protein. Two independent transformants were assayed in triplicate (for a minimum of six replicates) for each condition. Error bars represent the standard deviations of the means.
ICP-AES.
Inductively coupled plasma atomic emission spectroscopy (ICP-AES) was performed as described in reference 10. In brief, wild-type (BY4743) cells were grown from an OD600 of 0.5 to saturation in YPD either with no additions or with the addition of 100 μM BPS, 500 μM BCS, 25 μM curcumin, or 50 μM curcumin. Cells were then harvested by filtration, washed with water, resuspended in 500 μl of 30% nitric acid, and digested overnight at 65°C. The cell digests were centrifuged at 12,000 × g for 10 min, and the supernatants were transferred to new tubes and used in ICP-AES analysis on a Varian Vista ICP-AES (Varian Inc., Palo Alto, CA). Data analysis was performed as indicated in reference 10 for the following elements: Ca, Co, Cu, Fe, K, Mg, Mn, Na, Ni, P, S, and Zn. All element measurements were corrected for differences in cell mass. This correction assumes that treated and untreated cells differ only in cell mass, which appears to be the case, as most elements assessed did not change significantly between experimental and control cells (data not shown). The mean iron and copper contents for 5 experimental replicates are presented as percentages relative to the contents in untreated (YPD) control cells. Error bars represent the standard errors of the means.
Yeast growth curves, cell morphology, and viability analysis.
Wild-type (BY4743) cells were grown to saturation in YPD, washed, and inoculated at an OD600 of 0.5 into YPD either with no additives or with 80 μM BPS, 50 μM curcumin, or 100 μM curcumin, each with and without 50 μM Fe2SO4. Three independent cultures were monitored for each condition, and cells were grown in a Tecan GENios plate reader with orbital shaking at room temperature (from 23 to 27°C), with OD600 readings taken every 30 min. A path length correction factor was then applied to Tecan GENios OD600 readings, and these data are reported. For bud-type assessments, cells were harvested after 7 h of growth and fixed in 3.7% formaldehyde and their bud morphologies were scored. The morphologies of 200 cells were assessed for each of three replicate cultures (for a total of 600 cells in 3 replicates). Data are the means of these values, and the error bars represent the standard deviations of the means.
In addition to cell morphology, cells used in the growth curve analysis were scored for viability after 7 h of incubation with the additives indicated above. This analysis was performed to determine if the growth lag observed for certain additions was due to cell death. Twenty microliters of each culture was harvested by centrifugation and resuspended in 3.7% methylene blue in 100 mM phosphate-buffered saline (PBS; adjusted to pH 7), and the cells were scored for viability. No fewer than 100 cells were scored per culture, and cells staining dark blue were considered metabolically inactive and therefore inviable. As a control in this experiment, an additional sample was taken from the YPD-grown cultures and heat treated for 10 min at 70°C to kill all cells. As expected, 100% of cultures treated this way stained dark blue and were unable to form colonies. Viability was further confirmed for all cultures by assessing the numbers of CFU on YPD after 7 h of incubation with the indicated additives.
RESULTS AND DISCUSSION
Yeast mutants with defects in iron and copper homeostasis are hypersensitive to curcumin.
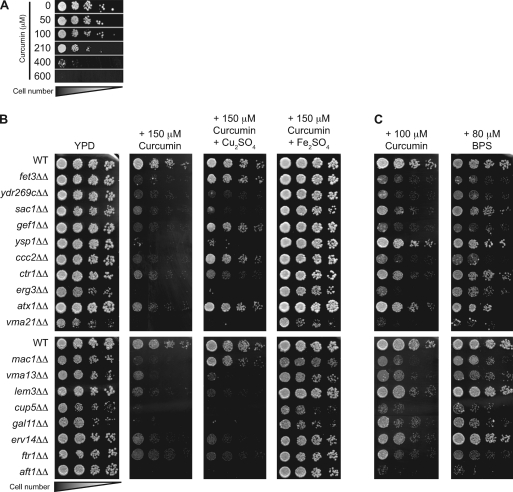
Curcumin was tested at a range of concentrations and found to inhibit growth of wild-type yeast in a dose-dependent manner (Fig. 1 A). To determine the mechanism by which growth was inhibited, we sought to identify mutant yeast strains with increased sensitivity to curcumin. First, a competitive growth assay (11), in which a genomic collection of ∼4,700 homozygous diploid deletion strains (39) was grown in a single culture using medium that contained curcumin, was used. After 5 generations of growth, the relative abundance of each strain in the pool was assessed by hybridization to a DNA microarray that contained sequence identifiers unique to each deletion strain (see Materials and Methods). Mutants whose growth is inhibited by curcumin were underrepresented in this pool. Analysis of these data identified 42 mutant strains that displayed a significant fitness defect during growth in curcumin-containing media (see Table S1 in the supplemental material). Growth of the 30 most sensitive strains was then tested individually on solid medium containing 150 μM curcumin (Fig. 1B; see also Table S1 in the supplemental material); however, the pho86Δ, ypk1Δ, and vma5Δ strains were excluded because they grew poorly on YPD alone. Of the strains tested, 18 were hypersensitive to 150 μM curcumin in the solid-medium growth assay and are hereafter referred to as curcumin-hypersensitive strains (Fig. 1B). The degree of hypersensitivity in the initial liquid growth assay did not strongly correlate with the degree of growth inhibition observed on solid medium containing curcumin. This could be due to the differences between the growth in liquid and solid media, the differences between competitive and clonal growth, and/or the length of exposure to curcumin: in the competitive growth assay, cells were exposed to curcumin for only 5 generations, compared to prolonged curcumin exposure for 2 to 4 days in the solid-medium growth assay (i.e., ∼20 generations of growth are required to form a colony). The combination of both assays allowed for a more accurate assessment of hypersensitivity.
