Abstract
Many Ras GTPases localize to membranes via C-terminal farnesylation and palmitoylation, and localization regulates function. In Candida albicans, a fungal pathogen of humans, Ras1 links environmental cues to morphogenesis. Here, we report the localization and membrane dynamics of Ras1, and we characterize the roles of conserved C-terminal cysteine residues, C287 and C288, which are predicted sites of palmitoylation and farnesylation, respectively. GFP-Ras1 is localized uniformly to plasma membranes in both yeast and hyphae, yet Ras1 plasma membrane mobility was reduced in hyphae compared to that in yeast. Ras1-C288S was mislocalized to the cytoplasm and could not support hyphal development. Ras1-C287S was present primarily on endomembranes, and strains expressing ras1-C287S were delayed or defective in hyphal induction depending on the medium used. Cells bearing constitutively activated Ras1-C287S or Ras1-C288S, due to a G13V substitution, showed increased filamentation, suggesting that lipid modifications are differentially important for Ras1 activation and effector interactions. The C. albicans autoregulatory molecule, farnesol, inhibits Ras1 signaling through adenylate cyclase and bears structural similarities to the farnesyl molecule that modifies Ras1. At lower concentrations of farnesol, hyphal growth was inhibited but Ras1 plasma membrane association was not altered; higher concentrations of farnesol led to mislocalization of Ras1 and another G protein, Rac1. Furthermore, farnesol inhibited hyphal growth mediated by cytosolic Ras1-C288SG13V, suggesting that farnesol does not act through mechanisms that depend on Ras1 farnesylation. Our findings imply that Ras1 is farnesylated and palmitoylated, and that the Ras1 stimulation of adenylate cyclase-dependent phenotypes can occur in the absence of these lipid modifications.
INTRODUCTION
Ras GTPases are highly conserved signaling proteins that play central roles in key physiological processes, such as growth, morphology, and survival in eukaryotes from yeast to humans and additionally regulate virulence in a variety of plant and human fungal pathogens (20, 44, 51, 55). In Candida albicans, one of the most prominent human fungal pathogens (56), Ras1 regulates a diverse array of phenotypes critical for both commensal and pathogenic lifestyles within the host (44, 72). Morphological plasticity, characterized by its ability to grow in filamentous or yeast forms in response to in vivo stimuli, is an important contributor to pathogenesis (67). Growth at 37°C in combination with chemical signals, such as serum and N-acetylglucosamine (GlcNAc) (10, 19), Hsp90 depletion (63), or growth within a matrix (17, 47), promotes Ras1 signaling through the cyclic AMP (cAMP) or Cek1 mitogen-activated protein (MAP) kinase signaling cascades (10, 17, 19, 44, 58), initiating the hyphal growth program. These pathways also regulate the expression of a number of cell wall adhesins and secreted proteases that mediate interactions between C. albicans and the host (30, 34, 49). Consequently, strains lacking functional Ras1 are unable to efficiently undergo filamentation and are attenuated in virulence (19, 44, 45, 72, 77).
Ras proteins cycle between inactive GDP-bound and active GTP-bound states and can functionally interact with effectors in the latter conformation. The intrinsic GTPase activity of Ras proteins is slow, thus the ratio of Ras-GTP to Ras-GDP is mediated by GTPase-activating proteins (GAPs) that facilitate GTP hydrolysis and guanine nucleotide exchange factors (GEFs) that catalyze the exchange of GDP to GTP (6). Cells lacking the Ras1-GAP, Ira2, phenocopy cells bearing Ras1 with a G13V mutation that stabilizes the GTP-bound conformation, rendering cells hyperfilamentous and more sensitive to heat stress than wild-type cells (10, 19, 44). The C. albicans genome contains a single gene encoding a Ras1 GEF, CDC25, and cells lacking Cdc25 are hypofilamentous (16, 63, 69). In the presence of serum, a functional interaction between Ras1 and adenylate cyclase (Cyr1) is required for the stimulation of cAMP synthesis (17) and the subsequent protein kinase A (PKA) induction of the yeast-to-hypha transition. Therefore, the initiation of hyphal growth in response to serum (44, 47), glucose (47), or muramyldipeptides, which are present in serum and directly activate Cyr1, also requires Ras1 (75).
Both mammalian and fungal Ras proteins are localized to the plasma membrane via posttranslational modifications at the C terminus (14, 29, 51, 54). In the budding yeast Saccharomyces cerevisiae, the cysteine in the highly conserved CAAX box (C, cysteine; A, aliphatic amino acid; X, any amino acid) is the site of farnesylation (22), and this modification promotes Ras association with the endoplasmic reticulum (ER) (3). Farnesylation is followed by the cleavage of the three C-terminal residues (7, 18) and the subsequent carboxymethylation of the farnesyl-cysteine (12). In both S. cerevisiae and Schizosaccharomyces pombe, the palmitoylation of a single cysteine residue adjacent to the farnesyl-cysteine enables efficient trafficking from endomembranes to the plasma membrane (14, 21, 54). Cryptococcus neoformans Ras1 has two adjacent cysteines, and the palmitoylation of either residue is sufficient for some plasma membrane localization (51).
Ras signaling in diverse organisms can be propagated from different cellular locations, such as the plasma membrane, endosomes, Golgi apparatus, endoplasmic reticulum, or mitochondria (28, 50), yielding distinct biological outputs (2, 9, 14, 51, 54). For example, in C. neoformans, Ras1 signaling from the plasma membrane is required for morphogenesis but not mating (51), while the reverse is true for S. pombe Ras1 (54). The anterograde and retrograde trafficking of Ras proteins between the endomembranes and plasma membrane occurs in part as a result of a cycle of palmitoylation and depalmitoylation (24, 59), but little is known about the regulation of these processes.
C. albicans secretes an acyclic, sesquiterpenoid alcohol, farnesol, that accumulates in culture supernatants and coordinates population-level behaviors (37, 42). Quorum-sensing (QS) systems permit single organisms within a population to sense the cell density through the accumulation of signaling molecules. These systems have been well characterized in many bacteria (39), and farnesol represents the first QS molecule identified in a eukaryotic organism (37, 52). Farnesol negatively regulates signaling through the Ras1-cAMP-PKA pathway (10, 13), thereby repressing the yeast-to-hypha transition (10) and biofilm formation (57) and derepressing certain stress response genes, including CTA1, which encodes catalase (10, 13, 15, 57, 74). While recent work indicates that farnesol directly inhibits the Ras1 effector, Cyr1 (27), structural similarities between farnesol and the farnesyl moiety that modifies Ras proteins leads to the appealing prediction that farnesol also perturbs Ras1 posttranslational modifications or the interactions of farnesylated Ras1 with membranes. A farnesyl-cysteine mimetic, S-trans, trans-farnesylthiosalicylic acid (FTS), promotes the removal of H-Ras, a mammalian homolog of Ras1, from membranes, resulting in decreased whole-cell H-Ras levels and activity without affecting the posttranslational processing of the protein (26).
In this report, we provide the first demonstration that GFP-Ras1 is functional in C. albicans and confirm that it is localized at the plasma membrane in C. albicans yeast (64, 77). We show that Ras1 also localizes to the plasma membranes of hyphae with no detectable areas of enrichment. C. albicans Ras1 variants with mutations of the cysteine residues in the C-terminal motif that contains lipid modification sites in other fungal Ras proteins were mislocalized. GFP-Ras1-C287S, bearing a mutation at the only predicted site of palmitoylation, was largely associated with endomembranes, while GFP-Ras1-C288S, lacking the predicted farnesylation site, was cytosolic. To determine the link between Ras1 localization and function, the phenotypes of ras1Δ/ras1Δ strains expressing these Ras1 variants were assessed. By combining the G13V substitution with mutations that alter Ras1 subcellular localization, we found that Ras1 localization is critical for activation but not essential for its interaction with the effector adenylate cyclase. Fluorescence recovery after photobleaching (FRAP) analysis of Ras1 indicated that its mobility in yeast was similar to that reported for Ras proteins in other organisms. Ras1 was more dynamic in yeast than in hyphae, and the differences were neither specific to protein near the growing hyphal tip nor dependent on the Ras1 activation state. Farnesol, at biologically relevant concentrations, did not alter Ras1 plasma membrane localization but did inhibit hyphal induction mediated by a cytosolic-targeted Ras1 mutant. These studies provide new insights into the regulation of the Ras1-Cyr1 signaling pathway.
MATERIALS AND METHODS
Strains and growth conditions.
