Abstract
Calcineurin is a calcium-calmodulin-activated serine/threonine-specific phosphatase that operates during cellular responses to stress and plays a prominent role in transcriptional control, whereas regulatory events beyond transcription are less well characterized. This study reveals a novel transcription-independent role of calcineurin during the temperature stress response in the human fungal pathogen Cryptococcus neoformans. The diffusely cytoplasmic calcineurin catalytic subunit Cna1 relocates to endoplasmic reticulum (ER)-associated puncta and the mother-bud neck when cells are subjected to 37°C. More than 50% of Cna1 puncta contain the P-body constituent decapping enzyme Dcp1 and the stress granule constituent poly(A)-binding protein Pub1. These results support a model in which calcineurin orchestrates thermal stress responses by associating with sites of mRNA processing.
INTRODUCTION
Calcineurin is a calcium-calmodulin-dependent serine/threonine-specific phosphatase consisting of a catalytic A subunit and a regulatory B subunit, both of which are essential for enzyme activity (2, 10). A well-established calcineurin response involves activation of transcription of a number of genes whose products promote cell proliferation, enable the cell to cope with environmental stress, and maintain calcium homeostasis. In multicellular eukaryotes, known substrates of calcineurin are members of the nuclear factor of the activated T cell (NFAT) family of transcription factors (9). Calcineurin is the target of the immunosuppressive drug cyclosporine (CsA) and FK506, which inhibit calcineurin and T cell activation.
The activity of calcineurin has been shown to be directed to specific cellular domains, such as calcium channels or the plasma membrane in mammals and the septa in fungi, where it dephosphorylates proteins that directly influence cell function, suggesting that posttranscriptional regulation mediated by calcineurin may be common (7, 17, 25, 33). Despite a growing list of calcineurin substrates, the mechanisms underlying calcineurin posttranscriptional functions are less well understood.
Signaling components of the calcineurin pathway are well conserved among fungi (39). In Saccharomyces cerevisiae calcineurin responds to high cation concentrations, cell wall stress, and prolonged exposure to mating pheromone by dephosphorylating the transcription factor Crz1 and triggering its nuclear translocation (11). Calcineurin is essential for the virulence of pathogenic fungi, including Cryptococcus neoformans (32, 36). Calcineurin is essential for growth of C. neoformans at temperatures above 35°C and in the presence of increased levels of CO2, alkaline pH, and high concentrations of cations. The substrates involved in calcinerin-mediated stress responses in Cryptococcus are largely unknown. A conserved member of the calcipressin family, Cbp1/Rcn1, has been shown to interact with calcineurin and is involved in mating in C. neoformans (19), but it plays no role in high-temperature growth. Isolation of multicopy suppressors of calcineurin mutants revealed CTS1, which encodes a leucine zipper-containing phospholipid-binding protein (13). Strains lacking CTS1 are temperature sensitive but are also hypersensitive to FK506, indicating that calcineurin and Cts1 function in parallel pathways. Importantly, no clear functional homologue of the CRZ1 gene, encoding the S. cerevisiae calcineurin-activated transcription effector, has been identified in C. neoformans, suggesting that the effects of calcineurin in C. neoformans could be mediated via different transcription factors or may be partly or entirely posttranscriptional.
Here we report for the first time that when cells are exposed to thermal stress, the calcineurin A catalytic subunit exhibits a dramatic change in localization from diffusely cytoplasmic to endoplasmic reticulum (ER)-associated puncta and the mother-bud neck. Calcineurin A colocalizes with the P-body (PB) constituent Dcp1 and a stress granule (SG) constituent, Pub1. These observations suggest that calcineurin operates during high-temperature stress by colocalizing with sites of mRNA processing.
MATERIALS AND METHODS
Strains, media, and growth conditions.