Fig. 1.
Yeast sensitivity to curcumin can be rescued by iron supplementation and is similar to yeast sensitivity to BPS, an iron chelator. (A) Yeast cells are sensitive to curcumin. Serial dilutions of WT (BY4743) cells were plated on YPD with or without curcumin at the concentrations indicated and grown for 2 days at 30°C. (B) Deletion of iron and copper homeostasis genes exacerbates yeast sensitivity to curcumin. WT (BY4743) and isogenic homozygous deletion strains (lacking the gene indicated) were plated on YPD without any additions or with 150 μM curcumin, either by itself, with 500 μM Cu2SO4, or with 50 μM Fe2SO4, and grown for 3 days at 30°C. (C) In parallel to the experiments shown in panel B, the same cultures were plated on YPD with either 100 μM curcumin or 80 μM BPS and grown for 3 days at 30°C.
Next, the functions of the genes deleted in the curcumin-hypersensitive strains were analyzed. Only 17 genes were identified in the 18 mutant strains, as two strains, the ccc2Δ and ydr269cΔ strains, delete overlapping open reading frames and thus both compromise Ccc2 function. Remarkably, 8 out of the 17 identified genes encode products that function in the transport and homeostasis of iron and copper ions (Table 1). Specifically, genes whose products mediate iron transport (FTR1 and FET3) and the regulation of gene expression under iron limitation conditions (AFT1) were identified (8, 40). Genes that encode proteins required for copper transport (CTR1) and the regulation of gene expression during copper limitation (MAC1) were also identified (6, 23). Interestingly, these processes are coupled in vivo (7), as an essential component of high-affinity iron uptake, Fet3p, is a copper-dependent ferro-oxidoreductase, and strains with deletions of genes whose products are required for the incorporation of copper ion into Fet3 (CCC2, ATX1, and GEF1) are sensitive to curcumin (13, 18, 42). Other identified genes include those that disrupt vacuolar acidification (VMA3, VMA13, and VMA21), a process required for intracellular iron utilization (9, 37). In addition, several mutants that disrupt the lipid composition of membranes and thus have wide-ranging defects in membrane function and trafficking are hypersensitive to curcumin (the sac1Δ, erg3Δ, and lem3Δ strains). Overall, these findings suggest that strains with defects in iron and/or copper homeostasis are hypersensitive to curcumin.
Table 1.
Deleted genes identified in curcumin-hypersensitive strains and their functions
| Biological process | Gene(s) |
|---|---|
| Transport or homeostasis of iron or copper ions | |
| Iron ion import/homeostasis | FET3, FTR1, AFT1 |
| Copper ion import/homeostasis | CTR1, MAC1 |
| Copper and iron homeostasis | ATX1, GEF1, CCC2a |
| Other function | |
| Cell signaling | SAC1 |
| RNA polymerase II-mediated | |
| transcription | GAL11 |
| Vesicle-associated secretion | ERV14 |
| Vacuolar acidification | VMA3, VMA13, VMA21 |
| Phospholipid translocation | LEM3 |
| Ergosterol biosynthesis | ERG3 |
| Programmed cell death | YSP1 |
The ydr269cΔ strain also disrupts ccc2.
Differential rescue of curcumin hypersensitivity by copper and iron.
To further define the mechanism by which curcumin inhibits yeast growth, we tested the ability of curcumin-hypersensitive deletion strains to grow when iron or copper supplements were added to the growth medium. Importantly, addition of iron or copper to YPD medium at the concentrations employed had no effect on cell growth in the absence of curcumin (data not shown). Addition of 50 μM iron significantly improved the growth of all curcumin-sensitive strains, whereas addition of a 10-fold-larger amount of copper (500 μM) was able to rescue growth of only a small subset of strains (the fet3Δ, gef1Δ, ccc2Δ, atx1Δ, and mac1Δ strains) (Fig. 1B).