For a list of all strains and plasmids used in this study, refer to Table 1. C. albicans strains were streaked from frozen glycerol stocks at −80°C onto YPD (1% yeast extract, 2% peptone, 2% glucose) or YNB (0.67% yeast nitrogen base, 1% glucose) plates every 10 to 14 days and maintained at room temperature. The medium was supplemented with 80 μg/ml uridine (Sigma) or 20 μg/ml histidine (Sigma) as needed. Stock solutions of trans, trans farnesol (50 mM; Sigma) in acidified ethyl acetate or dimethylsulfoxide (DMSO) were prepared daily (10). Escherichia coli strain DH5α, used for routine cloning, and E. coli strain BL21, used for the heterologous expression of RAS1, were grown on LB. Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added at 40 μM to induce protein expression.
Table 1.
Yeast strains and plasmids used in these studies
| Strain or plasmid | Genotype | Reference |
|---|---|---|
| Strains | ||
| SC5314 | Prototrophic clinical isolate | 25 |
| AH81 | ura3::λimm434/ura3::λimm434 ras1::hisG/ras1::hisG | 14 |
| CaAP13 | Same as AH81 but with ras1::hisG/ras1::hisG::URA3 | 14 |
| CaAP14 | Same as AH81 but with ras1::hisG/ras1::hisG::RAS1-URA3 | This study |
| CaAP15 | Same as AH81 but with ras1::hisG/ras1::hisG::ras1-G13V-URA3 | This study |
| CaAP22 | Same as AH81 but with ras1::hisG/ras1::hisG::ras1-C287S-URA3 | This study |
| CaAP24 | Same as AH81 but with ras1::hisG/ras1::hisG::ras1-G13VC287S-URA3 | This study |
| CaAP23 | Same as AH81 but with ras1::hisG/ras1::hisG::ras1-C288S-URA3 | This study |
| CaAP25 | Same as AH81 but with ras1::hisG/ras1::hisG::ras1-G13VC288S-URA3 | This study |
| CaAP30 | Same as AH81 but with ras1::hisG/ras1::hisG::GFP-RAS1-URA3 | This study |
| BWP17 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 76 |
| PY1306 | ura3::λimm434/ura3::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG RP10::ARG4 | This study |
| PY1314 | Same as PY1306 but with RP10::ARG4-GFP-RAS1 | This study |
| PY1728 | Same as PY1306 but with RP10::ARG4-GFP-ras1-G13V | This study |
| PY1720 | Same as PY1306 but with RP10::ARG4-GFP-ras1-C287S | This study |
| PY1723 | Same as PY1306 but with RP10::ARG4-GFP-ras1-C288S | This study |
| PY205 | rac1Δ::URA3/rac1Δ::HIS1 arg4Δ::hisG/arg4Δ::hisG RP10::ARG4-GFP-RAC1 | 4 |
| Plasmids | ||
| pDH240 | pYPB1-ADHpL-CaRAS1 | 59 |
| pLJ57 | pYPB1-ADHpL-CaRAS1G13V | 59 |
| pAP13 | pRAS1-pL-URA3 | 14 |
| pAP14 | pAP13pRAS1-RAS1-URA3 | This study |
| pAP15 | pAP13pRAS1-ras1-G13V-URA3 | This study |
| pAP22 | pAP13pRAS1-ras1-C287S-URA3 | This study |
| pAP23 | pAP13pRAS1-ras1-C288S-URA3 | This study |
| pAP24 | pAP13pRAS1-ras1-G13VC287S-URA3 | This study |
| pAP25 | pAP13pRAS1-ras1-G13VC288S-URA3 | This study |
| pAP30 | pAP13pRAS1-GFP-RAS1-URA3 | This study |
| pEA | pExpArg | 5 |
| pEA-GFP-Ras1 | pExpArg-pADHGFP-RAS1 | This study |
| pEA-GFP-Ras1-G13V | pExpArg-pADHGFP-ras1-G13V | This study |
| pEA-GFP-Ras1-C287S | pExpArg-pADHGFP-ras1-C287S | This study |
| pEA-GFP-Ras1-C288S | pExpArg-pADHGFP-ras1-C288S | This study |
| pEA-GFP-Ras1-G13VC288S | pExpArg-pADHGFP-ras1-G13VC288S | This study |
Plasmid and strain construction.
Strains overexpressing N-terminally tagged green fluorescent protein (GFP) fusion proteins from the ADH1 promoter were constructed by transforming BWP17 with the indicated plasmids. Plasmids pEA-GFP-RAS1, pEA-GFP-Ras1-C287S, and pEA-GFP-Ras1-C288S were constructed by first amplifying the RAS1 open reading frame (ORF) from pDH240 (pYPB1-ADHpL-CaRAS1) (58) with primers CaRas1pup7, CaRas1 MluIm, CaRas1C287S MluIm, or CaRas1C288S MluIm. The fragments were digested with RsrII and MluI and ligated into similarly digested pExp-PADH R GFPRAC1 (36). pEA-GFP-Ras-G13V was similarly constructed with primers CaRas1pup7 and CaRas1 MluIm, with the exception that amplification was from pLJ57 (pYPB1-ADHpL-CaRAS1G13V) (58). The plasmids were targeted to the RP10 locus by digestion with StuI, and integration was verified by PCR.
For the generation of strains expressing RAS1 variants in single copies from the endogenous promoter, AH81 (ras1Δ/ras1Δ ura3Δ/ura3Δ) (13) was transformed with PacI-digested plasmids encoding the RAS1 promoter, variant allele, and terminator sequences followed by the URA3 coding region. The vector-only (ras1V), ras1/RAS1, and ras1/ras1-G13V strains were generated by the transformation of AH81 with pAP13 (13), pAP14, and pAP15, respectively. pAP14 and pAP15 were constructed by amplifying the RAS1 and ras1-G13V open reading frames from pDH240 (pYPB1-ADHpL-CaRAS1) and pLJ57 (pYPB1-ADHpL-CaRAS1G13V) (58), respectively, with primers XhoIRAS1F and BamHIRAS1R. Strains expressing the C-terminal variant RAS1 alleles in single copy at the endogenous promoter were generated by transforming AH81 with pAP22, pAP23, pAP24, and pAP25. pAP22 (ras1-C287S) and pAP23(ras1-C288S) were constructed by the PCR amplification of an 876-bp fragment from pDH240 (pYPB-ADHpL-CaRAS1) (58) with primers RAS1XhoIF with BamHIC287SR and BamHIC288SR, respectively. pAP24 (ras1-G13VC287S) and pAP25 (ras1-G13VC288S) were similarly constructed by the PCR amplification of an 876-bp fragment from plasmid pLJ57 (pYPB1-ADHpL-CaRAS1G13V) (58). All PCR products were digested with XhoI and BamHI and ligated into similarly digested pAP13 (13).
To construct a ras1Δ/ras1Δ strain expressing GFP-RAS1 from the endogenous RAS1 promoter, plasmid pAP30 was linearized with PacI and transformed into AH81 (13). GFP-RAS1 was amplified with primers XhoIGFPF and BamHIRAS1R from pEA-GFP-Ras1, digested with XhoI and BamHI, and ligated into similarly digested pAP13 to create pAP30.
For all C. albicans clones, integration at the endogenous RAS1 locus was confirmed by PCR with primers RAS15up and RAS1intdel5′ followed by the restriction digestion of the product with XhoI, and at least two independent clones were assessed for each strain. Plasmid sequences are listed in Table 2.
Table 2.
Primers used in these studies
| Primer | Sequence |
|---|---|
| CaRas1pup7 | ATCGGACCGTGATGTTGAGAGAATATAAATTAGTTGTTG |
| CaRas1 MluIm | ATACGCGTCTCAAACAATAACACAACATCCATTCTTTG |
| CaRas1C287S MluIm | ATACGCGTCTCAAACAATAACACAACTTCCATTCTTTGATTTAGAGC |
| CaRas1C288S MluIm | ATACGCGTCTCAAACAATAACACTACATCCATTCTTTGATTTAGAGC |
| RAS1XhoI F | CTCGAGATGTTGAGAGAATATAAATTAGTTGTTGTTGTTGG |
| BamHIRAS1R | CTCGGATCCTCAAACAATAACACAACATCCATT |
| BamHIC287S R | CTCGGATCCTCAAACAATAACAGAACATCCATT |
| BamHIC288S R | CTCGGATCCTCAAACAATAACACAAGATCCATT |
| RAS1 5up | TTGGCTTTGTAACAGCAACA |
| RAS1 intdel5′ | ATATTGGTCTTGACCTTGTT |
| XhoIGFPF | CCCTCGAGGCGTTTATTAAAATGTCTAAAG |
Assessment of C. albicans morphology.
