Abstract
The ability of the pathogenic fungus Candida albicans to switch cellular morphologies is important for infection and virulence. Recent studies have revealed that C. albicans yeast cells can switch to filamentous growth under genotoxic stress in a manner dependent on the DNA replication/damage checkpoint. Here, we have investigated the functions of Pph3 (orf19.4378) and Psy2 (orf19.3685), whose orthologues in Saccharomyces cerevisiae mediate the dephosphorylation of the DNA damage checkpoint kinase Rad53 and the histone variant H2AX during recovery from DNA damage. Deleting PPH3 or PSY2 causes hypersensitivity to DNA-damaging agents, including cisplatin, methylmethane sulfonate (MMS), and UV light. In addition, pph3Δ and psy2Δ cells exhibit strong filamentous growth under genotoxic stress. Flow cytometry analysis shows that the mutant cells have lost the ability to adapt to genotoxic stress and remain arrested even after the stress is withdrawn. Furthermore, we show that Pph3 and Psy2 are required for the dephosphorylation of Rad53, but not H2AX, during DNA damage recovery. Taken together, these results show that C. albicans Pph3 and Psy2 have important roles in mediating genotoxin-induced filamentous growth and regulating Rad53 dephosphorylation.
INTRODUCTION
Candida albicans is a pleiomorphic fungal pathogen that causes superficial infections of the skin, vagina, and oral epithelia and life-threatening systemic infections in immunocompromised patients (12, 31, 38). The fungus can exist in three morphological forms, including budding yeast, pseudohyphae, and hyphae (9, 45). The ability to switch between the different growth forms has proved to be important for infection and virulence (22, 30, 52). Many environmental factors can induce hyphal growth, such as appropriate temperature, neutral pH, and serum (19, 45). Several signal transduction pathways regulate the yeast-to-hypha growth transition, among which the cyclic AMP (cAMP) and mitogen-activated protein kinase (MAPK) pathways play central roles. The two pathways target transcription factors Efg1 and Cph1, respectively, to activate the expression of hypha-specific genes, leading to filamentous growth (27, 29, 30, 39, 44).
A series of recent studies reported that perturbing the cell cycle progression by various means causes filamentous growth in a manner independent of the cAMP and MAPK pathways (1–6, 8, 13, 42). For example, C. albicans yeast cells treated with the DNA replication inhibitor hydroxyurea, the DNA-alkylating agent methylmethane sulfonate (MMS), or UV radiation exhibited significant cell elongation (3, 42). Depletion of either the G1 cyclin Cln3 or one of the mitotic cyclins Clb2 and Clb4 also induces constitutive pseudohyphal growth (5, 8, 13), and switching off two cell cycle regulatory genes, CDC4 and CDC5, has similar effects (2, 4). One of the most compelling pieces of evidence for direct involvement of the cell cycle machinery is the identification of the G1 cyclin Hgc1 as a specific regulator of C. albicans hyphal development (52). However, the molecular mechanism by which interference with the cell cycle triggers filamentous growth remains unknown. A rapidly emerging theme of control seems to intimately involve the cell cycle checkpoints, based on a significant body of evidence obtained in studies in both Saccharomyces cerevisiae and Candida albicans. These checkpoints are regulatory pathways that control the order and timing of cell cycle transitions and ensure that critical events, such as bud formation, DNA replication, and chromosome segregation, are completed with high fidelity (24, 34). When these events are impaired, the checkpoints temporarily arrest cell cycle progression to provide time for repairing damage and correcting mistakes. In S. cerevisiae, defective bud construction activates the Swe1-mediated morphogenesis checkpoint to temporarily arrest cell cycle progression, leading to bud elongation (11, 50). Several DNA damage/replication checkpoint proteins, such as Mec1 and Rad53, have been found to be involved in the filamentous growth induced by DNA replication stress (25, 42). In C. albicans, depletion of Rad52 triggers the DNA damage checkpoint, leading to constitutive filamentation (1), and deleting RAD53 completely abolishes the filamentous growth caused by genotoxic stresses (42). Interestingly, the spindle assembly checkpoint is required for the filamentous growth triggered by the disruption of microtubules (6).