C. neoformans strains and plasmids used in this study are listed in Table 1. All media were prepared as described previously (1, 20).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Reference |
|---|---|---|
| Strains | ||
| KN99 | a | 31 |
| KK8 | acna1::NEO | 29 |
| LK214 | acna1::NEO GFP-CNA1:NAT | This study |
| LK248 | α GFP-CNA1:NATmCH-CBP1:NEO | This study |
| LK254 | α cbp1::NAT mCH-CBP1:NEO | This study |
| LK282 | α GFP-HEM15 mCH-CNA1:NEO | This study |
| LK289 | α GFP-DCP1:NAT mCH-CNA1:NEO | This study |
| LK293 | acnb1::NAT mCH-CNA1:HYG | This study |
| LK294 | acbp1::NAT mCH-CNA1:NEO | This study |
| LK296 | α DsRed-SKL GFP-CNA1:NAT | This study |
| LK300 | acdc12::NEO GFP-CNA1:NAT | This study |
| LK303 | acna1::NEO GFP-CNA1:NAT mCH-PUB1 | This study |
| Plasmids | ||
| pLKB39 | GFP-CNA1:NAT | This study |
| pLKB54 | mCH-CBP1:NEO | This study |
| pLKB60 | mCH-CNA1:NEO | This study |
| pLKB61 | mCH-CNA1:HYG | This study |
| pLKB88 | mCH-PUB1:HYG | This study |
| pXW11 | GFP-DCP1:NAT | 37 |
All strains expressing N-terminally tagged fluorescent proteins were made by ectopic integration of a plasmid encoding a fluorescent chimera expressed from the histone promoter (green fluorescent protein [GFP]) or the GPD1 promoter (mCherry). Integrations were performed by biolistic transformation (12), and positive clones were screened based on drug resistance and a fluorescent signal. Strain LK214 was generated by introducing a plasmid encoding GFP-Cna1 (pLKB39) into a cna1Δ strain (KK8) (29). The resulting strain, LK214, was used to construct LK303 by integrating plasmid pLKB88, which encodes mCherry-Pub1. A strain expressing GFP-Hem15 and mCherry-Cna1 (LK282) was generated by integrating plasmid pLKB60 into strain AI132 (24). A strain expressing DsRed-SKL and GFP-Cna1 (LK296) was made by integrating plasmid pLKB39 into strain AI100 (23). Strain LK300 was constructed by integrating pLKB39 into a cdc12Δ strain (LK174) (30). Similarly pLKB60 was integrated into a cbp1Δ strain (K. Kojima, unpublished data) to generate LK294, and pLKB61 was integrated into a cnb1Δ strain (29) to generate LK293. Strain LK289, expressing GFP-Dcp1 and mCherry-Cna1, was constructed by integrating pXW11 (37) and pLKB60 into strain H99. Strain LK254 was generated by integrating pLKB54 (mCherry-Cbp1) into a cbp1Δ strain (K. Kojima, unpublished data). Strain LK248 was generated by integrating pLKB39 and pLKB54 into strain H99.
Generation of fluorescent protein chimeras.
All plasmid constructs contained between 300 and 500 bp of the 3′ untranslated region (UTR) of the respective gene. To construct GFP chimeras expressed from a histone H3 promoter, plasmid pCN19 (kindly provided by Connie Nichols and Andrew Alspaugh, Duke University) was used, and the PCR products of the respective genes were cleaved with BamHI. To isolate mCherry chimeras, plasmid pLKB49 was first produced by introducing a STOP-less sequence encoding mCherry cleaved with PacI/FseI into plasmid pXL1 cleaved with the same restriction enzymes (38). The resulting plasmid, pLKB49, contains the neomycin resistance gene and can be used to express mCherry-tagged chimeras from a constitutive GPD1 promoter. The PCR product of the respective gene was cleaved with NheI/PacI and cloned into pLKB49 cleaved with the same enzymes. An analogous plasmid, pLKB55, was constructed with the hygromycin resistance marker (antibiotic resistance genes were swapped using BamHI/EcoRV sites). To amplify the CNA1 open reading frame (ORF) (CNAG_ 04796) to generate pLKB39, primers JOHE20523 (CAAGGATCCGCTATGGCTTCCCCAGCCACTCAG) and JOHE20524 (CTAGGATCCTGACCTTCTTCGAAGACTTGC) were used. To generate pLKB60 and pLKB61, primers JOHE23842 (GCAGCTAGCTGCTTCCCCAGCCACTCAG) and JOHE23685 (GACTTAATTAAAGCGACCAATGGAGTGTGACG) were used. To amplify the PUB1 ORF (CNAG_ 04441) to generate pLKB88, primers JOHE26122 (GCAGCTAGCTTCCGTCGAAACCGCTACATC) and JOHE26123 (GACTTAATTAAAGTAGTATGGATTGAGACCTC) were used. To amplify the CBP1 ORF (CNAG_00802) to generate pLKB54, primers JOHE23852 (GCAGCTAGCTACCATAAGCTCACCGCAAAG) and JOHE23853 (GACTTAATTAACGTCCTCATGCTTGTGGCTAC) were used.
Microscopy.
For imaging yeast cells, ∼0.5 μl of cell suspension was placed on a slide containing a thin complete growth medium 2% agar patch and covered with a coverslip. Bright-field, differential interference contrast (DIC), and fluorescence microscopy images were captured with either a Zeiss Axioskop 2 Plus fluorescence microscope (Carl Zeiss, Thornwood, NY) equipped with an AxioCam MRm digital camera, a Zeiss Axioscope equipped with an Orca cooled charge-coupled device (CCD) camera (Hamamatsu, Bridgewater, NJ) and interfaced with MetaMorph software (Universal Imaging, Silver Spring, MD), or a Zeiss Axio Observer Z1 equipped with Pecon XL S1 incubator chamber and QuantEM back-thinned EM-CCD camera.
To perform Z-section analysis and three-dimensional (3D) reconstruction, the Deltavision system (Olympus IX-71 base), equipped with a Coolsnap HQ2 high-resolution CCD camera, was used. Images were processed using Photoshop (Adobe Systems, San Jose, CA).
To visualize nuclei, cells were briefly fixed with ice-cold 4% paraformaldehyde (phosphate-buffered saline [PBS] solution) for 5 min, washed with PBS, stained with Hoechst 33258 (Invitrogen) for 30 min, and washed with PBS prior to imaging.
Other methods.