We next compared the curcumin sensitivity profile to those of BPS, a known iron chelator, and BCS, a known copper chelator. The 100 μM curcumin sensitivity profile was strikingly similar to the 80 μM BPS sensitivity profile (Fig. 1C), with the erg3Δ strain as the single exception, showing sensitivity to curcumin and not BPS. Similar experiments comparing growth on 100 μM curcumin to growth on as much as 500 μM BCS identified only three of the candidates, the vma21Δ, vma13Δ, and cup5Δ strains, as BCS sensitive (data not shown). Taken together, these observations suggest that curcumin inhibits yeast growth primarily through the limitation of iron availability.
Competitive growth assays similar to the one described here for curcumin have been performed using 75 to 100 μM BPS (21). This BPS concentration range results in a yeast sensitivity profile similar to that observed for 150 μM curcumin (Fig. 1C and data not shown), which was the curcumin concentration employed in our competitive growth assays. Strikingly, 18 of 42 of our curcumin-sensitive candidates were also identified in the BPS sensitivity screen (21). While the curcumin-hypersensitive strains identified in this study were well represented in the BPS hypersensitivity profile, the BPS competitive growth assays described by Jo et al. (21) identified approximately 3-fold-more sensitive strains. This difference could be due to the fact that cells were exposed to BPS for 15 generations as opposed to the 5 generations used in the curcumin screen. Furthermore, a less stringent fitness cutoff was used when assigning BPS-sensitive strains. Taken together, these data further support the interpretation that curcumin-mediated growth inhibition is due to iron starvation.
Curcumin accumulates in yeast endoplasmic reticulum membranes.
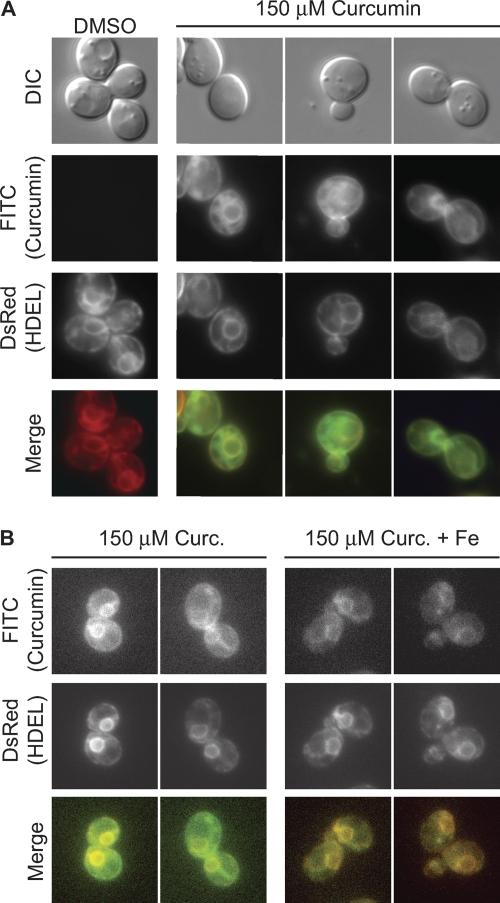
Chelators can deplete the exogenous pool of metal ions from the growth medium, enter cells and deplete the intracellular reserves of metal ions, or act both intra- and extracellularly to alter metal ion pools. Curcumin is able to bind iron in solution, demonstrating that it is an effective extracellular iron chelator (2). To determine if curcumin can also traverse the yeast cell wall and plasma membrane to act intracellularly, we exploited the natural fluorescent properties of curcuminoids. Curcumin autofluorescence can be excited at ∼455 nm and emits at approximately 540 nm (19, 24). We treated cells containing an HDEL-DsRed (red fluorescent protein from Discosoma striata) marker, which uniformly stains both cortical and perinuclear endoplasmic reticulum (ER) membranes (16), with either dimethyl sulfoxide (DMSO) or 150 μM curcumin for 5 h and visualized curcumin fluorescence using a filter set for green fluorescent proteins (GFPs) that overlaps with the curcumin excitation and emission wavelengths (Fig. 2). Curcumin fluorescence colocalized with the HDEL-DsRed marker in 69% of cells (n = 103) examined, indicating that curcumin not only enters yeast cells but also accumulates within the ER membrane (Fig. 2A). While there is clear colocalization of curcumin with the ER membrane, curcumin may also localize to additional intracellular membranes, accounting for the background of curcumin fluorescence that does not colocalize with the ER HDEL marker (Fig. 2A). These observations are consistent with demonstrations of curcumin accumulation in intracellular membranes, including the ER, of mammalian cells (19, 24). These findings further suggest that curcumin may impact intracellular iron pools in addition to chelation of extracellular iron.
Fig. 2.
Curcumin enters yeast cells and accumulates in ER membranes. (A) Wild-type yeast cells expressing HDEL-DsRed (from strain VHY87, described in reference 16) were treated with either DMSO (left column) or 150 μM curcumin and visualized by fluorescence microscopy using the GFP filter set for curcumin and the red fluorescent protein (RFP) filter set for HDEL-DsRed. Colocalization of curcumin fluorescence with the ER marker was observed for 69% of the cells examined (n = 103). (B) Cells from VHY87 expressing the HDEL-DsRed marker were treated with 150 μM curcumin, either with or without the addition of 50 μM FeSO4, and visualized as described for panel A.