Reversible protein phosphorylation is a highly conserved regulatory mechanism involved in many steps of the DNA damage response. While events activated by kinases have been studied rather extensively, little is known about how cells deactivate the damage response once genotoxic stress is overcome or withdrawn. Clearly, inactivation of the cell cycle checkpoints is required for cells to resume cell cycle progression. Previous studies have indicated that the PP2A-like protein phosphatase Pph3 plays an important role in the DNA damage response pathway in S. cerevisiae: deleting PPH3 or PSY2, a gene encoding a regulatory subunit of Pph3, increases sensitivity to cisplatin and MMS (23, 51). One of the earliest events in a eukaryotic cell's response to DNA damage is serine phosphorylation of the histone variant H2AX at the C-terminal SQE motif, leading to assembly of phospho-H2AX (γH2AX)-containing nucleosomes that recruit DNA repair and signaling proteins (20). Work in S. cerevisiae has identified a complex containing Pph3 that regulates the phosphorylation status of γH2AX in vivo and efficiently dephosphorylates γH2AX in vitro (26). Genetic and biochemical evidence is available that Pph3 and Psy2 form a complex (Pph3-Psy2) that binds and dephosphorylates activated Rad53 during adaptation to and recovery from MMS-mediated DNA damage, and in the absence of Pph3-Psy2, Rad53 dephosphorylation and the resumption of DNA synthesis are delayed (35). In addition to Pph3, the PP2C-type phosphatases Ptc2 and Ptc3 also participate in Rad53 deactivation under similar conditions (28, 46, 47).
Although the deactivation of DNA checkpoints has been extensively investigated in S. cerevisiae, it remains totally unknown whether PP2A-related phosphatases have a role in DNA damage recovery and damage-induced filamentous growth in C. albicans. Here, we describe the identification and functional characterization of C. albicans PPH3 (CaPPH3) and PSY2. We found that deletion of PPH3 or PSY2 significantly increased the cell's sensitivity to MMS, cisplatin, and UV. Interestingly, after removing the DNA-damaging agents, both pph3Δ and psy2Δ mutants continued to grow in filamentous form and could not reenter the cell cycle for a long time before losing viability, while wild-type cells switched back to normal yeast growth. In addition, Rad53 dephosphorylation was blocked during recovery from DNA damage in the mutants, suggesting a role for Pph3 and Psy2 in deactivating Rad53. These results provide new insights into the molecular mechanisms underlying the deactivation of the DNA damage checkpoint and filamentous growth in C. albicans response to genotoxic stresses.
MATERIALS AND METHODS
Strains and culture conditions.
All C. albicans strains used in this study are listed in Table 1. C. albicans was routinely grown at 30°C in YPD medium (2% yeast extract, 1% peptone, and 2% glucose), in GMM (2% glucose and 6.79 g/liter yeast nitrogen base without amino acids), or in GMM supplemented with the required nutrients. For hyphal growth, yeast cells were inoculated into YPD medium containing 20% serum. Solid media contained 2% agar.
Table 1.
C. albicans strains used in this study
| Strain | Relevant genotype | Source |
|---|---|---|
| SC5314 | Wild type; clinical isolate | 21 |
| BWP17 | ura3/ura3 his1::hisG/his1::hisG arg4::hisG/arg4::hisG | 17 |
| CAI4 | ura3/ura3 | 21 |
| SJL1 | BWP17 PPH3/pph3Δ::ARG4 | This study |
| SJL2 | BWP17 pph3Δ::ARG4/pph3Δ::HIS1 | This study |
| SJL2.1 | BWP17 pph3Δ::ARG4/pph3Δ::HIS1 PPH3-URA3 | This study |
| SJL3 | BWP17 pph3Δ::ARG4/pph3Δ::HIS1 URA3 | This study |
| SJL4 | BWP17 PSY2/psy2Δ::ARG4 | This study |
| SJL5 | BWP17 psy2Δ::ARG4/psy2Δ::HIS1 | This study |
| SJL5.1 | BWP17 psy2Δ::ARG4/psy2Δ::HIS1 PSY2-URA3 | This study |
| SJL6 | BWP17 psy2Δ::ARG4/psy2Δ::HIS1 URA3 | This study |
| SJL7 | BWP17 pph3Δ::ARG4/pph3Δ::HIS1 RAD53-Myc-URA3 | This study |
| SJL8 | BWP17 psy2Δ::ARG4/psy2Δ::HIS1 RAD53-Myc-URA3 | This study |
| SJL9 | BWP17 RAD53-Myc-URA3 | This study |
C. albicans gene deletion, rescue, and epitope tagging of proteins.
C. albicans deletion mutants were constructed by sequentially deleting the two copies of the target gene from BWP17. The deletion cassettes were constructed by flanking a selectable marker gene (ARG4 or HIS1) with the AB and CD fragments, which correspond to the 5′ and 3′ untranslated regions (UTRs) of the target gene, respectively. The first copy was replaced by using the ARG4 cassette, and the second copy was deleted by using the HIS1 cassette. Homozygous deletion mutants were verified by PCR.