For analysis of cell viability, 10-fold serial dilutions from overnight liquid cultures were performed and 2 μl of each dilution was spotted on an appropriate medium as indicated in the figure legends. The largest spotted amount of cells was 104 cells. SDS-PAGE and immunoblotting were performed as described previously (30). To detect GFP-Cna1, an anti-GFP polyclonal antibody (Santa Cruz Biotech, Santa Cruz, CA) was used at a 1:1,000 dilution. The PSTAIR antibody (Abcam, Cambridge, MA) was used at a 1:2,000 dilution as a loading control.
The statistical analysis was performed using a Poisson regression model. A chi-square test was used for the analysis of deviance. All tests of hypotheses were at a 5% significance level.
RESULTS
Calcineurin localizes to cytoplasmic puncta and the bud neck at 37°C.
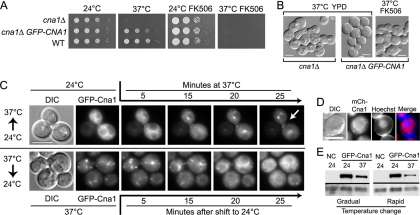
Calcineurin promotes the growth of C. neoformans at 37°C by unknown mechanisms. We hypothesized that calcineurin elicits this function at a posttranscriptional level by accessing its targets/substrates in specific cellular domains. To test this hypothesis, we examined the localization of the calcineurin catalytic subunit. Cna1 was tagged with GFP or the dsRed derivative mCherry at the N terminus and expressed from a constitutive promoter. Expression of the GFP-Cna1 fusion protein restored growth of the cna1Δ mutant at 37°C (Fig. 1 A). Calcineurin mutant cells grown at 37°C became elongated and exhibited a multivesicular phenotype. GFP-Cna1 restored wild-type cellular morphology at 37°C (Fig. 1B). Moreover, a strain expressing only GFP-Cna1 in place of wild-type Cna1 was sensitive to the immunosuppressive drug FK506 at 37°C, indicating that GFP-Cna1 is subject to inhibition similar to the wild type (Fig. 1A and B).
Fig. 1.
Calcineurin catalytic subunit Cna1 localizes to cytoplasmic puncta and the mother-bud neck at 37°C. (A and B) GFP-Cna1 restores growth of a cna1Δ mutant at 37°C, and the GFP-Cna1-expressing cells are sensitive to FK506 (1 μg/ml) at 37°C. (A) A spot dilution assay with three congenic wild-type (WT), cna1Δ, and cna1Δ GFP-CNA1 strains was performed as indicated. (B) Expression of GFP-Cna1 in a cna1Δ mutant restores wild-type cell morphology at 37°C. (C) GFP-Cna1-expressing cells were initially incubated 24°C and put on a microscope slide preheated to 37°C. The slide was examined immediately under a microscope equipped with an environmental chamber set to 37°C (upper panels). After 45 min of incubation, the temperature was shifted back to 24°C, and the cells were imaged again at 5-min intervals (lower panels). An arrow indicates localization to the mother-bud neck. (D) mCherry-Cna1 forms puncta that surround the nucleus. Cells expressing mCherry-Cna1 were fixed and stained with the Hoechst nuclear dye. (E) Western blot analysis of cell lysates from cells expressing GFP-Cna1 and a control strain (NC) incubated at 24°C and 37°C for 30 min. Equal amounts of total protein were loaded. The temperature was changed either gradually (left gel) or rapidly (right gel). Membranes were probed with an antibody against GFP (top) and a PSTAIR antibody as a control (bottom). The scale bars in panels B, C, and D represent 5 μm.
The GFP-Cna1-expressing strain was initially incubated at 24°C, and cells were placed on a slide (preheated to 37°C) and analyzed using a microscope equipped with an environmental chamber that was set to 37°C. This allowed for a relatively rapid temperature change. GFP-Cna1 was mostly cytoplasmic and largely excluded from the nucleus and the vacuole in cells imaged initially at time zero (Fig. 1C). Strikingly, after exposure of the cells to 37°C, the GFP-Cna1 localization was altered and a significant proportion was now observed in cytoplasmic puncta and at the mother-bud neck. This change was rapid, as puncta containing GFP-Cna1 and the accumulation at the bud neck were visible within 15 min of the temperature shift. This new localization of Cna1 persisted at 37°C. After 30 min at 37°C all of the cells showed punctate localization of GFP-Cna1, and ∼70% of all budded cells, irrespective of the size of the bud, contained GFP-Cna1 at the bud neck (208 cells were counted). Most of the GFP-Cna1 puncta were localized in a region surrounding the nucleus as evidenced by staining mCherry-Cna1-expressing cells with the nucleus-specific Hoechst stain (Fig. 1D). When the temperature was shifted back to 24°C after 45 min of incubation at 37°C, within ∼30 min the cytoplasmic puncta dispersed and GFP-Cna1 was again pan-cytoplasmic (Fig. 1C). Western blot analysis of GFP-Cna1 indicates that both rapid and gradual temperature changes from 24 to 37°C lead to a significant decrease in the GFP-Cna1 protein abundance and that the effect seems to be more pronounced during rapid temperature change (Fig. 1E). This suggests that the fluorescent puncta at 37°C represent sites of GFP-Cna1 enrichment specific to conditions of thermal stress. Furthermore, we cannot exclude at this point that Cna1 is also associated with these sites at 24°C but that this is obscured by pan-cytoplasmic localization under these conditions.