Cells for inoculation were cultured in either YNB or YPD with the appropriate supplement for 16 to 18 h at 30°C and washed once with distilled H2O prior to transfer to hypha-inducing medium. Liquid induction was carried out by inoculating cells from overnight cultures into 0.2% YNBNP (0.67% yeast nitrogen base, 25 mM potassium phosphate buffer, 5 mM N-acetyl-d-glucosamine, 0.2% glucose) (10) or YNBP, pH 7 (0.67% yeast nitrogen base, 25 mM potassium phosphate buffer, 0.2% glucose), at 37°C. Serum-inducing experiments were carried out on exponentially growing cells in the presence of 50% fetal calf serum (FCS). Trans, trans farnesol (Sigma), DMSO, or ethyl acetate was added to media prior to cell addition at the indicated concentrations. Cell morphology in liquid hypha-inducing conditions was assessed in triplicate on multiple days by following published methods (48, 73) using a Zeiss Axiovert inverted microscope equipped with a 63× long-working-distance objective and Axiovision software. For each replicate, more than 600 cells were counted, and the percentages of cells growing as yeast, pseudohyphae, and hyphae were determined from an average of three replicates. Embedded filamentation assays were carried out in YPS (1% yeast extract, 2% peptone, 2% sucrose, 2% agar) as previously published (8) in triplicate on different days. Fifty μM trans, trans farnesol (Sigma) or ethyl acetate was added to agar prior to cell addition. Colonies were imaged with a Nikon SMZ1500 stereomicroscope and MetaVue imaging software, version 5.0r1 (Universal Imaging Corp.). Images were adjusted with Adobe PhotoshopCS, version 8.0.
Fluorescence microscopy and FRAP analysis.
Confocal imaging was conducted on a Quorum Wave FX-X1 spinning disk confocal system (Quorum Technologies Inc., Guelph, Canada) with a Plan-Apo 60× objective/1.4 numerical aperture. Z-stacks of images were deconvolved and processed by the classical maximum likelihood estimation (CMLE) algorithm using a calculated point spread function with Huygens Essentials software (Scientific Volume Imaging B.V.). Fluorescent images shown represent central slices from deconvolved Z-stacks unless otherwise indicated. Analysis of cell membranes by fluorescence recovery after photobleaching (FRAP) was carried out essentially as described previously (4) on a Zeiss LSM510 Meta confocal microscope with a Plan-Apo 63× objective (numerical aperture, 1.4) with bleaching performed at 100% laser intensity on a 1.1-μm2 circular area of the cell membrane. Data analysis was conducted as previously described (4).
Western blot analysis and cell fractionation.
Lysate preparation and fractionation were conducted as previously published, with several modifications (66). Whole-cell lysates were prepared by resuspending cells in HB buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 10% sucrose) with protease inhibitors (Roche) and disrupting cells with glass beads in a Bio-Spec bead beater with six rounds of 50-s disruptions at 4°C and 1-min rests on ice. Protein concentrations were determined by the Bradford assay. A total of 15 μg protein diluted in SDS loading buffer was separated by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) with the iBlot system (Invitrogen), and detected with monoclonal anti-Ras clone 10 (1.5 μg/ml; Millipore) or anti-GFP (Roche), followed by secondary detection with goat anti-mouse (Pierce) and enhanced chemiluminescent visualization (Pierce).
For membrane fractionation, lysates were subjected to ultracentrifugation in a Beckman Coulter TLA100 Ultracentrifuge with a TLA100.3 rotor at 100,000 × g for 1 h at 4°C. The cytosolic fraction (S100) was transferred to a fresh tube, and the membrane fraction (P100) was washed three times with HB plus protease inhibitors (Roche), resuspended in membrane detergent lysis buffer (MDLB; 10 mM Tris-HCl, pH 7.4, 1% Triton-X, 0.1% SDS, 1.5 mM NaCl) and protease inhibitors (Roche), and incubated with rocking for 1 h at 4°C to solubilize. To assess the effects of farnesol on Ras1 stability in the membrane fraction, membranes were incubated in the presence of ethyl acetate, 3.75 mM trans, trans farnesol (Sigma), or 1% Triton X-100 (Fisher) for 1 h on ice, centrifuged at 100,000 × g for 1 h at 4°C, and then solubilized as described above.
Growth rate analysis.
Cells were grown overnight in YPD, diluted to an optical density at 600 nm (OD600) of 0.05 in YPD in 96-well plates, and incubated at 30°C with shaking prior to each read at OD600 in a SpectraMax M5e (Molecular Diagnostics). Data were examined and plotted in Excel 2007 (Microsoft).
Iodine detection of farnesol.
One ml of 100 mM phosphate buffer was supplemented with trans, trans farnesol (Sigma) and incubated in borosilicate glass or polystyrene. Samples were extracted twice with a 1:1 dilution of ethyl acetate and reduced under nitrogen. Dried samples were resuspended in 20 μl ethyl acetate, spotted on Silica Gel 60 F-254 TLC plates (Selecto Scientific), and detected by exposure to iodine vapor. Images were adjusted with Adobe PhotoshopCS, version 8.0, and densitometry analysis was performed with ImageJ (1).
RESULTS
GFP-Ras1 is localized to the plasma membrane during hyphal growth.
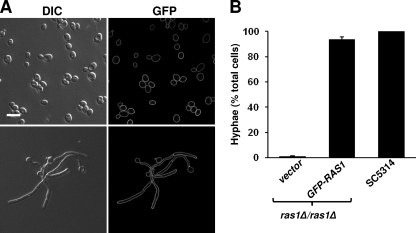
Because some proteins involved in polarized growth, such as Cdc42, Cdc24, and Spa2, show distinct localization patterns in yeast and hyphae (5, 32, 76), we sought to determine if Ras1 localization differed in yeast and hyphal cells. Consistent with results published by Zhu et al. (77), an N-terminal GFP-Ras1 fusion expressed from the ADH1 promoter in the wild-type background (PY1314) localized to the plasma membranes of yeast cells (Fig. 1A). In hyphal cells grown in YNBNP at 37°C, we observed that GFP-Ras1 was uniformly distributed throughout the plasma membrane of the mother cell, the filament periphery, and at septa with no obvious enrichment at discrete sites, such as the growing tip (Fig. 1A). Similar localization patterns were observed when GFP-RAS1 was expressed under the endogenous RAS1 promoter in a RAS1 deletion background (see Fig. S1 in the supplemental material), demonstrating that homogeneous plasma membrane localization was not an artifact of overexpression. The GFP-Ras1 was functional, as it fully complemented hyphal growth defects of the ras1Δ/ras1Δ strain at levels similar to that of the wild-type control, SC5314 (Fig. 1B).
Fig. 1.
GFP-Ras1 localizes to the plasma membranes of yeast and hyphae. (A) Cells with two endogenous RAS1 alleles overexpressing GFP-RAS1 from the ADH1 promoter (PY1314) as yeast in YPD overnight cultures (top) and hyphae after 3 h hyphal induction in YNBNP at 37°C (bottom) were visualized by confocal microscopy. Scale bar, 10 μM. (B) ras1Δ/ras1Δ cells expressing the empty vector or GFP-RAS1 from the endogenous RAS1 promoter were induced to form hyphae in YNPNP at 37° for 3 h, at which time the percent hyphae was quantified and compared to the amount of the wild-type strain, SC5314. Data are expressed as the means (±SD) of triplicate experiments conducted on different days. DIC, differential interference contrast.
Ras1 plasma membrane localization is determined by C-terminal cysteines.
The plasma membrane localization of other fungal Ras proteins is mediated by farnesyl and palmitoyl modifications at the C terminus (11, 14, 51, 54). The alignment of C. albicans Ras1 with Saccharomyces cerevisiae Ras2 and Schizosaccharomyces pombe Ras1 indicated the conservation of both the cysteine within the CAAX farnesylation motif (C288) and a single adjacent cysteine residue (C287), which is the site of palmitoylation in these fungi. There are no other cysteine residues in the 143 amino acids preceding cysteine 287, indicating the presence of a single predicted palmitoylation site on Ras1.
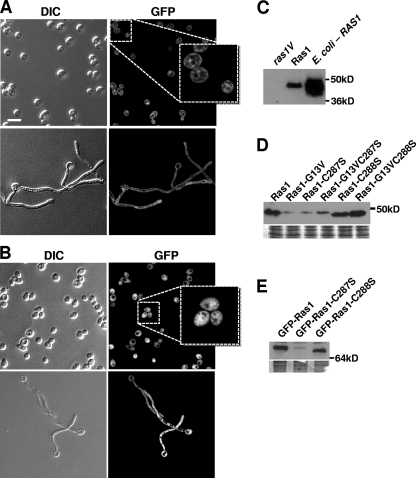
To assess the contributions of the C-terminal cysteines in Ras1 localization, GFP-RAS1 alleles encoding C288S and C287S variants were constructed and expressed under the control of the ADH1 promoter in the wild-type strain (Fig. 2). The levels of GFP-Ras1 were greater than those of endogenous Ras1 in these cells (see Fig. S2A in the supplemental material). Cells bearing either allele formed hyphae with kinetics similar to those of the parental strain carrying an empty vector, indicating that the variant proteins did not hinder the activity of endogenous Ras1 (data not shown). GFP-Ras1-C287S mislocalized primarily to endomembrane structures (Fig. 2A) with dramatically lower levels at the plasma membrane in both yeast and hyphae compared to GFP-Ras1 plasma membrane levels (Fig. 1A and 2A). GFP-Ras1-C288S remained restricted to the cytosol, excluded from vacuoles, in both yeast and hyphae (Fig. 2B).