For gene rescue, the entire open reading frame (ORF), together with the promoter (∼1,000 bp), was cloned into the CIp10 plasmid at KpnI and ClaI sites, followed by the GAL4 3′ UTR. The construct was linearized by StuI, which is in the RP10 sequence of CIp10, and then introduced into the gene deletion strains.
For C-terminal Myc tagging of Rad53, the DNA fragment for the Myc epitope was ligated in frame to the 3′ end of the target gene in a CIp10 plasmid. After linearizing the plasmid at a unique restriction site within the coding sequence, the plasmid was transformed into strains SJL2 and SJL5, yielding C-terminally Myc-tagged proteins.
Cisplatin, MMS, and UV sensitivity and recovery assays.
Tests of sensitivity to DNA-damaging agents on solid and liquid media were carried out as described previously (42). For the damage recovery experiments, cells were grown in YPD medium at 30°C overnight and diluted in fresh YPD medium at a concentration of 5 × 106 cells/ml. After 2 h of growth at 30°C, MMS or cisplatin was added to a final concentration of 0.02% or 1 mM, respectively. After 2 h of drug treatment, cells were harvested, washed twice with distilled water, and resuspended in fresh YPD medium without drugs for further growth.
Protein extraction, Western blotting, and protein dephosphorylation.
Cells were harvested by centrifugation, and ∼100 mg of cells was resuspended in 200 μl of ice-cold RIPA buffer (8). After adding an equal volume of acid-washed glass beads (Sigma-Aldrich), the cells were broken by four rounds of 45 s of beating at 5,000 rpm in a MicroSmash MS-100 bead beater (Tomy Medico, Minato-ku, Japan) with 2 min of cooling on ice between rounds. The supernatant was collected after centrifugation of the cell lysate at 13,000 rpm for 20 min at 4°C. The protein concentrations of the supernatant were determined using the bicinchoninic acid (BCA) protein assay (Galen). For Western blots, 30 μg of protein was separated by 8% or 12% SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Millipore). The membrane was immersed in Tris-buffered saline containing 0.1% Tween 20 (TBST) and 5% milk for 1 h at room temperature, followed by 1 h in TBST containing 1% milk and the primary antibody and then in TBST containing 1% milk and the second antibody conjugated with hydrogen peroxidase. The target protein was visualized by using an enhanced-chemiluminescence (ECL) system. The Hta-P antibody was purchased from Upstate Millipore and the anti-Cdc28 (PSTAIRE) antibody from Santa Cruz. Protein dephosphorylation was carried out as described previously (7). Lambda phosphatase was purchased from New England BioLabs (catalog no. P07535).
Microscopy and flow cytometry.
Staining of nuclei and chitin was carried out as previously described (52). A Zeiss 510 metamicroscope and cell observer system (Carl Zeiss MicroImaging, Germany) were used for imaging. Flow cytometry was carried out as described previously (42).
RESULTS
Identification of CaPPH3 and CaPSY2.
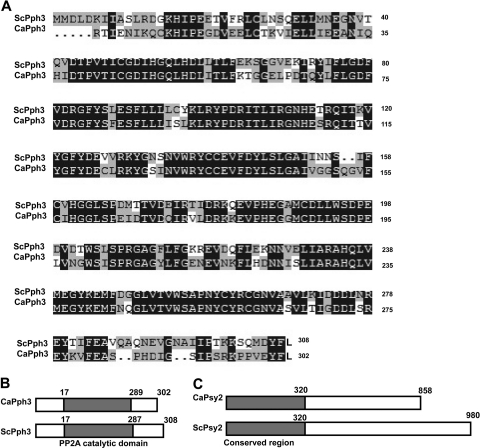
The C. albicans genome (http://www.candidagenome.org) contains candidate orthologues of S. cerevisiae PPH3 (ScPPH3) and ScPSY2. CaPPH3 (orf19.4378) encodes a protein of 302 amino acids (aa), which is 6 aa shorter than ScPph3. The two proteins share ∼70% identity (Fig. 1 A), and each contains a highly conserved PP2A-like catalytic domain (Fig. 1B), strongly suggesting that CaPPH3 is orthologous to ScPPH3. CaPSY2 (orf19.3685) encodes a 980-aa protein, which is 122 aa longer than ScPsy2. Although the two proteins share low overall sequence identity, a conserved region with 68% similarity is found in the N termini of the proteins (Fig. 1C).