To test how long GFP-Cna1 remains associated with the puncta at 37°C, the microscope slide containing the cells was initially incubated at 24°C for ∼1 h and then placed in the environmental chamber set to 37°C, and cells were imaged every 5 min for over 2 h (Fig. 2). Similar to the case in the previous experiment, in which the temperature change was more rapid, GFP-Cna1 became punctate at 37°C. In contrast to the case in the previous experiment, no visible bud neck localization of GFP-Cna1 was observed. Interestingly, after 45 min of exposure to 37°C, the GFP-Cna1 signal returned to being cytoplasmic. The growth of cells (judged based on the size of buds) did not occur for at least the initial 1 h during the incubation at 37°C, presumably due to cell cycle arrest in response to thermal stress. After 90 min at 37°C, growth was evident based on the appearance of new buds and an increase in bud sizes. Taken together, these data suggest that Cna1 localizes to ER-associated puncta when cells are exposed to thermal stress and remains at the puncta until cells adjust to the high temperature and growth resumes. Furthermore, whereas a rapid temperature change results in bud neck localization of GFP-Cna1, a more gradual change in temperature does not.
Fig. 2.
The localization of GFP-Cna1 to cytoplasmic puncta at 37°C is transient. GFP-Cna1-expressing cells were placed on a slide containing a thin complete growth medium 2% agar patch, covered with a coverslip, and initially incubated at 24°C. Subsequently, the slide was examined under a microscope equipped with an environmental chamber set to 37°C. The punctate signal of GFP-Cna1 was visible after 5 min at 37°C and persisted for no more than 45 min. Cells imaged at the 45-min time point show only cytoplasmic signal of GFP-Cna1. No bud growth was observed until the 90-min time point. The bar represents 5 μm.
To identify the minimal temperature increase that triggers these changes in Cna1 localization and to further test whether localization of Cna1 depends on the rapidity of the temperature change, cells were exposed to three different temperatures, 32°C, 34°C, and 37°C, for 30 min using a heating block to allow a very rapid temperature change (see Fig. S1 in the supplemental material). Incubation at 32°C for 30 min resulted in punctate localization of GFP-Cna1, although this was not as prominent as at 37°C (21% of cells exposed to 32°C for 30 min showed more than three puncta of GFP-Cna1, versus 96% of cells exposed to 37°C for 30 min). However, no bud neck localization of GFP-Cna1 was observed at 32°C. Samples incubated for 30 min at 34°C showed more pronounced punctate localization of GFP-Cna1 (44% of cells with more than three puncta), and again no bud neck localization was observed. Strikingly, samples that were sequentially exposed to a heating block that was set to these three temperatures for 30 min each (32°C followed by 34°C and finally 37°C) showed only 21% of cells with more than three puncta, and no bud neck localization was observed (see Fig. S1 in the supplemental material). These data suggest that an abrupt change in temperature to 37°C results in localization of Cna1 to the bud neck and to multiple puncta, whereas a gradual increase in temperature above 32°C triggers less pronounced changes in Cna1 localization and does not result in visible localization to the mother-bud neck. These results suggest that cells may adapt to the temperature change, with less reorganization of Cna1, when the change is gradual.
The localization of Cnb1 and Cbp1, known Cna1-associated proteins, was examined. The regulatory B subunit Cnb1 is essential for calcineurin activity. In order to examine the localization of Cnb1 during thermal stress, an mCherry-Cnb1 chimera protein was generated. mCherry-Cnb1 remained cytoplasmic during a temperature shift to 37°C (data not shown). However, mCherry-Cnb1 did not restore growth of a cnb1Δ mutant strain at 37°C, indicating that the mCherry-Cnb1 chimera protein is not functional, possibly due to a lack of interaction with Cna1 (data not shown).
Calcipressins interact with calcineurin and modulate its activity and physiological functions in yeast and mammals (15, 22, 34). In C. neoformans, the calcipressin Cbp1 functions as a specificity modulator or targeting subunit of calcineurin-dependent hyphal elongation during mating, and unlike calcineurin, it is dispensable for growth at 37°C (16). An mCherry-Cpb1 chimera protein restored bilateral mating of a cbp1Δ strain, indicating that it is functional (data not shown). The localization of mCherry-Cbp1 was examined during a temperature change to 37°C. At 24°C, mCherry-Cbp1 was cytoplasmic. While GFP-Cna1 became punctate after cells were shifted to 37°C, mCherry-Cbp1 remained cytoplasmic (see Fig. S2 in the supplemental material). This result demonstrates that Cbp1 does not colocalize with Cna1 in cytoplasmic puncta during thermal stress.
Localization of Cna1 at 37°C does not depend on calcineurin activity, the regulatory B subunit, or calcipressin.