Fig. 2.
Ras1-C287S and Ras1-C288S are mislocalized in yeast and hyphae. Wild-type cells overexpressing GFP-ras1-C287S (PY1720) (A) or GFP-ras1-C288S (PY1723) (B) were isolated as yeast from YPD overnight cultures or hyphae following 3 h of hyphal induction in YNBNP at 37°C and visualized by confocal microscopy. Scale bar, 10 μm. Insets in panels A and B show mislocalization of Ras1. (C) Lysates from C. albicans ras1Δ/ras1Δ cells expressing the empty vector (lane 1), RAS1 (lane 2), or E. coli BL21 expressing C. albicans RAS1 (lane 3) were analyzed by immunoblotting with an antibody directed to Ras. (D) Lysates from ras1Δ/ras1Δ cells expressing (from left to right) RAS1, ras1-G13V, ras1-C287S, ras1-G13VC287S, ras1-C288S, or ras1-G13VC288S in single copies from the endogenous promoter were analyzed as described for panel C. A Coomassie-stained gel serves as the loading control. Cells were cultured in YNBNP for 3 h. (E) Lysates from wild-type cells overexpressing GFP-RAS1, GFP-ras1-C287S, or GFP-ras1-C288S under the ADH1 promoter in the BWP17 background were analyzed by immunoblotting with an antibody directed to GFP.
Mutation of C-terminal cysteines affects Ras1 electrophoretic mobility and abundance.
Lipid modifications can alter the mobility of Ras proteins (18, 33). Western blot analysis was used to examine the migration patterns of the untagged variant proteins in the ras1Δ/ras1Δ background. In cells expressing native Ras1 (Fig. 2C, lane 2), the anti-Ras antibody detected a single band with an apparent molecular mass that was 14 kDa larger than the predicted mass of Ras1 (32 kDa). We confirmed that the 46-kDa band was full-length Ras1 by comparing its migration to that of Ras1 in lysates of E. coli heterologously expressing RAS1 (Fig. 2C, lane 3), verifying that Ras1 is not detected in lysates of the ras1V strain (Fig. 2C, lane 1). Furthermore, Ras1 levels were similar in CAF2 cells with two endogenous copies of RAS1 and ras1Δ/RAS1 cells (see Fig. S2B in the supplemental material). The analysis of lysates from cells expressing the variant proteins demonstrated that Ras1 and Ras1-C287S display similar electrophoretic mobilities, while Ras1-C288S migrated slightly more rapidly (Fig. 2D). The migration of the GFP-Ras1 variants also was slower than predicted by the calculated molecular mass, and again GFP-Ras1-C288S migrated slightly more rapidly than the Ras1 or Ras1-C287S derivatives (Fig. 2E).
The Ras1 variant protein levels were reproducibly different from those in comparable strains bearing nonmutated Ras1 regardless of whether the allele was expressed from the endogenous promoter in the ras1Δ/ras1Δ background (Fig. 2D) or expressed from the ADH1 promoter in the wild-type background (Fig. 2E). Both untagged and GFP-Ras1-C287S proteins were less abundant than the equivalent Ras1 proteins (Fig. 2D and E), while Ras1-C288S and GFP-Ras1-C288S were present at comparable or higher levels than Ras1 (Fig. 2D and E), revealing that C-terminal modifications to Ras1 affect protein abundance and/or stability.
Both Ras1 C-terminal cysteines contribute to Ras1 function in hyphal growth.
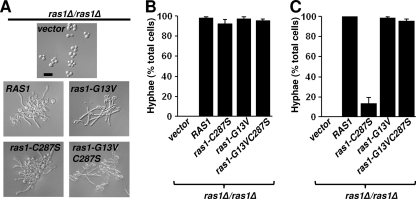
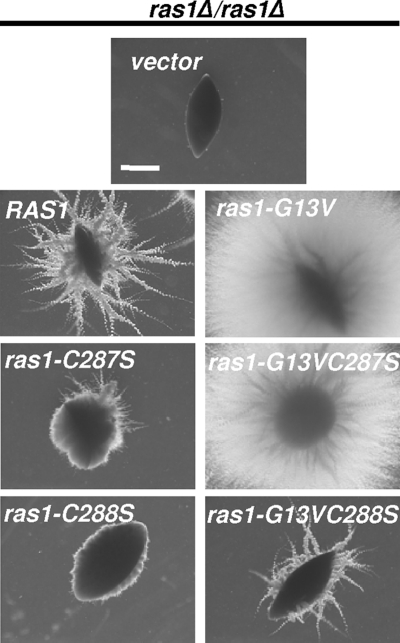
To determine if mislocalized C. albicans Ras1-C287S and Ras1-C288S were capable of supporting filamentation in the absence of wild-type Ras1, quantitative morphological studies were performed on cells expressing the RAS1 variant alleles in single copies from the endogenous RAS1 locus in the ras1Δ/ras1Δ background. The growth defects of the RAS1 deletion strain have been noted previously by our laboratory and others (10, 44, 77), and the reintegration of a single copy of the RAS1 allele rescued the growth rate defects. The complemented strain doubled every 1.8 ± 0.04 h, while the ras1V strain doubled every 2.6 ± 0.08 h. Cells expressing ras1-C287S also doubled every 1.9 ± 0.03 h, which is comparable to cells expressing the wild-type allele. However, the growth rate of ras1-C288S-expressing cells (2.23 ± 0.05 h) was more similar to that of the ras1V reference strain. As previously observed (10, 19, 44), the ras1 reference strain expressing the empty vector remained as yeast in hypha-inducing medium, and this filamentation defect was complemented by RAS1 (Fig. 3A); 98% ± 1.1% of cells expressing RAS1 formed hyphae in YNBNP (Fig. 3B). The ras1-C287S-expressing strain formed hyphae at levels similar to those of cells expressing RAS1 (Fig. 3A and B). It was noted, however, that cultures of cells expressing RAS1 contained hyphae of relatively equal length, while Ras1-C287S hyphae ranged from newly emerging germ tubes to hyphae equal in length to those in cells expressing RAS1 (data not shown). A stronger defect in filamentation was observed in cells expressing ras1-C287S when hyphal growth was assessed in medium lacking GlcNAc (YNBP, pH 7, at 37°C) (Fig. 3C). Under these conditions, hypha development likely is mediated through a combination of temperature, release from quorum sensing (15), and transfer to a neutral-pH medium (46). Ras1 also is required for the filamentation of cells embedded in a matrix (17, 47), which occurs at lower temperatures (25°C) in the absence of chemical inducers and has been shown to involve both cAMP-dependent and MAP kinase-dependent pathways (17, 47, 61). Colonies formed by cells expressing ras1-C287S developed fewer and shorter radial hyphae at the periphery compared to colonies formed by RAS1-expressing cells, while ras1V control colonies lacked any radial hyphae (Fig. 4). Cells expressing the ras1-C288S allele remained largely in the yeast morphology, forming only 2% ± 0.4% hyphae in YNBNP (Fig. 5A). When embedded in agar, these cells formed colonies composed of yeast (Fig. 4).
Fig. 3.
Ras1-C287S variants are capable of hyphal growth. ras1Δ/ras1Δ cells expressing the empty vector, RAS1, ras1-G13V, ras1-C287S, or ras1-G13VC287S from the endogenous RAS1 promoter were incubated overnight in YPD (A) and then cultured in YNBNP for 3 h at 37°C, at which time DIC images were captured (scale bar, 20 μM), and (B) the percent hyphae were determined. (C) Cells incubated in YPD overnight from panel A were cultured in YNBP for 3 h at 37°C, at which time the percentages of hyphae were determined. Data were expressed as the means (±SD) from triplicate experiments conducted on different days.
Fig. 4.
Ras1 variants are altered in embedded filamentation. ras1Δ/ras1Δ cells expressing the empty vector, RAS1, ras1-G13V, ras1-C287S, ras1-G13VC287S, ras1-C288S, or ras1-G13VC288S in single copies from the endogenous RAS1 promoter were embedded in YP-2% sucrose (YPS) for 5 days at 25°C. Images of colonies were acquired at 10× magnification and are representative of >300 colonies from three biological replicates. Scale bar, 1.0 mm.