To test whether addition of iron to the growth medium suppresses curcumin sensitivity by preventing curcumin uptake into cells, cells grown in curcumin and iron were examined by fluorescence microscopy. Curcumin also colocalized with the ER membrane in iron-treated cells, indicating that it still enters cells under conditions of iron supplementation (Fig. 2B). Curcumin fluorescence was reduced in iron-treated cells. However, because iron decreases the fluorescence intensity of curcumin-containing solutions in a dose-dependent manner (data not shown), fluorescence per se cannot be used to compare intracellular curcumin concentrations under these two conditions. Nonetheless, our findings indicate that curcumin permeates yeast cells grown in the presence or absence of supplemental iron and are consistent with the hypothesis that curcumin acts intracellularly to chelate iron and induce iron starvation.
Curcumin treatment results in reduced intracellular iron.
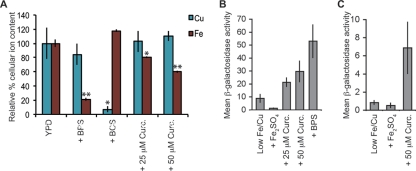
We tested the effect of curcumin on cellular iron levels directly by using inductively coupled plasma atomic emission spectroscopy (ICP-AES). Wild-type cells were treated with curcumin, BPS, or BCS (see Materials and Methods). The majority of elements assessed this way did not change as a result of these treatments (data not shown). As expected, the iron content of cells treated with 100 μM BPS was dramatically reduced (80% less iron than cells grown in YPD). Curcumin also significantly decreased cellular iron levels, with cells grown in the presence of 25 μM and 50 μM curcumin containing 20 and 40% less iron, respectively, than cells grown in YPD (Fig. 3 A). Treatment with BCS reduced cellular copper content by 90% from that of cells grown in YPD; however, consistent with our earlier findings, curcumin treatment did not significantly alter copper levels from those of YPD-grown cells (Fig. 3A).
Fig. 3.
Curcumin depletes cellular iron and activates iron starvation-induced transcription. (A) WT (BY4743) cells were treated with 100 μM BPS, 500 μM BCS, or the indicated concentration of curcumin. The mean cellular iron and copper levels relative to those of untreated cells in YPD (as determined by ICP-AES measurement; see Materials and Methods) are shown for a minimum of five experimental replicates. Error bars represent the standard errors of the means. Paired Student's t tests were performed to compare YPD control cultures and each of the treatment conditions. *, P value of <0.05; **, P value of <0.0005. (B) WT (BY4741) cells containing the PFET3-lacZ reporter were grown in low-Fe/Cu medium and treated for 12 h with 100 μM Fe2SO4, 10 μM BPS, or the indicated concentrations of curcumin. Protein extracts were prepared, and β-galactosidase activity was determined for a minimum of two transformants, each of which was assayed in triplicate. The mean β-galactosidase activities and standard deviations are reported. (C) WT (BY4741) cells containing the PFRE2-lacZ reporter were grown and assayed for β-galactosidase activity as described for panel B. The mean β-galactosidase activities are presented, and the error bars are the standard deviations of the means.
The physiological relevance of this cellular iron depletion was next examined by measuring the transcriptional response induced by curcumin. Under conditions of iron starvation, the Aft1 transcription factor upregulates genes responsible for high-affinity iron transport (41). Two targets of Aft1 transcription regulation are fet3, the high-affinity iron transporter, and fre2, a gene encoding an iron reductase that functions as the rate-limiting step in high-affinity iron import (1, 43). Transcription of fet3 can also be induced by Mac1p, a transcription factor that activates gene expression in response to copper starvation (14); however, regulation of fre2 is independent of copper starvation (28). Following 50 μM curcumin treatment, fet3 and fre2 expression increased 3-fold and 6-fold, respectively (Fig. 3B and C). Because the curcumin-mediated induction levels of fre2 and fet3 are similar, the most parsimonious explanation is that curcumin induces predominantly an iron starvation response. These data suggest that the growth of yeast in the presence of curcumin leads to a decrease in available iron and, as a result, induces the iron starvation transcriptional response.
Curcumin slows yeast cell cycle progression with no accompanying decrease in cell viability.