To test whether changes in calcineurin localization depend upon its activity, cells expressing mCherry-Cna1 were treated with the calcineurin inhibitor FK506 at 1 μg/ml. FK506-treated and control cells showed similar changes in localization of mCherry-Cna1 upon a shift to 37°C (Fig. 3A), indicating that the catalytic activity of Cna1 is not required for its relocalization. A requirement for the regulatory Cnb1 subunit of calcineurin (14) was tested in a cnb1Δ mutant expressing mCherry-Cna1. Similarly, mCherry-Cna1 was expressed in a strain deleted for the CBP1 gene, encoding calcipressin (19). We hypothesized that Cnb1 may be required for the localization of Cna1 at 37°C, while Cbp1 may be dispensable. However, neither Cnb1 nor Cbp1 was required for the change in localization of mCherry-Cna1 (Fig. 3B). These data demonstrate that the calcineurin catalytic subunit Cna1 associates with specific regions within the cytoplasm during growth at 37°C in a manner that is not dependent on its catalytic activity, its regulatory subunit Cnb1, or the calcipressin Cbp1.
Fig. 3.
Localization of Cna1 to the cytoplasmic puncta and the mother-bud neck does not depend on calcineurin catalytic activity, Cnb1, Cbp1, or septins. (A) Cells expressing GFP-Dcp1 and mCherry-Cna1 were treated with 1 μg/ml FK506 or dimethyl sulfoxide (DMSO) vehicle at 24°C in synthetic complete liquid medium for 2 h. Subsequently, cells were mounted on an agarose patch-containing slide and placed on a heating block preset to 37°C. Samples were incubated for 30 min prior to microscopy. (B) cnb1Δ, cbp1Δ, and cdc12Δ mutant strains expressing fluorescently tagged Cna1 were grown and analyzed as for panel A except that no FK506 treatment was performed. Arrows indicate localization of Cna1 to the mother-bud neck. The scale bars represent 5 μm.
Localization of Cna1 to the mother-bud neck does not depend on the septins.
In S. cerevisiae, the septins form a complex at the mother-bud neck that serves as a scaffold for a number of signaling proteins (18). To test if the localization of Cna1 to the mother-bud neck at 37°C depends on the septins, GFP-Cna1 was expressed in a strain in which CDC12, encoding one of the major components of the septin complex, was deleted. In the C. neoformans cdc12Δ mutant, other septins do not localize to the bud neck, and yet this mutant is viable at 24°C (30). GFP-Cna1 localization at 37°C in the cdc12Δ mutant strain was indistinguishable from that in the wild-type cells (Fig. 3B), providing evidence that Cna1 localizes to the bud neck in a septin-independent manner.
Cna1 colocalizes with PBs and SGs during thermal stress.
We next investigated the identity of the puncta to which GFP-Cna1 relocates during thermal stress. We excluded the possibility that the Cna1 puncta represent mitochondria or peroxisomes, which were previously shown to exhibit a punctate appearance in Cryptococcus. Cells coexpressing mCherry-Cna1 with GFP-Hem15 as a mitochondrial marker (24) or with GFP-Cna1 with DsRed-SKL as a peroxisomal marker (23) exhibited little or no colocalization of calcineurin with these punctate intracellular organelles at 37°C (see Fig. S3 in the supplemental material).
Given that the calcineurin-enriched puncta are localized near the nucleus, it is possible that they may be associated with the ER. Recent findings indicate that during stress there is an increase in the mRNA-processing bodies, called P-bodies (PBs), that form ER-associated foci (27, 28). In addition, a variety of stresses induce the formation of stress granules (SGs), where mRNAs are sequestered to prevent their translation until the stress is alleviated (6). The decapping protein Dcp1, a P-body component, was tagged with GFP, and the stress granule protein Pub1, a poly(A)-binding protein, was fused to mCherry. Strains were generated in which mCherry-Cna1 is coexpressed with GFP-Dcp1 or GFP-Cna1 is coexpressed with mCherry-Pub1. At 24°C, GFP-Dcp1 formed a few foci per cell (Fig. 4A and 5A); in contrast, mCherry-Pub1 did not form foci at 24°C, and it was cytoplasmic and slightly enriched in the nucleus (Fig. 4B). Upon a rapid temperature shift to 37°C, the number of GFP-Dcp1 foci increased (P < 0.001), and mCherry-Pub1 formed cytoplasmic puncta (Fig. 4 and 5). Strikingly, fluorescently tagged Cna1 colocalized with both GFP-Dcp1 and mCherry-Pub1 in cytoplasmic foci and at the mother-bud neck at 37°C (Fig. 4). While most of the GFP-Dcp1 puncta were colocalized with mCherry-Cna1, the distribution of mCherry-Cna1 was broader, and a significant proportion of the mCherry-Cna1 fluorescent signal did not show colocalization with GFP-Dcp1 (Fig. 4A and 5A). Similarly, a significant proportion of GFP-Cna1 did not colocalize with mCherry-Pub1 (Fig. 4B and 5B).
Fig. 4.
(A and B) Cna1 colocalizes with the P-body component Dcp1 (A) and with the stress granule constituent Pub1 (B) during a rapid temperature shift to 37°C. Cells were initially grown at 24°C. Subsequently, the slide containing the cells was placed on a heating block preset to 37°C and the cells were imaged after 30 min of incubation at 37°C. An arrow indicates localization to the mother-bud neck. (C) mCherry-Cna1 localizes to elaborate membrane-like structures and puncta, whereas GFP-Dcp1 forms more defined foci where it colocalizes with mCherry-Cna1. Z-section microscopy analysis was performed using the DeltaVision system. 3D views of cells shown in the DIC image were constructed based on Z-section images taken with two different channels to visualize mCherry-Cna1 and GFP-Dcp1. A pseudocolor merged image shows regions of colocalization. The scale bars represent 5 μm.