Fig. 5.
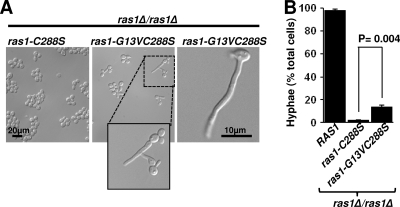
Farnesylation is required for Ras1 function. (A) ras1Δ/ras1Δ cells expressing ras1-C288S or ras1-G13VC288S in single copies from the endogenous RAS1 promoter were grown overnight in YPD and then cultured in YNBNP at 37°C for 4 h, at which time DIC images of representative populations of ras1-C288S (left) and ras1-G13VC288S (middle) cells were captured. The inset in middle shows a pseudohypha formed by ras1-G13VC288S. The right panel shows a hypha formed by ras1-G13VC288S. (B) ras1Δ/ras1Δ cells expressing RAS1, ras1-C288S, or ras1-G13VC288S in single copies from the endogenous RAS1 promoter were grown overnight in YPD and then cultured in YNBNP at 37°C for 3 h, at which time the percent hyphae was determined. Data are expressed as the means (±SD) from triplicate experiments conducted on different days.
Constitutive activation of Ras1 can partially suppress defects due to C-terminal mutations.
To determine if Ras1 variants defective in lipid modifications were altered in their ability to support hyphal growth because of defects in activation, defects in effector interactions, or both, we constructed alleles that combined the G13V mutation with either the C287S or C288S mutation and expressed these under the endogenous promoter in the ras1Δ/ras1Δ background. In both YNBNP, which contains GlcNAc, and YNBP at 37°C, strains expressing RAS1 or ras1-G13V underwent robust filamentation (Fig. 3). The Ras1-G13V strain exhibited phenotypes indicative of increased Ras1-cAMP signaling, including sensitivity to heat shock and reduced glycogen accumulation (data not shown) (10, 19, 68). While Ras1-C287S developed hyphae of heterogeneous lengths, Ras1-G13VC287S cells formed hyphae at levels similar to those of the Ras1 strain in YNBNP (data not shown and Fig. 3A and B). Furthermore, in contrast to cells bearing Ras1-C287S, of which only 13.0% ± 5.7% of the cell population developed hyphae in YNBP, Ras1-G13VC287S cells underwent robust hyphal growth, with 98.7% ± 1.2% of cells forming hyphae (Fig. 3C). In addition, strains expressing either the ras1-G13V or ras1-G13VC287S allele formed hyperfilamentous colonies in embedded conditions compared to colonies formed by the Ras1 strain (Fig. 4). Immunoblot analysis of cells cultured in hypha-inducing conditions revealed that Ras1-G13VC287S is only slightly more abundant than Ras1-C287S (Fig. 2D) but still is less abundant than Ras1.
When cells expressing ras1-G13VC288S under the endogenous promoter as the sole copy of RAS were compared to the ras1-C288S strain, a significant increase in filamentation was observed (P = 0.004) (13.6% ± 1.4% of cells formed true hyphae) (Fig. 5B). In contrast to cells expressing ras1-C288S, which grew at rates similar to those of the ras1V control, the ras1-G13VC288S allele restored growth rates (1.89 ± 0.07 h) to nearly the levels of RAS1-expressing cells. Under embedded conditions, the colonies formed by Ras1-C288S cells lacked hyphae at the periphery, whereas all of the colonies formed by Ras1-G13VC288S cells formed radial filaments (Fig. 4). Immunoblot analysis of cell lysates from cells grown in YNBNP for 3 h at 37°C revealed little to no increase in Ras1-G13VC288S compared to the level of Ras1-C288S (Fig. 2D).
Differences in Ras1 mobility at the plasma membrane of yeast and hyphae are independent of Ras1 activation.
The G13V substitution in Ras1-C287S and Ras1-C288S restored function to different degrees and enabled some cells to grow as hyphae (Fig. 3A, 4, and 5), indicating that the proper localization of Ras1 is essential for Ras1 activation but not absolutely required for subsequent effector interactions. While GFP-Ras1-G13V localized to the periphery of the cell, GFP-Ras1-G13VC288S was localized in the cytoplasm in a manner similar to that of GFP-Ras1-C288S (data not shown), revealing that the G13V substitution in the C288S allele was not sufficient to rescue the localization defects. Repeated attempts to generate a strain overexpressing GFP-Ras1-G13VC287S were unsuccessful, suggesting the toxicity of this protein when it is overexpressed. Thus, we could not compare its localization pattern to that of GFP-Ras1-C287S.
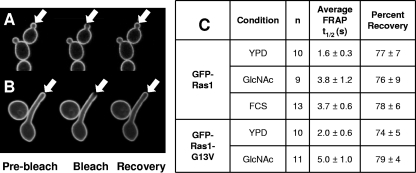
As an alternative approach to assess the consequences of the G13V substitution on Ras1 membrane interactions, we explored the effects of the constitutive activation on Ras1 dynamics in the membrane environment using FRAP in yeast and hyphae. First, the percent recoveries and FRAP t1/2 values were determined for GFP-Ras1 and GFP-Ras1-G13V at the bud sites in yeast (Fig. 6A). No significant differences were observed for either metric (Fig. 6C) despite the fact that cells expressing GFP-ras1-G13V displayed phenotypes consistent with increased cAMP signaling, as discussed above. These results suggest that Ras1 interactions at the plasma membrane were not radically altered upon Ras1 activation.
Fig. 6.
GFP-Ras1 dynamics at the plasma membrane of yeast and hyphae. Wild-type cells overexpressing GFP-RAS1 (PY1314) or GFP-ras1-G13V (PY1728) were used for FRAP analyses. Images of budding yeast cells (A) or hyphae (B) were taken by confocal microscopy prior to, during, and following photobleaching to assess fluorescence recovery. Arrows indicate bleached areas. (C) Quantification of fluorescence recoveries in wild-type cells expressing GFP-RAS1 or GFP-ras1-G13V in yeast and hyphae, as indicated.
Although GFP-Ras1 did not localize discretely to the hyphal tip (Fig. 1A), we speculated that interactions at the filament tip during hyphal growth alter its mobility and thus the reliance on lipid modifications for localization. A comparison of the FRAP t1/2 of GFP-Ras1 at the sites of growth at the hyphal apex (Fig. 6B) and at the yeast bud tip (Fig. 6A) showed that the percent recoveries were similar, yet there was a significant decrease in mobility at the hyphal tip relative to that of mobility in yeast cells (P < 0.0001) (Fig. 6C). The observed reduction in FRAP t1/2 at the filament tip was similar whether induction was carried out in YNBNP or in serum (Fig. 6C). The decrease in mobility was not regionally specific, as GFP-Ras1 mobility at the hyphal tip (3.8 ± 1.2 s) was the same as that observed at sites on the mother blastospore (3.8 ± 0.6 s), suggesting that factors at the site of polarized growth were not profoundly altering Ras1 dynamics in hyphae. The decreased mobility of GFP-Ras1 in hyphae likely was not due to an increase in Ras1 activation, as the average FRAP t1/2 for GFP-Ras1-G13V also was significantly slower in hyphae than in yeast (P < 0.0001) (Fig. 6C). Taken together, the GFP localization and the FRAP analyses indicated that C. albicans Ras1 associates with the plasma membrane via lipid anchors, similarly to Ras2 in S. cerevisiae (53, 71).
Farnesol blocks filamentation without altering Ras1 plasma membrane distribution.