Fig. 1.
Sequence and domain comparison of CaPph3 and CaPsy2 with S. cerevisiae orthologues. (A) Alignment of the CaPph3 amino acid sequence with that of ScPph3. Dark shading indicates identical residues, and light shading indicates similar residues. (B and C) Domain organizations of CaPph3 (B) and CaPsy2 (C) compared with their S. cerevisiae counterparts.
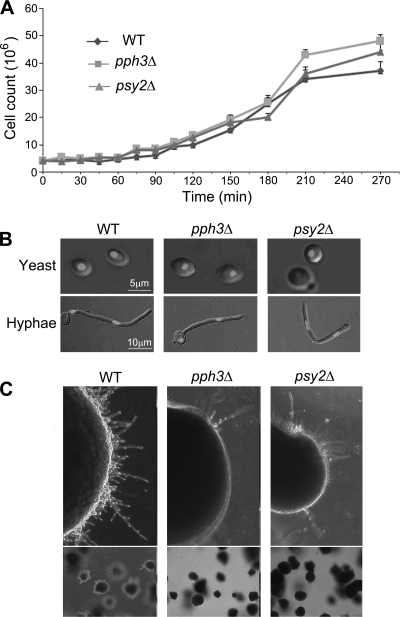
To investigate the functions of Pph3 and Psy2 in C. albicans, we constructed PPH3 and PSY2 deletion mutants by sequentially replacing the entire coding region of the two copies of the target gene with the selectable markers ARG4 and HIS1. The pph3Δ and psy2Δ mutants grew at rates similar to those of the wild-type strains (SC5314 and BWP17) in both liquid and solid YPD medium under unperturbed conditions (Fig. 2 A and 3). Like wild-type cells, both mutants grew as yeast at 30°C in liquid YPD medium and switched to hyphal growth at 37°C in the presence of serum (Fig. 2B). On solid media, the mutants grew colonies with appearances indistinguishable from those of wild-type colonies (data not shown). However, when grown under embedded conditions, the colonies of the pph3Δ or psy2Δ mutant exhibited less filamentous growth at the edge than wild-type colonies (Fig. 2C), indicating that Pph3 and Psy2 are involved in cell morphology under certain conditions.
Fig. 2.
Effects of PPH3 and PSY2 deletion on growth rate and cell morphology. (A) Cells (5 × 105) of wild-type (WT) (SC5314, BWP17, and BWP17 ura+ strains gave similar results in the experiments described in this study), pph3Δ (SJL3), and psy2Δ (SJL6) strains (all strains used are listed in Table 1) were inoculated into liquid YPD medium in starting cultures. The cells were counted microscopically every 15 min. The experiment was done in triplicate, and the average of each time point was used to generate the growth curves. The error bars indicate 1 standard deviation (SD). (B) Cell morphology. Cells from the same strains as in panel A were grown in YPD medium at 30°C for 16 h or in YPD medium containing 20% serum at 37°C for 3 h. Nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole). (C) pph3Δ (SJL3) and psy2Δ (SJL6) mutants showed decreased filamentation under embedded conditions. Cells were embedded in YPD agar and grown at 30°C for 48 h.