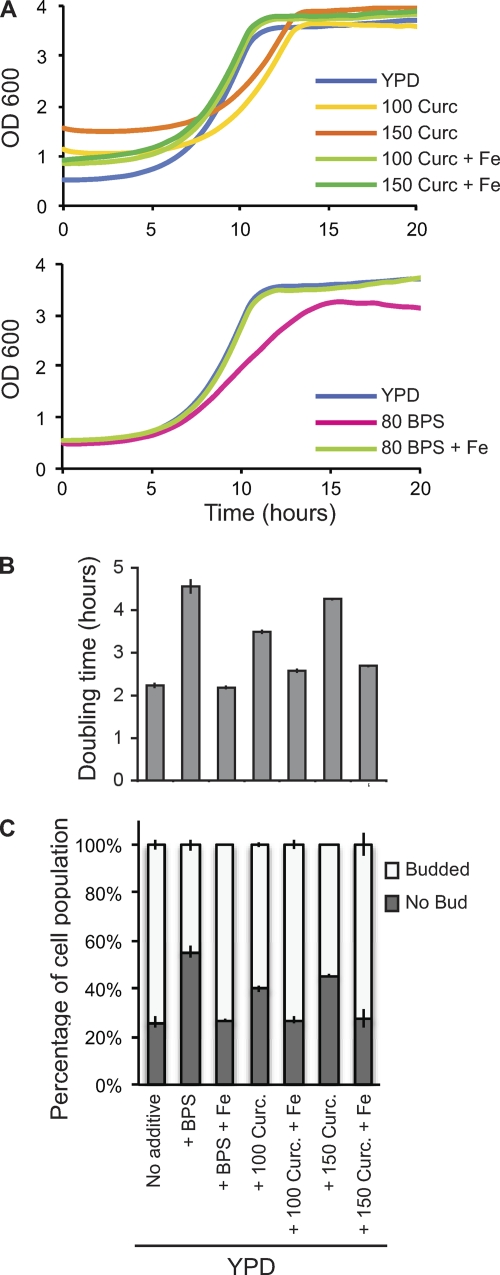
Next, we examined the kinetics of curcumin-induced yeast cell growth inhibition. Yeast cells were grown to mid-log phase and treated with curcumin or BPS in the presence or absence of additional iron, and the growth rate (i.e., doubling time) was monitored. As expected, addition of curcumin or BPS lowered the yeast growth rate (Fig. 4 A), with treatments of either 80 μM BPS or 150 μM curcumin increasing the doubling time approximately 2-fold (Fig. 4B). Addition of iron to the BPS- or curcumin-treated cells rescued this growth inhibition, shortening the doubling times to levels similar to those observed for untreated control cells grown in YPD (Fig. 4A and B).
Fig. 4.
Curcumin treatment causes a cell cycle delay in yeast that is alleviated by iron supplementation. (A) WT (BY4743) cells were grown overnight to saturation and inoculated at an OD600 of 0.5 into fresh YPD either with no supplement or with 100 μM curcumin, 150 μM curcumin, or 80 μM BPS, each with (indicated as “+ Fe”) or without 50 μM Fe2SO4, and optical density measurements were obtained every 30 min using a Tecan GENios microtiter plate reader. Addition of curcumin to the growth medium results in a shift in absorbance of approximately 0.6 OD600 for 100 μM curcumin and 1.0 OD600 for 150 μM curcumin; therefore, growth below these densities cannot be measured accurately for this concentration of drug. The growth curves presented are the averages of measurements for three independent cultures. (B) The cell doubling times during logarithmic growth were determined for the cultures used in panel A. Values are the averages of the doubling times determined for three independent cultures, and error bars represent the standard errors of the means. (C) Cells were grown as described for panel A, and at 7 h postinoculation (during growth curve analysis), samples were taken and bud morphologies were scored for three independent cultures, with a minimum of 200 cells counted for each.
After ∼10 h of treatment with either BPS or curcumin, at which point control cells in YPD had doubled twice, cell viability and cell cycle progression were assessed for the various conditions. Viability of cells grown in the presence of curcumin or BPS was monitored using the vital stain methylene blue (see Materials and Methods) and by assessing the number of CFU of washed cells plated on YPD. At 10 h posttreatment, 100% cell viability was observed for all cultures (data not shown), consistent with the hypothesis that these compounds cause a delay in cell cycle progression rather than cell death. To assess cell cycle progression, we analyzed the morphologies of yeast cells exposed to curcumin and BPS. In G1 phase, cells are unbudded, while cells in S, G2, and M phases of the cell cycle possess buds of characteristic sizes (reviewed in reference 25). During mid-log-phase growth in YPD, 25% of cells were unbudded and 75% of cells were budded (Fig. 4C). Exposure to curcumin or BPS caused an increase in the fraction of unbudded cells, which is commonly observed for treatments that increase the length of G1 (Fig. 4C). Curcumin exposure caused a dose-dependent increase in the proportion of unbudded cells to 40% and 45% for 100 μM and 150 μM curcumin, respectively. BPS treatment increased the unbudded fraction to 55%. Taken together, these observations demonstrate that curcumin treatment lengthens the time required to complete the cell cycle and suggest that the duration of G1 is increased.