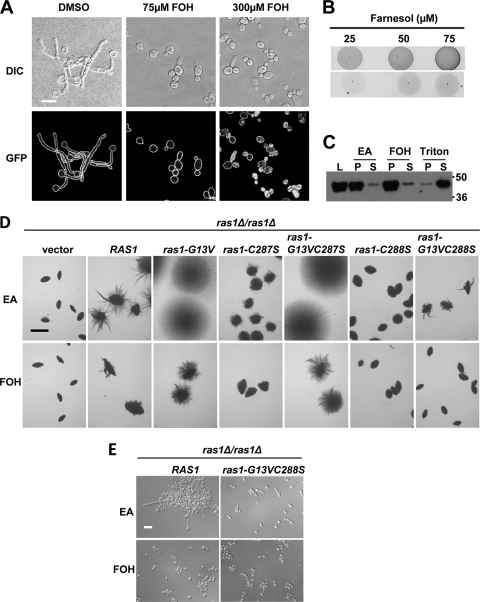
Farnesol, an autoregulatory molecule with structural similarity to the farnesyl molecule that modifies Ras proteins, inhibits Ras1-dependent cAMP signaling in both yeast and hyphae (10, 13), and recent work by Shareck et al. (64) showed that conjugated linoleic acid inhibited the yeast-to-hypha transition and concomitantly led to a decrease in plasma membrane-associated GFP-Ras1 in treated cells. Therefore, we sought to determine if farnesol alters the plasma membrane association of Ras1. Hypha formation in YNBNP was repressed by 75 μM farnesol in cells expressing GFP-RAS1 from the endogenous promoter as the sole copy of RAS1 (Fig. 7A). Like the hyphae in control cultures with vehicle alone (DMSO or ethyl acetate), the yeast cells in farnesol-containing medium still exhibited the uniform distribution of GFP-Ras1 throughout the plasma membrane (Fig. 7A and data not shown). At higher concentrations of farnesol (300 μM), GFP-Ras1 was no longer located exclusively at the plasma membrane and was found in punctate patches within a subset of cells (Fig. 7A). Similar observations were made of GFP-Rac1, which localizes to the plasma membrane by lipid anchors (see Fig. S3 in the supplemental material). We observed that the concentration of farnesol necessary to affect Ras1 signaling and to inhibit hypha induction depended on the composition of the culture vessel. For example, farnesol shows the dose-dependent inhibition of filamentation at 25 to 200 μM in polystyrene plates (35), although concentrations of farnesol (200 to 300 μM) can be toxic to cells (43, 65). The examination of farnesol availability in liquid medium incubated in either borosilicate glass or polystyrene chambers revealed, on average, 2.4-fold (± 0.6) less farnesol in the aqueous phase in polystyrene vessels (Fig. 7B).
Fig. 7.
Effects of farnesol on Ras1 localization and filamentation of cells expressing Ras1 C-terminal variants. (A) ras1Δ/ras1Δ cells expressing GFP-RAS1 in single copies from the endogenous promoter were incubated for 3 h in YNBNP at 37°C in the presence of the vehicle (DMSO) and 75 or 300 μM farnesol (FOH). The mislocalization of GFP-Ras1 was assessed by confocal microscopy. Scale bar, 10 μm. (B) The indicated concentrations of farnesol were incubated in phosphate buffer in borosilicate glass (top) or polystyrene (bottom), and farnesol was detected by iodine vapor. (C) Membranes from wild-type cells were treated with the vehicle, high concentrations of farnesol, or the nonionic detergent Triton X-100, and the repartitioning of the protein was assessed by immunoblot with anti-Ras. L, whole-cell lysate; P, pellet fraction; S, soluble fraction. (D) ras1Δ/ras1Δ cells expressing the variant RAS1 alleles in single copies at the native RAS1 promoter were embedded in YP-2% sucrose (YPS) with vehicle or 50 μM farnesol for 5 days at 25°C. Images of colonies were acquired at 10× magnification and are representative of >300 colonies from three biological replicates. EA, ethyl acetate vehicle; FOH, farnesol (scale bar, 2.0 mm). (E) ras1Δ/ras1Δ cells expressing RAS1 or ras1-G13VC288S alleles in single copies at the native promoter were cultured in YNBNP at 37°C in vehicle (EA) or farnesol (FOH) for 3 h and visualized with DIC microscopy. Scale bar, 20 μM.
The in vivo relocalization of GFP-Ras1 (and GFP-Rac1) by high concentrations of farnesol (Fig. 7A) may be the result of farnesol insertion into the lipid bilayer, causing the displacement of the Ras1 or Rac1 C-terminal lipids from the plasma membrane. However, farnesol did not change the amount of Ras1 associated with the membrane fraction in vitro even when total membrane fractions were incubated with very high concentrations (3.75 mM) of farnesol. In contrast, the treatment of membranes with the detergent Triton X-100 led to the repartitioning of Ras1 from the pellet to the soluble pool (Fig. 7C). To further support our data indicating that farnesol does not alter Ras1 signaling by changing its localization, we assessed the farnesol sensitivity of the cells expressing the Ras1 variant proteins in both embedded and liquid conditions. The limited filamentation seen in the strain expressing ras1-G13VC288S (Fig. 4 and 5) was repressed by farnesol in both embedded (Fig. 7D) and liquid growth conditions (Fig. 7E). Furthermore, the filamentation of cells expressing ras1-C287S and ras1-G13VC287S was inhibited by farnesol, and these cells were as sensitive to farnesol as the appropriate reference strains expressing RAS1 or ras1-G13V (Fig. 7C and data not shown). Taken together, these data suggest that Ras1 is lipid modified at C-terminal cysteine residues, and that farnesol does not act by inhibiting Ras1 posttranslational lipid modifications, nor does it act solely by affecting the interactions between Ras1 lipid modifications and the plasma membrane.
DISCUSSION
Evidence reported here suggests that C. albicans Ras1 is modified by palmitoylation and farnesylation at C287 and C288, respectively. Ras1 was detected only at the plasma membranes of both yeast and hyphae, and FRAP analysis revealed that Ras1 dynamics were comparable to those for S. cerevisiae Ras2 (t1/2 = 3.6 s) (71), which is anchored to the membrane by farnesyl and palmitoyl moieties (14) and thus is highly mobile. For comparison, the S. cerevisiae integral membrane protein, Pma1, is much less dynamic (t1/2 = 576 s) (71). In other species, the mutation of the cysteine corresponding to Cys-288 abolishes sequential farnesylation and palmitoylation and, consequently, any membrane association. C. albicans Ras1 likely is similarly modified, as Ras1-C288S is cytoplasmic. Ras1-C287S was localized mainly to internal membranes, suggesting that this protein is farnesylated but lacks the subsequent palmitoyl modification that is necessary for the plasma membrane localization of Ras in other fungi (14, 51, 54).
Compared to S. cerevisiae Ras2 and C. neoformans Ras1, which require palmitoylation for some activities unless the protein is overexpressed or contains a hyperactivating amino acid substitution (11, 51), C. albicans Ras1 palmitoylation is not absolutely required for its role in growth rate or morphogenesis. Microscopic analysis of Ras1-C287S shows fluorescence associated with the cell periphery (Fig. 2A), which indicates that there is a pool of plasma membrane-associated Ras1 that is sufficient to support filamentation. Alternatively, Ras1-C287S may be associated with the membrane compartments just below the plasma membrane, such as the endoplasmic reticulum, as was observed in S. pombe (54), and signaling may occur from this compartment. Distinguishing between the two compartments will require resolution at the level of electron microscopy or the colocalization of Ras1-C287S with plasma membrane- and ER-specific markers.
Our data suggest that the lipidation of Ras1 is more important for facilitating Ras1 activation, and thus interactions with its GEF, Cdc25, than for promoting interactions with downstream effectors. First, combining the G13V substitution with C287S created a variant that fully complemented the filamentation defects of the ras1Δ/ras1Δ strain, whereas cells expressing the ras1-C287S allele exhibited delayed or defective hyphal induction (Fig. 3C). Second, cells bearing Ras1-G13VC288S, but not Ras1-C288S, grew at rates equal to those of cells expressing the wild-type allele, and a subset of these cells was capable of hyphal induction in response to chemical and physical stimuli (Fig. 4 and 5). Ghaemmaghami and colleagues (23) detected, on average, 319 Cdc25 molecules per S. cerevisiae cell and nearly 2 × 104 Ras2 proteins per cell, indicating that factors that affect Ras1 interactions with Cdc25 are key in the control of Ras1 activity. Ras1-C287S also was present at lower levels than in similarly expressed Ras1, regardless of the promoter driving expression (Fig. 2D and E), suggesting that enzymes that control the Ras1 palmitoylation state are responsible for controlling Ras1 levels or that the Ras1-C287S protein is less stable than Ras1. Differences in abundance also may influence the phenotypes of the Ras1-C287S strain, although Ras1-G13VC287S also was present at lower levels yet had activity comparable to that of the Ras1-G13V variant (Fig. 2D, 3, and 4). Future studies that further examine the localization of Cdc25 and Cyr1, the main Ras1 effectors for hyphal growth under these conditions, will provide insight into whether Ras1 signals from internal membranes in wild-type cells, as has been shown for other Ras proteins (9, 54).