Fig. 5.
Quantification of the numbers of Dcp1, Pub1, and Cna1 foci. (A) Cells expressing mCherry-Cna1 and GFP-Dcp1 were grown either at 24°C or at 37°C for 30 min. Cultures were treated with 1 μg/ml FK506 or with DMSO vehicle (control). For each drug treatment, the numbers of fluorescent mCherry-Cna1 and GFP-Dcp1 foci in each cell from the same cell population were counted. A minimum of 100 cells were counted for each condition. The size of the box is determined by the 25th and 75th percentiles, the whiskers represent the 5th and 95th percentiles, and the vertical line and the diamond show the median and the mean, respectively. At 24°C, mCherry-Cna1 did not form foci. (B) A quantification analogous to that in panel A was performed for cells expressing GFP-Cna1 and mCherry-Pub1 grown at 37°C for 30 min with FK506 (n = 90) or with DMSO vehicle (n = 60). At 24°C, neither GFP-Cna1 nor mCherry-Pub1 formed foci.
To further analyze the colocalization between mCherry-Cna1 and GFP-Dcp1, a DeltaVision microscope and postimaging 3D reconstruction software were employed. Samples were imaged in 25 Z-sections spanning a distance of ∼5 μm to cover most of the cell body. The 3D reconstruction confirmed colocalization between mCherry-Cna1 and GFP-Dcp1 (Fig. 4C). This analysis also showed that whereas mCherry-Cna1 formed a more elaborate net of membrane-like signals, GFP-Dcp1 was mostly visible in more defined foci, where it colocalized with mCherry-Cna1. One possible interpretation is that during thermal stress Cna1 associates with general ER-derived membranes and forms foci of higher intensity that also contain Dcp1.
To test whether the localization of Dcp1 and Pub1 depended on calcineurin catalytic activity, cells expressing either GFP-Dcp1 or mCherry-Pub1 were examined after treatment with the calcineurin inhibitor FK506. At 24°C, a statistically significant (P = 0.0003) modest increase of GFP-Dcp1 foci was observed when cells were treated with FK506 at a concentration sufficient to fully inhibit calcineurin based on previous studies (32) (Fig. 5A). No significant difference was detected in the number of GFP-Dcp1 or mCherry-Pub1 foci per cell in cells treated with FK506 compared to control cells at 37°C (Fig. 5).
To test if the relocalization of Cna1 and colocalization with Dcp1 were specific to the temperature stress or whether these features were a general response to other stresses, localization of mCherry-Cna1 and GFP-Dcp1 in cells exposed to other stresses, including 1 M LiCl, 1.25 M KCl, and 2 M sorbitol, was tested. All three conditions resulted in punctate localization of mCherry-Cna1 and partial colocalization with GFP-Dcp1 (Fig. 6). Thus, association of Cna1 with P-bodies is a common response to diverse forms of cellular stress.
Fig. 6.
mCherry-Cna1 colocalizes with the P-body marker GFP-Dcp1 during exposure to a variety of stresses. Cells expressing mCherry-Cna1 and GFP-Dcp1 were initially grown at 24°C in complete synthetic medium. Subsequently, the medium was replaced with medium supplemented with KCl, LiCl, or sorbitol as indicated and cells were incubated at 24°C for 10 min prior to microscopic analysis. Arrows in the merged images indicate cytoplasmic puncta where mCherry-Cna1 and GFP-Dcp1 colocalize. The scale bars represent 5 μm.
DISCUSSION
Cells respond to stress by adjusting the abundance and activity of proteins through the regulation of transcription, translation, or degradation. Stress results in the attenuation of general protein synthesis, and translation of some mRNAs is maintained or elevated. Calcineurin has been implicated in the eukaryotic stress response mainly as a regulator of the transcriptional program. However, several substrates of calcineurin have been identified, indicating that calcineurin also regulates multiple processes in a transcription-independent manner.
In C. neoformans, calcineurin is essential for growth at elevated temperature, but no transcription factor that is known to be regulated by calcineurin has been identified in this pathogen. Although it is plausible that calcineurin regulates transcription in response to stress in Cryptococcus, our findings suggest that it functions in a transcription-independent manner.
We show here that the catalytic subunit of calcineurin (Cna1) accumulates at multiple foci surrounding the nucleus in response to temperature elevation. This perinuclear localization and colocalization with Dcp1, a component of P-bodies previously shown to associate with the ER in S. cerevisiae (28), suggest that Cna1 foci may be associated with the ER. The Cna1 puncta may be new localization sites for Cna1 or sites of enrichment. Indeed, even at 24°C a slightly brighter signal of GFP-Cna1 is observed in areas surrounding the nucleus, consistent with ER, than in the cytoplasm. A rapid temperature raise causes a significant decrease in Cna1 protein levels. Thus, we cannot exclude the possibility that Cna1 is always associated with the punctate sites observed at 37°C but at relatively lower levels and that stress results in its enrichment at puncta and protein degradation at remaining locations.