To determine if iron starvation was the cause of this increased doubling time, iron was added to the medium to counter the effect of curcumin on cellular iron levels. Supplementation with 50 μM iron in cultures with 100 μM or 150 μM curcumin or 80 μM BPS completely alleviated the growth inhibition, resulting in restoration of the cell morphology distributions to those observed for YPD-grown cells (Fig. 4C). Thus, BPS- or curcumin-induced iron starvation increases yeast doubling time, and this is, at least in part, due to a prolonged G1 phase. Treatment of yeast with other metal chelators has been reported to induce a G1 growth arrest that can be rescued by the addition of exogenous metal ions to the growth medium (22).
Curcumin and cultured human cells.
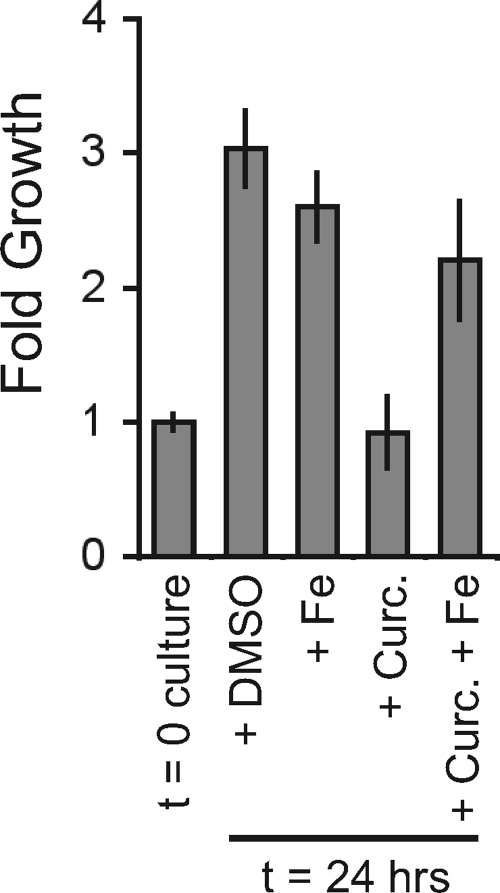
Exposure of certain human cancer cells to curcumin results in a G1 cell cycle arrest (30), and treatment of human cells with other iron chelators also induces a reversible G1 cell cycle arrest (3, 29). To extend the observations made in yeast, we further explored the effects of curcumin on cell growth by treating human osteosarcoma U2OS cells with curcumin and monitoring cell density relative to that of a DMSO drug vector control. The density of control cultures increased 3-fold in 24 h, whereas the curcumin-treated culture density did not increase (Fig. 5). Addition of iron to the curcumin-treated cells suppressed this growth defect and allowed for population doubling in 24 h (Fig. 5). These data suggest that curcumin's effects are conserved and that it inhibits growth of both yeast and human cells by limiting iron availability.
Fig. 5.
Curcumin-mediated inhibition of U2OS cells is alleviated by the addition of iron. Three independent cultures of U2OS cells were grown for 24 h in Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal calf serum (FCS) with the addition of DMSO (as a vehicle control), 100 μM FeCl3 (Fe), and/or 7.5 μM curcumin. (This is an approximately 20× lower concentration than what we routinely used for yeast. The difference in human versus yeast cell sensitivity to curcumin may be due to the yeast cell wall or the multiple drug efflux pumps present in yeast cells.) Cells were then trypsinized (100 μl of 0.05% trypsin; 5 min at room temperature [RT]), resuspended in DMEM and FCS, harvested by centrifugation, and resuspended in PBS. Cell density was determined for each culture by counting the cells in three fields with a hemocytometer. Fold growth presented is relative to the density of the starting culture (t = 0), and error bars represent the standard deviations of the means.
An understanding of curcumin's mechanism of action on human cells has been elusive, due in part to the myriad effects associated with curcumin treatment (reviewed in reference 15). We used yeast as a model system to explore the mechanism of action of curcumin, and our results strongly suggest that curcumin inhibits eukaryotic cell growth predominantly by chelating intracellular iron. Yeast cells with defects in iron homeostasis are sensitive to curcumin, and curcumin delays cell cycle progression in an iron-dependent manner. These findings suggest that the potent chemotherapeutic effects of curcumin may be explained by iron depletion of cancer cells, a process which inhibits their proliferation. Iron chelation may also explain the diverse physiological effects of curcumin. Many cellular enzymes require iron as a cofactor, and several signaling pathways are also affected by iron availability. Curcumin penetrates both yeast and mammalian cells and accumulates in intracellular membranes, including the ER. This accumulation in intracellular compartments may selectively limit the availability of essential iron cofactors, impairing specific enzymatic functions and altering cellular signaling to produce the diverse physiological effects associated with curcumin treatment.
Supplementary Material
ACKNOWLEDGMENTS
We thank Caroline Philpott and Dennis Winge for kindly providing plasmids used in this work. We thank the Environmental Measurement: Gas-Solution Analytical Center at Stanford University for its assistance with the ICP-AES measurements. We also acknowledge Aaron Goldman for key technical assistance.