Our FRAP analyses demonstrated that Ras1 was less dynamic at the plasma membrane in hyphae than budding yeast, and that these differences were not due solely to changes in the level of activated Ras1, as Ras1-G13V showed similar differences (Fig. 6). The reduced mobility of Ras1 in hyphae could be due to a change in membrane fluidity during morphogenesis or differences in interactions of Ras1 and elements within the plasma membrane, such as lipid rafts or protein assemblages, which are distinct between these two morphologies. We favor the latter hypothesis, since lipid-anchored Rac1, which also is uniformly distributed within the plasma membranes of yeast and hyphae, exhibits no differences in mobility within the membranes of yeast and hyphae (4). During the yeast-to-hypha transition, C. albicans cells undergo massive rearrangement of the cytoskeleton, with the formation of cortical actin clusters at hyphal tips and longitudinally oriented actin filaments throughout the growing hypha (67). The S. cerevisiae adenylate cyclase-associated protein, Srv2, links changes in the actin cytoskeleton to Ras-cAMP signaling (25). In C. albicans, hyphal growth in response to serum requires both complex formation between Cyr1-Srv2-G-actin (78) and physical interactions between Ras1 and Cyr1 (17). Therefore, our findings that Ras1 localized homogenously throughout the plasma membranes of hyphae (Fig. 1A) and was equally dynamic at the hyphal tip and at sites opposite to the hyphal apex within the mother cell indicate that the Ras1-effector interactions at the plasma membrane are transient. This model is consistent with the finding that Ras1 cannot be copurified with the Cyr1-Srv2-G-actin complex in C. albicans (78).
Farnesol inhibits Ras1-cAMP signaling in both yeast and hyphae (10, 13). Our data demonstrate that farnesol, at biologically active concentrations, does not impair Ras1-farnesylcysteine association with the plasma membrane, since both Ras1 and Rac1 remain homogenously distributed within the plasma membranes of cells amended with lower biologically active farnesol concentrations. Under some conditions, C. albicans growth is not affected by the concentrations of farnesol tested (42); however, there is evidence that 300 μM farnesol triggers apoptosis in a subset of cells (65). These differences likely are due to a variety of factors, including carbon source availability, culture growth phase (42), and the composition of the culture vessels. The effects of high concentrations of farnesol on GFP-Ras1 localization were not specific to Ras1, as GFP-Rac1, which is associated with the plasma membrane by lipid anchors (4), also was mislocalized. We speculate that the cells exhibiting the mislocalization of the GFP-tagged proteins are experiencing a global response, perhaps apoptosis, in response to toxic levels of farnesol. In further support of recently published work that shows that farnesol inhibits Cyr1 activity (27), cells bearing cytosolic Ras1-G13VC288S still are sensitive to the farnesol-mediated inhibition of filamentation. Farnesol, however, may have other effects on cells that relate to its ability to inhibit growth and trigger apoptosis and to influence other signaling pathways (10, 13, 27, 38, 41, 43, 60, 65, 70, 74).
Candida species are the most common causes of mycosis in individuals with underlying disease, and candidiasis and candidemia are associated with mortality and morbidity rates ranging from 10 to 50% (56). The evolution of antifungal-resistant strains (62) and the growing incidence of C. albicans antifungal-resistant biofilms on implanted medical devices (40) that provide a mode of entry for other pathogens (31) underscore the need for novel strategies to combat this pathogen. Because Ras1 is a key modulator of hyphal growth and virulence (19, 44, 45), a better understanding of its regulation in response to host-associated stimuli and its interaction with effectors will surely provide insight and improved understanding of elements of C. albicans biology that can be exploited as drug targets.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anne Lavanway for excellent microscopy support. This work was generously supported in part by the Ruth L. Kirschstein National Research Service Awards 5T32AI007519 and 5T32GM008704 (A.E.P.) and NIH K22 DE016542 (D.A.H.). M.B. was supported by the Centre National de la Recherche Scientifique and the French National Research Agency ANR-09-BLAN-0299-01, and O.B. was supported by funding from FRM-BNP Paribas.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Abramoff M., Magelhaes P., Ram S. 2004. Image processing with ImageJ. Biophotonics Int. 11:36–42 [Google Scholar]
- 2. Ashery U., Yizhar O., Rotblat B., Kloog Y. 2006. Nonconventional trafficking of Ras associated with Ras signal organization. Traffic 7:119–126 [DOI] [PubMed] [Google Scholar]
- 3. Bartels D. J., Mitchell D. A., Dong X., Deschenes R. J. 1999. Erf2, a novel gene product that affects the localization and palmitoylation of Ras2 in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:6775–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bassilana M., Arkowitz R. A. 2006. Rac1 and Cdc42 have different roles in Candida albicans development. Eukaryot. Cell 5:321–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bassilana M., Hopkins J., Arkowitz R. A. 2005. Regulation of the Cdc42/Cdc24 GTPase module during Candida albicans hyphal growth. Eukaryot. Cell 4:588–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boguski M. S., McCormick F. 1993. Proteins regulating Ras and its relatives. Nature 366:643–654 [DOI] [PubMed] [Google Scholar]
- 7. Boyartchuk V. L., Ashby M. N., Rine J. 1997. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science 275:1796–1800 [DOI] [PubMed] [Google Scholar]
- 8. Brown D. H., Jr., Giusani A. D., Chen X., Kumamoto C. A. 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34:651–662 [DOI] [PubMed] [Google Scholar]
- 9. Chiu V. K., et al. 2002. Ras signalling on the endoplasmic reticulum and the Golgi. Nat. Cell Biol. 4:343–350 [DOI] [PubMed] [Google Scholar]
- 10. Davis-Hanna A., Piispanen A. E., Stateva L. I., Hogan D. A. 2008. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol. Microbiol. 67:47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deschenes R. J., Broach J. R. 1987. Fatty acylation is important but not essential for Saccharomyces cerevisiae RAS function. Mol. Cell. Biol. 7:2344–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deschenes R. J., Stimmel J. B., Clarke S., Stock J., Broach J. R. 1989. RAS2 protein of Saccharomyces cerevisiae is methyl-esterified at its carboxyl terminus. J. Biol. Chem. 264:11865–11873 [PubMed] [Google Scholar]
- 13. Deveau A., Piispanen A. E., Jackson A. A., Hogan D. A. 2010. Farnesol induces hydrogen peroxide resistance in Candida albicans yeast by inhibiting the Ras-cyclic AMP signaling pathway. Eukaryot. Cell 9:569–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dong X., et al. 2003. Palmitoylation and plasma membrane localization of Ras2p by a nonclassical trafficking pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:6574–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Enjalbert B., Whiteway M. 2005. Release from quorum-sensing molecules triggers hyphal formation during Candida albicans resumption of growth. Eukaryot. Cell 4:1203–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Enloe B., Diamond A., Mitchell A. P. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang H. M., Wang Y. 2006. RA domain-mediated interaction of Cdc35 with Ras1 is essential for increasing cellular cAMP level for Candida albicans hyphal development. Mol. Microbiol. 61:484–496 [DOI] [PubMed] [Google Scholar]
- 18. Farh L., Mitchell D. A., Deschenes R. J. 1995. Farnesylation and proteolysis are sequential, but distinct steps in the CaaX box modification pathway. Arch. Biochem. Biophys. 318:113–121 [DOI] [PubMed] [Google Scholar]
- 19. Feng Q., Summers E., Guo B., Fink G. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181:6339–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fortwendel J. R., et al. 2005. A fungus-specific ras homolog contributes to the hyphal growth and virulence of Aspergillus fumigatus. Eukaryot. Cell 4:1982–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fujiyama A., Tamanoi F. 1986. Processing and fatty acid acylation of RAS1 and RAS2 proteins in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 83:1266–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fujiyama A., Tsunasawa S., Tamanoi F., Sakiyama F. 1991. S-farnesylation and methyl esterification of C-terminal domain of yeast RAS2 protein prior to fatty acid acylation. J. Biol. Chem. 266:17926–17931 [PubMed] [Google Scholar]
- 23. Ghaemmaghami S., et al. 2003. Global analysis of protein expression in yeast. Nature 425:737–741 [DOI] [PubMed] [Google Scholar]
- 24. Goodwin J. S., et al. 2005. Depalmitoylated Ras traffics to and from the Golgi complex via a nonvesicular pathway. J. Cell Biol. 170:261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gourlay C. W., Ayscough K. R. 2006. Actin-induced hyperactivation of the Ras signaling pathway leads to apoptosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:6487–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haklai R., et al. 1998. Dislodgment and accelerated degradation of Ras. Biochemistry 37:1306–1314 [DOI] [PubMed] [Google Scholar]
- 27. Hall R. A., et al. 2011. The quorum sensing molecules farnesol/homoserine lactone and dodecanol operate via distinct modes of action in Candida albicans. Eukaryot. Cell 10:1034–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hancock J. F. 2003. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 4:373–384 [DOI] [PubMed] [Google Scholar]