One mode of gene expression regulation in eukaryotes is mRNA compartmentalization and processing. P-bodies (PBs) are cytoplasmic sites where untranslated mRNAs and translational repressors, including the decapping enzyme Dcp1, accumulate (5). Our recent findings show that RNAi machinery also associates with PBs in Cryptococcus (37). In response to stress, the number of PBs increases as cellular translation is inhibited. Stress results in the formation of a second class of RNA aggregates called stress granules (SGs), which are “triage” sites for mRNA resulting from translation inhibition (6). In this study, we show that the number of PBs significantly increases when cryptococcal cells are exposed to temperature stress and this is accompanied by the redistribution of Cna1 from the cytoplasm to puncta that colocalize with PBs and SGs based on colocalization with two markers (Dcp1 and Pub1). This suggests that Cna1 associates with PBs and SGs following a temperature shift. We propose that in C. neoformans undergoing thermal stress, PBs, SGs, and Cna1 accumulate at the ER. A detailed comparison of the localizations of Cna1 and Dcp1 suggests that whereas Cna1 associates with ER membranes, PBs are present only at specific sites within the ER, and these locations are also enriched with Cna1. Association of the calcineurin catalytic subunit with the ER has not been directly described, but previous studies with S. cerevisiae and higher eukaryotes are consistent with this possibility. Hph1, a tail-anchored integral membrane protein that localizes to the ER and is involved in the cellular response to stress, is a known substrate of calcineurin (21).
Recently, Kilchert et al. described that defects in the secretory pathway and high calcium concentrations induce the formation of multiple PBs in S. cerevisiae. Induction of PBs in response to elevated Ca2+ required calmodulin, but calcineurin activity was not required (28). Interestingly, Kilchert et al. showed that calmodulin does not localize to PBs upon stress. It will be of interest to examine the localization of calmodulin in Cryptococcus and calcineurin in S. cerevisiae to test if the roles of calcineurin and calmodulin are conserved or diverged between these two fungal species. Future studies will further reveal the relationship between calcineurin, PBs, and SGs in Cryptococcus.
One possible role of calcineurin at the cytoplasmic puncta at 37°C is regulating protein synthesis. In multicellular eukaryotes, calcineurin may either promote or inhibit translation, depending upon the cell cycle stage, tissue type, or protein identity (3, 8, 26, 35). A recent study in mammalian and Xenopus cells by Bollo et al. shows that calcineurin associates with the ER-resident kinase PERK and promotes its autophosphorylation, leading to an increase in phosphorylation of eIF2 alpha and the attenuation of protein synthesis in response to ER stress (4). Thus, it seems likely that a role for calcineurin in protein synthesis may be conserved in C. neoformans.
In summary, this study for the first time establishes stress-related changes in localization of calcineurin from the cytoplasm to P-bodies and stress granules associated with the ER. We hypothesize that the change in localization of calcineurin contributes to the increased survival of cells during thermal stress, although this requires further study. The sites where calcineurin localizes during thermal stress may also contain a number of stress-related proteins participating in a variety of stress-regulated responses. Further studies should explore this intriguing possibility.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joanne Kingsbury and Cecelia Shertz for critical reading of the manuscript. We thank Anna Panorska (University of Nevada, Reno) for help with the statistical analysis.
These studies were supported by NIH/NIAID R01 grants AI42159 and AI50438 and NCI R01 grant CA154499.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Alspaugh J. A., Perfect J. R., Heitman J. 1997. Cryptococcus neoformans mating and virulence are regulated by the G-protein alpha subunit GPA1 and cAMP. Genes Dev. 11:3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aramburu J., Heitman J., Crabtree G. R. 2004. Calcineurin: a central controller of signalling in eukaryotes. EMBO Rep. 5:343–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Biswas A., et al. 2011. Opposing action of casein kinase 1 and calcineurin in nucleo-cytoplasmic shuttling of mammalian translation initiation factor eIF6. J. Biol. Chem. 286:3129–3138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bollo M., et al. 2010. Calcineurin interacts with PERK and dephosphorylates calnexin to relieve ER stress in mammals and frogs. PLoS One 5:e11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchan J. R., Nissan T., Parker R. 2010. Analyzing P-bodies and stress granules in Saccharomyces cerevisiae. Methods Enzymol. 470:619–640 [DOI] [PubMed] [Google Scholar]
- 6. Buchan J. R., Parker R. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36:932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cameron A. M., et al. 1995. Calcineurin associated with the inositol 1,4,5-trisphosphate receptor-FKBP12 complex modulates Ca2+ flux. Cell 83:463–472 [DOI] [PubMed] [Google Scholar]
- 8. Cao Q., Kim J. H., Richter J. D. 2006. CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat. Struct. Mol. Biol. 13:1128–1134 [DOI] [PubMed] [Google Scholar]
- 9. Crabtree G. R., Olson E. N. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109(Suppl.):S67–S79 [DOI] [PubMed] [Google Scholar]