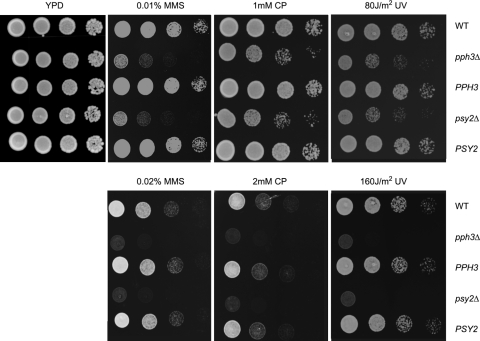
Fig. 3.
pph3Δ and psy2Δ mutant cells are hypersensitive to DNA-damaging agents. Cells of WT (SC5314 or BWP17), pph3Δ (SJL3), and psy2Δ (SJL6) strains and the rescued PPH3 (SJL2.1) and PSY2 (SJL5.1) strains were serially diluted, spotted onto YPD plates containing different concentrations of cisplatin (CP) and MMS, and incubated at 30°C for 24 h. For UV treatment, serially diluted cells were spotted onto YPD plates and then irradiated with UV (80 or 160 J/m2), followed by incubation at 30°C for 24 h. All experiments were repeated using at least 3 independent clones for each mutant.
CaPPH3 and CaPSY2 are required for resistance to DNA-damaging agents.
Recently, PPH3 and PSY2 have been identified in a genome-wide screen of S. cerevisiae genes for hypersensitivity to the DNA-damaging agents cisplatin and oxaliplatin (51). Deletion of PPH3 or PSY2 also led to a moderate increase in sensitivity to MMS (23). Consistently, we found that C. albicans pph3Δ and psy2Δ mutants are more sensitive than wild-type cells to DNA-damaging agents, including cisplatin, MMS, and UV (Fig. 3). When a copy of the wild-type PPH3 or PSY2 gene was reintroduced into the respective gene deletion mutant, sensitivity to genotoxins was fully rescued (Fig. 3). Taken together, these data indicate that the protein phosphatase Pph3 and its regulatory subunit Psy2 play evolutionarily conserved roles in regulating cellular response to DNA damage.
PPH3 and PSY2 have roles in cell cycle arrest and filamentous growth induced by DNA damage.
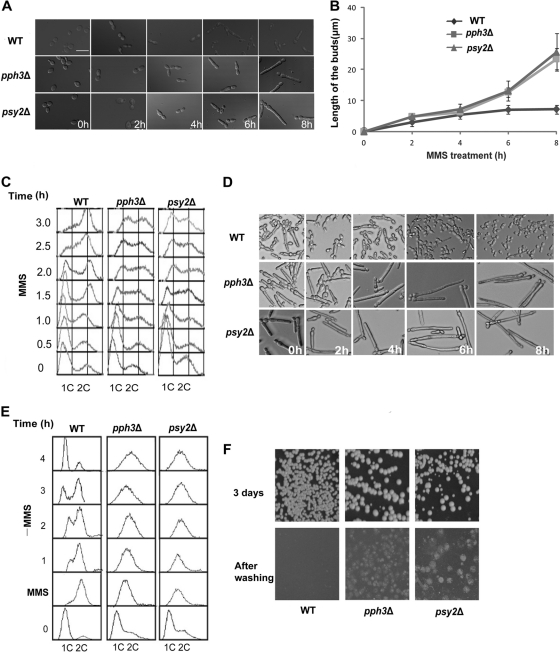
In the past few years, a significant number of publications reported that C. albicans can switch to filamentous growth when treated with genotoxins (3, 42). To investigate the roles of Pph3 and Psy2 in response to DNA damage in C. albicans, we first examined the phenotypes of the pph3Δ and psy2Δ mutants during constant exposure to MMS. We found that when treated with 0.02% MMS, both wild-type and mutant cells switched to a pseudohyphal form of growth. However, the mutant cells were much more elongated than wild-type cells after 4 h (Fig. 4 A), and a similar phenotype was also observed when cells were grown on solid media (data not shown). We measured the lengths of the filaments (n = 30) and found that the average length of pph3Δ and psy2Δ cells was about 25 μm, in contrast to 8 μm for wild-type cells, after exposure to MMS for 8 h (Fig. 4B). Flow cytometry analysis of MMS-treated yeast cells showed that while nearly all the wild-type cells entered G2/M phase at 3 h, a large fraction of pph3Δ and psy2Δ cells were still in S phase (Fig. 4C), indicating that the Pph3 complex is necessary for cells to go through S phase in the presence of MMS. We next examined pph3Δ and psy2Δ cells for morphogenesis and cell cycle progression during recovery from MMS-induced DNA damage. Interestingly, after washing off MMS, wild-type cells reentered the cell cycle and generated round cells from the tips of the filamentous cells, whereas the majority of pph3Δ and psy2Δ cells remained arrested and continued to elongate for at least 8 h (Fig. 4D and E). Methylene blue viability staining of cells at 8 h after washing off MMS did not show a significant increase in positively stained cells in the mutants (data not shown), further indicating that the mutant cells were alive at this time point. However, when we spread the treated cells onto plates without genotoxins, we found that the CFU were dramatically reduced, by ∼90% and ∼95%, for the mutant cells after treatment with 0.02% MMS for 6 h and 8 h, respectively. The results indicate that although the MMS-treated mutant cells remained alive for at least 8 h, the majority of them eventually lost viability and were unable to form visible colonies on plates. In contrast, few wild-type cells lost viability. Moreover, after removing MMS, pph3Δ and psy2Δ cells plated onto YPD solid medium showed strong invasive growth. After 3 days of incubation, we washed the cells off