- 29. Hancock J. F., Magee A. I., Childs J. E., Marshall C. J. 1989. All ras proteins are polyisoprenylated but only some are palmitoylated. Cell 57:1167–1177 [DOI] [PubMed] [Google Scholar]
- 30. Harcus D., Nantel A., Marcil A., Rigby T., Whiteway M. 2004. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell 15:4490–4499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harriott M. M., Noverr M. C. 2009. Candida albicans and Staphylococcus aureus form polymicrobial biofilms: effects on antimicrobial resistance. Antimicrob. Agents Chemother. 53:3914–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hazan I., Liu H. 2002. Hyphal tip-associated localization of Cdc42 is F-actin dependent in Candida albicans. Eukaryot. Cell 1:856–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He B., et al. 1991. RAM2, an essential gene of yeast, and RAM1 encode the two polypeptide components of the farnesyltransferase that prenylates a-factor and Ras proteins. Proc. Natl. Acad. Sci. U. S. A. 88:11373–11377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hogan D. A., Sundstrom P. 2009. The Ras/cAMP/PKA signaling pathway and virulence in Candida albicans. Future Microbiol. 4:1263–1270 [DOI] [PubMed] [Google Scholar]
- 35. Hogan D. A., Vik A., Kolter R. 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54:1212–1223 [DOI] [PubMed] [Google Scholar]
- 36. Hope H., Bogliolo S., Arkowitz R. A., Bassilana M. 2008. Activation of Rac1 by the guanine nucleotide exchange factor Dck1 is required for invasive filamentous growth in the pathogen Candida albicans. Mol. Biol. Cell 19:3638–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hornby J. M., et al. 2001. Quorum sensing in the dimorphic fungus Candida albicans is mediated by farnesol. Appl. Environ. Microbiol. 67:2982–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kebaara B. W., et al. 2008. Candida albicans Tup1 is involved in farnesol-mediated inhibition of filamentous-growth induction. Eukaryot. Cell 7:980–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keller L., Surette M. G. 2006. Communication in bacteria: an ecological and evolutionary perspective. Nat. Rev. Microbiol. 4:249–258 [DOI] [PubMed] [Google Scholar]
- 40. Kojic E. M., Darouiche R. O. 2004. Candida infections of medical devices. Clin. Microbiol. Rev. 17:255–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kruppa M., et al. 2004. The two-component signal transduction protein Chk1p regulates quorum sensing in Candida albicans. Eukaryot. Cell 3:1062–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Langford M. L., Atkin A. L., Nickerson K. W. 2009. Cellular interactions of farnesol, a quorum-sensing molecule produced by Candida albicans. Future Microbiol. 4:1353–1362 [DOI] [PubMed] [Google Scholar]
- 43. Langford M. L., Hasim S., Nickerson K. W., Atkin A. L. 2010. Activity and toxicity of farnesol towards Candida albicans are dependent on growth conditions. Antimicrob. Agents Chemother. 54:940–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leberer E., et al. 2001. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol. Microbiol. 42:673–687 [DOI] [PubMed] [Google Scholar]
- 45. Lo H. J., et al. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 46. Lotz H., Sohn K., Brunner H., Muhlschlegel F. A., Rupp S. 2004. RBR1, a novel pH-regulated cell wall gene of Candida albicans, is repressed by RIM101 and activated by NRG1. Eukaryot. Cell 3:776–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maidan M. M., et al. 2005. The G protein-coupled receptor Gpr1 and the Galpha protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 16:1971–1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Merson-Davies L. A., Odds F. C. 1989. A morphology index for characterization of cell shape in Candida albicans. J. Gen. Microbiol. 135:3143–3152 [DOI] [PubMed] [Google Scholar]
- 49. Monge R. A., Roman E., Nombela C., Pla J. 2006. The MAP kinase signal transduction network in Candida albicans. Microbiology 152:905–912 [DOI] [PubMed] [Google Scholar]
- 50. Mor A., Philips M. R. 2006. Compartmentalized Ras/MAPK signaling. Annu. Rev. Immunol. 24:771–800 [DOI] [PubMed] [Google Scholar]
- 51. Nichols C. B., Ferreyra J., Ballou E. R., Alspaugh J. A. 2009. Subcellular localization directs signaling specificity of the Cryptococcus neoformans Ras1 protein. Eukaryot. Cell 8:181–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nickerson K. W., Atkin A. L., Hornby J. M. 2006. Quorum sensing in dimorphic fungi: farnesol and beyond. Appl. Environ. Microbiol. 72:3805–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Niv H., Gutman O., Kloog Y., Henis Y. I. 2002. Activated K-Ras and H-Ras display different interactions with saturable nonraft sites at the surface of live cells. J. Cell Biol. 157:865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Onken B., Wiener H., Philips M. R., Chang E. C. 2006. Compartmentalized signaling of Ras in fission yeast. Proc. Natl. Acad. Sci. U. S. A. 103:9045–9050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Park G., et al. 2006. Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell 18:2822–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pfaller M. A., Diekema D. J. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53 [DOI] [PubMed] [Google Scholar]
- 57. Ramage G., Saville S. P., Wickes B. L., Lopez-Ribot J. L. 2002. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl. Environ. Microbiol. 68:5459–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rocha C. R., et al. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rocks O., et al. 2005. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science 307:1746–1752 [DOI] [PubMed] [Google Scholar]
- 60. Román E., et al. 2009. The Cek1 MAPK is a short-lived protein regulated by quorum sensing in the fungal pathogen Candida albicans. FEMS Yeast Res. 9:942–955 [DOI] [PubMed] [Google Scholar]
- 61. Sánchez-Martínez C., Perez-Martin J. 2002. Gpa2, a G-protein alpha subunit required for hyphal development in Candida albicans. Eukaryot. Cell 1:865–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shapiro R. S., Robbins N., Cowen L. E. 2011. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev. 75:213–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shapiro R. S., et al. 2009. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr. Biol. 19:621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shareck J., Nantel A., Belhumeur P. 2011. Conjugated linoleic acid inhibits hyphal growth in Candida albicans by modulating Ras1p cellular levels and downregulating TEC1 expression. Eukaryot. Cell 10:565–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shirtliff M. E., et al. 2009. Farnesol-induced apoptosis in Candida albicans. Antimicrob. Agents Chemother. 53:2392–2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sithanandam G., Ramakrishna G., Diwan B. A., Anderson L. M. 1998. Selective mutation of K-ras by N-ethylnitrosourea shifts from codon 12 to codon 61 during fetal mouse lung maturation. Oncogene 17:493–502 [DOI] [PubMed] [Google Scholar]
- 67. Sudbery P., Gow N., Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317–324 [DOI] [PubMed] [Google Scholar]
- 68. Tanaka K., et al. 1992. A dominant activating mutation in the effector region of RAS abolishes IRA2 sensitivity. Mol. Cell. Biol. 12:631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Uhl M. A., Biery M., Craig N., Johnson A. D. 2003. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 22:2668–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Uppuluri P., Mekala S., Chaffin W. L. 2007. Farnesol-mediated inhibition of Candida albicans yeast growth and rescue by a diacylglycerol analogue. Yeast 24:681–693 [DOI] [PubMed] [Google Scholar]
- 71. Vinnakota K. C., Mitchell D. A., Deschenes R. J., Wakatsuki T., Beard D. A. 2010. Analysis of the diffusion of Ras2 in Saccharomyces cerevisiae using fluorescence recovery after photobleaching. Phys. Biol. 7:026011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wächtler B., Wilson D., Haedicke K., Dalle F., Hube B. 2011. From attachment to damage: defined genes of Candida albicans mediate adhesion, invasion and damage during interaction with oral epithelial cells. PLoS One 6:e17046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Warenda A. J., Konopka J. B. 2002. Septin function in Candida albicans morphogenesis. Mol. Biol. Cell 13:2732–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Westwater C., Balish E., Schofield D. A. 2005. Candida albicans-conditioned medium protects yeast cells from oxidative stress: a possible link between quorum sensing and oxidative stress resistance. Eukaryot. Cell 4:1654–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Xu X.-L., et al. 2008. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe 4:28–39 [DOI] [PubMed] [Google Scholar]
- 76. Zheng X. D., Wang Y. M., Wang Y. 2003. CaSPA2 is important for polarity establishment and maintenance in Candida albicans. Mol. Microbiol. 49:1391–1405 [DOI] [PubMed] [Google Scholar]
- 77. Zhu Y., et al. 2009. Ras1 and Ras2 play antagonistic roles in regulating cellular cAMP level, stationary-phase entry and stress response in Candida albicans. Mol. Microbiol. 74:862–875 [DOI] [PubMed] [Google Scholar]
- 78. Zou H., Fang H. M., Zhu Y., Wang Y. 2010. Candida albicans Cyr1, Cap1 and G-actin form a sensor/effector apparatus for activating cAMP synthesis in hyphal growth. Mol. Microbiol. 75:579–591 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.