- 10. Crabtree G. R., Schreiber S. L. 2009. Ca2+-calcineurin-NFAT signaling. Cell 138:210, 210 e211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cyert M. S. 2003. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311:1143–1150 [DOI] [PubMed] [Google Scholar]
- 12. Davidson R. C., et al. 2000. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet. Biol. 29:38–48 [DOI] [PubMed] [Google Scholar]
- 13. Fox D. S., Cox G. M., Heitman J. 2003. Phospholipid-binding protein Cts1 controls septation and functions coordinately with calcineurin in Cryptococcus neoformans. Eukaryot. Cell 2:1025–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fox D. S., et al. 2001. Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol. Microbiol. 39:835–849 [DOI] [PubMed] [Google Scholar]
- 15. Fox D. S., Heitman J. 2002. Good fungi gone bad: the corruption of calcineurin. Bioessays 24:894–903 [DOI] [PubMed] [Google Scholar]
- 16. Fox D. S., Heitman J. 2005. Calcineurin-binding protein Cbp1 directs the specificity of calcineurin-dependent hyphal elongation during mating in Cryptococcus neoformans. Eukaryot. Cell 4:1526–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Frey N., Richardson J. A., Olson E. N. 2000. Calsarcins, a novel family of sarcomeric calcineurin-binding proteins. Proc. Natl. Acad. Sci. U. S. A. 97:14632–14637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gladfelter A. S., Pringle J. R., Lew D. J. 2001. The septin cortex at the yeast mother-bud neck. Curr. Opin. Microbiol. 4:681–689 [DOI] [PubMed] [Google Scholar]
- 19. Gorlach J., et al. 2000. Identification and characterization of a highly conserved calcineurin binding protein, CBP1/calcipressin, in Cryptococcus neoformans. EMBO J. 19:3618–3629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Granger D. L., Perfect J. R., Durack D. T. 1985. Virulence of Cryptococcus neoformans. Regulation of capsule synthesis by carbon dioxide. J. Clin. Invest. 76:508–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heath V. L., Shaw S. L., Roy S., Cyert M. S. 2004. Hph1p and Hph2p, novel components of calcineurin-mediated stress responses in Saccharomyces cerevisiae. Eukaryot. Cell 3:695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hilioti Z., Cunningham K. W. 2003. The RCN family of calcineurin regulators. Biochem. Biophys. Res. Commun. 311:1089–1093 [DOI] [PubMed] [Google Scholar]
- 23. Idnurm A., Giles S. S., Perfect J. R., Heitman J. 2007. Peroxisome function regulates growth on glucose in the basidiomycete fungus Cryptococcus neoformans. Eukaryot. Cell 6:60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Idnurm A., Heitman J. 2010. Ferrochelatase is a conserved downstream target of the blue light-sensing White collar complex in fungi. Microbiology 156:2393–2407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Juvvadi P. R., et al. 2008. Calcineurin localizes to the hyphal septum in Aspergillus fumigatus: implications for septum formation and conidiophore development. Eukaryot. Cell 7:1606–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kahl C. R., Means A. R. 2004. Calcineurin regulates cyclin D1 accumulation in growth-stimulated fibroblasts. Mol. Biol. Cell 15:1833–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kedersha N., et al. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kilchert C., Weidner J., Prescianotto-Baschong C., Spang A. 2010. Defects in the secretory pathway and high Ca2+ induce multiple P-bodies. Mol. Biol. Cell 21:2624–2638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kojima K., Bahn Y. S., Heitman J. 2006. Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology 152:591–604 [DOI] [PubMed] [Google Scholar]
- 30. Kozubowski L., Heitman J. 2009. Septins enforce morphogenetic events during sexual reproduction and contribute to virulence of Cryptococcus neoformans. Mol. Microbiol. 2010 75:658–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nielsen K., et al. 2003. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect. Immun. 71:4831–4841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Odom A., et al. 1997. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 16:2576–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oliveria S. F., Gomez L. L., Dell'Acqua M. L. 2003. Imaging kinase-AKAP79-phosphatase scaffold complexes at the plasma membrane in living cells using FRET microscopy. J. Cell Biol. 160:101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Parry R. V., June C. H. 2003. Calcium-independent calcineurin regulation. Nat. Immunol. 4:821–823 [DOI] [PubMed] [Google Scholar]
- 35. Sans M. D., Williams J. A. 2004. Calcineurin is required for translational control of protein synthesis in rat pancreatic acini. Am. J. Physiol. 287:C310–C319 [DOI] [PubMed] [Google Scholar]
- 36. Steinbach W. J., Reedy J. L., Cramer R. A., Jr., Perfect J. R., Heitman J. 2007. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat. Rev. Microbiol. 5:418–430 [DOI] [PubMed] [Google Scholar]
- 37. Wang X., et al. 2010. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 24:2566–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xue C., Bahn Y. S., Cox G. M., Heitman J. 2006. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans. Mol. Biol. Cell 17:667–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zelter A., Bencina M., Bowman B. J., Yarden O., Read N. D. 2004. A comparative genomic analysis of the calcium signaling machinery in Neurospora crassa, Magnaporthe grisea, and Saccharomyces cerevisiae. Fungal Genet. Biol. 41:827–841 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.