the agar surface and found that, unlike the colonies of wild-type cells, which were easily removed, pph3Δ and psy2Δ cells had grown into the agar, further demonstrating continuous filamentous growth of the mutant cells after the removal of MMS (Fig. 4F). These data indicate that cells lacking PPH3 or PSY2 elicit prolonged S-phase arrest in response to DNA damage by MMS and have great difficulty in resuming DNA synthesis and cell cycle progression even after the removal of the DNA-damaging agent. Together, the results showed that pph3Δ and psy2Δ cells are defective in recovery from the cell cycle arrest and filamentous growth induced by DNA damage. pph3Δ and psy2Δ cells exhibited similar responses when treated with cisplatin (data not shown).
Fig. 4.
pph3Δ and psy2Δ cells exhibit irreversible pseudohyphal growth and cell cycle arrest when treated with MMS. (A) WT (SC5314 or BWP17), pph3Δ (SJL3), and psy2Δ (SJL6) yeast cells were grown in YPD medium supplemented with 0.02% MMS at 30°C. Cells were collected for microscopic examination at timed intervals. Bar = 5 μm. (B) Comparison of filament lengths of cells treated with MMS. Filament length was measured using ImageJ (http://rsbweb.nih.gov/ij/index.html). The data points show the average of 30 cells. The experiment was repeated 3 times. The error bars indicate 1 SD. (C) Cells were treated as described for panel A and harvested at intervals for flow cytometry analysis. (D and E) The same cells used in panel A were incubated in YPD medium containing 0.02% MMS for 6 h before washing the cells with fresh YPD and inoculating the MMS-treated cells into MMS-free YPD medium. Aliquots were harvested at intervals for microscopy and flow cytometry analysis. (F) pph3Δ and psy2Δ cells invade agar during recovery from DNA damage. BWP17 and mutant (SJL3 and SJL6) cells that had been grown in YPD medium containing 0.02% MMS for 6 h were spread onto YPD agar plates and incubated at 30°C. As ∼90% of the mutant cells lost viability, after the MMS treatment, the cells were first concentrated by ∼10-fold before being spread onto the plates so that more colonies would grow. The plates were photographed after 3 days and then washed to remove cells on the agar surface and rephotographed.
Pph3 and Psy2 are not required for HTA dephosphorylation during recovery from genotoxic stress.
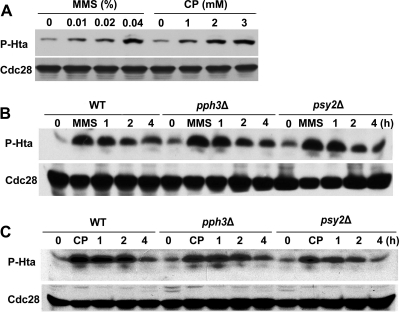
Phosphorylation of histone H2AX is one of the earliest events in the response to DNA double-strand breaks (DSB) (40). When cells have adapted to DNA damage or when the damage stress is removed, cells deactivate the DNA damage checkpoint in which γH2AX needs to be dephosphorylated (26). Recently, a Pph3 complex has been implicated as a γH2AX phosphatase in both mammal and yeast (15, 26, 32). To further understand the filamentous growth and the cell cycle defect of pph3Δ and psy2Δ mutants during DNA damage recovery, we sought to determine whether C. albicans Pph3 is also involved in γH2AX dephosphorylation. We first needed to identify the C. albicans histone H2AX, which had not been studied. The C. albicans genome contains candidate orthologues of ScH2AX. CaHta1 (orf19.1051) and CaHta2 (orf19.6924) are two histone H2A subtypes. The amino acid sequences of the two proteins are nearly identical, with only two differences. Both proteins contain the conserved C-terminal SQE motif. Comparison of the CaHta sequences with those of S. cerevisiae showed considerable evolutionary conservation, with 92% identity, suggesting that CaHta is orthologous to ScH2AX. Western blot analysis using an antibody specific for phospho-H2A showed that the amount of phosphorylated CaHta (P-Hta) in wild-type cells increased with increasing concentrations of cisplatin or MMS (Fig. 5 A), indicating that CaHta, like H2AX in mammalian cells and S. cerevisiae, becomes phosphorylated in response to DNA damage. Together, the high sequence conservation and the DNA damage-induced phosphorylation strongly support an orthologous relationship of CaHta and H2AX.