M.S.C. is supported by National Institutes of Health research grant GM-48728. T.P.S. is supported by NIH grant GM52022. C.N. and G.G. are supported by grants from NHGRI and the Canadian Institutes of Health (grant 84305 for C.N. and grant 81340 for G.G.).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Anderson G. J., et al. 1992. Ferric iron reduction and iron assimilation in Saccharomyces cerevisiae. J. Inorg. Biochem. 47:249–255 [DOI] [PubMed] [Google Scholar]
- 2. Bernabe-Pineda M., Ramirez-Silva M. T., Romero-Romo M. A., Gonzalez-Vergara E., Rojas-Hernandez A. 2004. Spectrophotometric and electrochemical determination of the formation constants of the complexes curcumin-Fe(III)-water and curcumin-Fe(II)-water. Spectrochim. Acta A Mol. Biomol. Spectrosc. 60:1105–1113 [DOI] [PubMed] [Google Scholar]
- 3. Brodie C., et al. 1993. Neuroblastoma sensitivity to growth inhibition by deferrioxamine: evidence for a block in G1 phase of the cell cycle. Cancer Res. 53:3968–3975 [PubMed] [Google Scholar]
- 4. Bultynck G., et al. 2006. Slm1 and Slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell. Biol. 26:4729–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bush J. A., Cheung K. J., Jr., Li G. 2001. Curcumin induces apoptosis in human melanoma cells through a Fas receptor/caspase-8 pathway independent of p53. Exp. Cell Res. 271:305–314 [DOI] [PubMed] [Google Scholar]
- 6. Dancis A., Haile D., Yuan D. S., Klausner R. D. 1994. The Saccharomyces cerevisiae copper transport protein (Ctr1p). Biochemical characterization, regulation by copper, and physiologic role in copper uptake. J. Biol. Chem. 269:25660–25667 [PubMed] [Google Scholar]
- 7. Dancis A., et al. 1994. Molecular characterization of a copper transport protein in S. cerevisiae: an unexpected role for copper in iron transport. Cell 76:393–402 [DOI] [PubMed] [Google Scholar]
- 8. De Silva D. M., Askwith C. C., Eide D., Kaplan J. 1995. The FET3 gene product required for high affinity iron transport in yeast is a cell surface ferroxidase. J. Biol. Chem. 270:1098–1101 [DOI] [PubMed] [Google Scholar]
- 9. Eide D. J., Bridgham J. T., Zhao Z., Mattoon J. R. 1993. The vacuolar H(+)-ATPase of Saccharomyces cerevisiae is required for efficient copper detoxification, mitochondrial function, and iron metabolism. Mol. Gen. Genet. 241:447–456 [DOI] [PubMed] [Google Scholar]
- 10. Eide D. J., et al. 2005. Characterization of the yeast ionome: a genome-wide analysis of nutrient mineral and trace element homeostasis in Saccharomyces cerevisiae. Genome Biol. 6:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Giaever G., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387–391 [DOI] [PubMed] [Google Scholar]
- 12. Giaever G., et al. 2004. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc. Natl. Acad. Sci. U. S. A. 101:793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Greene J. R., Brown N. H., DiDomenico B. J., Kaplan J., Eide D. J. 1993. The GEF1 gene of Saccharomyces cerevisiae encodes an integral membrane protein; mutations in which have effects on respiration and iron-limited growth. Mol. Gen. Genet. 241:542–553 [DOI] [PubMed] [Google Scholar]
- 14. Gross C., Kelleher M., Iyer V. R., Brown P. O., Winge D. R. 2000. Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J. Biol. Chem. 275:32310–32316 [DOI] [PubMed] [Google Scholar]
- 15. Hatcher H., Planalp R., Cho J., Torti F. M., Torti S. V. 2008. Curcumin: from ancient medicine to current clinical trials. Cell. Mol. Life Sci. 65:1631–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Heath V. L., Shaw S. L., Roy S., Cyert M. S. 2004. Hph1p and Hph2p, novel components of calcineurin-mediated stress responses in Saccharomyces cerevisiae. Eukaryot. Cell 3:695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hoon S., St. Onge R. P., Giaever G., Nislow C. 2008. Yeast chemical genomics and drug discovery: an update. Trends Pharmacol. Sci. 29:499–504 [DOI] [PubMed] [Google Scholar]
- 18. Huffman D. L., O'Halloran T. V. 2000. Energetics of copper trafficking between the Atx1 metallochaperone and the intracellular copper transporter, Ccc2. J. Biol. Chem. 275:18611–18614 [DOI] [PubMed] [Google Scholar]
- 19. Jaruga E., et al. 1998. Apoptosis-like, reversible changes in plasma membrane asymmetry and permeability, and transient modifications in mitochondrial membrane potential induced by curcumin in rat thymocytes. FEBS Lett. 433:287–293 [DOI] [PubMed] [Google Scholar]
- 20. Jiao Y., et al. 2006. Iron chelation in the biological activity of curcumin. Free Radic. Biol. Med. 40:1152–1160 [DOI] [PubMed] [Google Scholar]