Fig. 5.
PPH3 and PSY2 are dispensable for CaHta dephosphorylation during recovery from genotoxic stress. (A) BWP17 cells were incubated with various concentrations of MMS or cisplatin for 1 h and then harvested for immunoblot analysis using the anti-Hta-P antibody. Cdc28 was probed with anti-PSTAIRE antibody as a loading control. (B and C) Wild-type (BWP17), pph3Δ (SJL3), and psy2Δ (SJL6) cells were incubated in YPD medium containing 0.02% MMS or 2 mM cisplatin for 2 h. MMS and cisplatin were then washed out, and the cells were resuspended in YPD medium and incubated at 30°C. Cells were harvested at the indicated time points for immunoblot analysis using the Hta-P antibody. Cdc28 was probed with anti-PSTAIRE antibody as a loading control.
Next, we wished to explore whether Pph3 and Psy2 are required for Hta dephosphorylation during DNA damage recovery. In untreated pph3Δ and psy2Δ cells, we found that P-Hta was barely detectable, as in untreated wild-type cells, and the level of P-Hta increased similarly upon treatment with cisplatin or MMS in both the mutant and wild-type cells. After the genotoxins were washed off, P-Hta levels deceased gradually to near basal levels in ∼4 h at similar rates in wild-type and mutant cells (Fig. 5B and C). Thus, the results indicate that Pph3 and Psy2 are not required for HTA dephosphorylation during DNA damage recovery in C. albicans.
Pph3 and Psy2 are required for Rad53 dephosphorylation during DNA damage recovery.
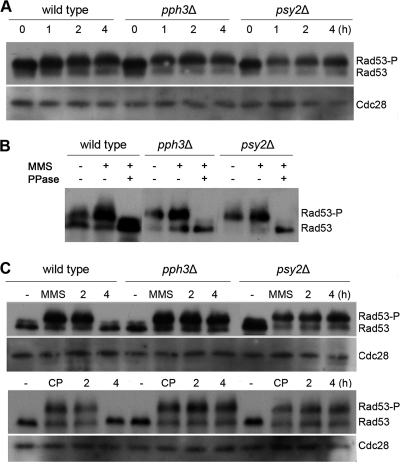
Activation of Rad53 is a critical response to DNA damage that results in the activation of gene transcription, stabilization of stalled replication forks, inhibition of late-origin firing, and delayed entry into mitosis (53). Previous work in S. cerevisiae showed that Pph3 and Psy2 are required for Rad53 deactivation during MMS treatment and recovery from MMS-caused DNA damage (35). Recently, Shi et al. (42) identified Rad53 in C. albicans. We therefore wished to examine whether PPH3 and PSY2 are required for Rad53 dephosphorylation during recovery from DNA damage in C. albicans. Using Myc-tagged Rad53 in Western blot analysis, we detected that wild-type, pph3Δ, and psy2Δ cells all accumulated partially phosphorylated Rad53 after 1 h of MMS or cisplatin treatment and hyperphosphorylated Rad53 at 4 h (Fig. 6 A and B). We next examined the phosphorylation state of Rad53 in cells during recovery from MMS and cisplatin treatment. After removing MMS or cisplatin, Rad53 was completely dephosphorylated at 4 h in wild-type cells, while it remained hyperphosphorylated at that time point in both pph3Δ and psy2Δ cells (Fig. 6C). These data demonstrate that Pph3 and Psy2 are required for Rad53 dephosphorylation during recovery from DNA damage.