- 21. Jo W. J., et al. 2009. Novel insights into iron metabolism by integrating deletome and transcriptome analysis in an iron deficiency model of the yeast Saccharomyces cerevisiae. BMC Genomics 10:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnston G. C., Singer R. A. 1978. RNA synthesis and control of cell division in the yeast S. cerevisiae. Cell 14:951–958 [DOI] [PubMed] [Google Scholar]
- 23. Jungmann J., et al. 1993. MAC1, a nuclear regulatory protein related to Cu-dependent transcription factors is involved in Cu/Fe utilization and stress resistance in yeast. EMBO J. 12:5051–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kunwar A., et al. 2008. Quantitative cellular uptake, localization and cytotoxicity of curcumin in normal and tumor cells. Biochim. Biophys. Acta 1780:673–679 [DOI] [PubMed] [Google Scholar]
- 25. Lew D. J. 2003. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 15:648–653 [DOI] [PubMed] [Google Scholar]
- 26. Lim G. P., et al. 2001. The curry spice curcumin reduces oxidative damage and amyloid pathology in an Alzheimer transgenic mouse. J. Neurosci. 21:8370–8377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lum P. Y., et al. 2004. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell 116:121–137 [DOI] [PubMed] [Google Scholar]
- 28. Martins L. J., Jensen L. T., Simon J. R., Keller G. L., Winge D. R. 1998. Metalloregulation of FRE1 and FRE2 homologs in Saccharomyces cerevisiae. J. Biol. Chem. 273:23716–23721 [DOI] [PubMed] [Google Scholar]
- 29. Pahl P. M., Horwitz L. D. 2005. Cell permeable iron chelators as potential cancer chemotherapeutic agents. Cancer Invest. 23:683–691 [DOI] [PubMed] [Google Scholar]
- 30. Park M. J., et al. 2002. Curcumin inhibits cell cycle progression of immortalized human umbilical vein endothelial (ECV304) cells by up-regulating cyclin-dependent kinase inhibitor, p21WAF1/CIP1, p27KIP1 and p53. Int. J. Oncol. 21:379–383 [PubMed] [Google Scholar]
- 31. Parsons A. B., et al. 2006. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell 126:611–625 [DOI] [PubMed] [Google Scholar]
- 32. Pierce S. E., Davis R. W., Nislow C., Giaever G. 2007. Genome-wide analysis of barcoded Saccharomyces cerevisiae gene-deletion mutants in pooled cultures. Nat. Protoc. 2:2958–2974 [DOI] [PubMed] [Google Scholar]
- 33. Quiles J. L., et al. 2002. Curcuma longa extract supplementation reduces oxidative stress and attenuates aortic fatty streak development in rabbits. Arterioscler. Thromb. Vasc. Biol. 22:1225–1231 [DOI] [PubMed] [Google Scholar]
- 34. Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 [DOI] [PubMed] [Google Scholar]
- 35. Shureiqi I., Baron J. A. 2011. Curcumin chemoprevention: the long road to clinical translation. Cancer Prev. Res. 4:296–298 [DOI] [PubMed] [Google Scholar]
- 36. Sidhu G. S., et al. 1998. Enhancement of wound healing by curcumin in animals. Wound Repair Regen. 6:167–177 [DOI] [PubMed] [Google Scholar]
- 37. Szczypka M. S., Zhu Z., Silar P., Thiele D. J. 1997. Saccharomyces cerevisiae mutants altered in vacuole function are defective in copper detoxification and iron-responsive gene transcription. Yeast 13:1423–1435 [DOI] [PubMed] [Google Scholar]
- 38. Thangapazham R. L., Sharma A., Maheshwari R. K. 2006. Multiple molecular targets in cancer chemoprevention by curcumin. AAPS J. 8:E443–E449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Winzeler E. A., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901–906 [DOI] [PubMed] [Google Scholar]
- 40. Yamaguchi-Iwai Y., Dancis A., Klausner R. D. 1995. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 14:1231–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yamaguchi-Iwai Y., Stearman R., Dancis A., Klausner R. D. 1996. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 15:3377–3384 [PMC free article] [PubMed] [Google Scholar]
- 42. Yuan D. S., et al. 1995. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc. Natl. Acad. Sci. U. S. A. 92:2632–2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yun C. W., Bauler M., Moore R. E., Klebba P. E., Philpott C. C. 2001. The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:10218–10223 [DOI] [PubMed] [Google Scholar]
- 44. Zbarsky V., et al. 2005. Neuroprotective properties of the natural phenolic antioxidants curcumin and naringenin but not quercetin and fisetin in a 6-OHDA model of Parkinson's disease. Free Radic. Res. 39:1119–1125 [DOI] [PubMed] [Google Scholar]
- 45. Zhang C., Browne A., Child D., Tanzi R. E. 2010. Curcumin decreases amyloid-beta peptide levels by attenuating the maturation of amyloid-beta precursor protein. J. Biol. Chem. 285:28472–28480 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.