Fig. 6.
Pph3 and Psy2 regulate the phosphorylation state of Rad53 in C. albicans. (A) Wild-type (SJL9), pph3Δ (SJL7), and psy2Δ (SJL8) cells were incubated in YPD medium containing 0.02% MMS. Cells were harvested at the indicated time points for immunoblot analysis with anti-Myc antibody. Cdc28 was probed with anti-PSTAIRE antibody as a loading control. (B) Rad53 hyperphosphorylation in MMS-treated cells. The lysate of SJL9 (wild type with RAD53-Myc), SJL7 (pph3Δ RAD53-Myc), and SJL8 (psy3Δ RAD53-Myc) cells that had been grown in the presence of (+) 0.02% MMS for 4 h was divided into 2 parts. One was treated with λ-phosphatase (PPase), and the other was mock treated with the reaction buffer alone. The two samples, along with untreated cell lysates, were then subjected to Western blot analysis using anti-Myc antibody. (C) Wild-type (SJL9), pph3Δ (SJL7), and psy2Δ (SJL8) cells were incubated in YPD medium containing 0.02% MMS or 2 mM cisplatin for 6 h. Untreated cells (−) were included as a control. MMS and cisplatin were washed out, and the cells were resuspended in YPD medium at 30°C. The cells were harvested at the indicated time points for immunoblot analysis with anti-Myc antibody. Cdc28 was probed with anti-PSTAIRE antibody as a loading control.
DISCUSSION
CaPPH3 and CaPSY2 are required for resistance to DNA-damaging agents.
We have identified the C. albicans PPH3 and PSY2 genes based on sequence identity to their S. cerevisiae homologues. In S. cerevisiae, Pph3 interacts with the regulatory subunits Psy2 and Psy4, and this complex dephosphorylates γH2AX during DNA damage and repair. These three proteins and their interactions are conserved through evolution from yeast to Drosophila to mammals (16). In S. cerevisiae, Pph3 and Psy2 play roles in Rad53 dephosphorylation during recovery from MMS or cisplatin (23, 51), while Psy4 participates only in recovery from cisplatin treatment. In mammalian cells, inhibition of PP4C expression, the orthologue of Pph3, also sensitizes cancer cells to cisplatin treatment (49). Consistently, our data showed that pph3Δ and psy2Δ cells are hypersensitive to DNA-damaging agents, including cisplatin, UV, and MMS. We also found that wild-type cells could adapt to the genotoxins after several hours. Even in the continuous presence of the genotoxins, they were able to exit from the DNA damage-induced S-phase arrest, reenter the cell cycle, and switch from filamentous growth back to yeast growth. In contrast, pph3Δ and psy2Δ cells remained arrested and continued to elongate for a long time before losing viability. This explains why PPH3 or PSY2 deletion increases sensitivity to genotoxins. These data indicate that the evolutionarily conserved protein phosphatase Pph3 complex plays an important role in the DNA damage response.
We searched for the homologue of ScPSY4 in the C. albicans genome database but did not find any significant match. Perhaps C. albicans has only one regulatory subunit for Pph3.
CaPPH3 and CaPSY2 are dispensable for Hta dephosphorylation.
In S. cerevisiae, one of the earliest events in cellular response to DNA damage is the phosphorylation of H2AX by the checkpoint kinases Tel1 and Mec1, which are orthologues of the mammalian ATM and ATR, respectively. The phosphorylation occurs at Ser129 in the C-terminal SQE motif, and the phosphorylated H2AX accumulates at the sites of DSB (20, 37, 40). This modification is important for maintaining cell cycle arrest and the repair of DNA damage by recruiting DSB recognition and repair factors to the break site, including DNA damage checkpoint proteins, chromatin remodelers, and cohesins (37, 40). In the C. albicans genome database, we identified candidate genes for the histone H2A variant H2AX. CaHta is highly similar to H2AX in both sequence and domain organization. Our data show that CaHta is also rapidly phosphorylated at the conserved SQE motif upon DNA damage and is dephosphorylated after removing the DNA-damaging agent. In S. cerevisiae, a protein complex containing the phosphatase Pph3 and its regulatory subunits Psy2 and Psy4 has been shown to regulate the H2AX phosphorylation level in vivo and to efficiently dephosphorylate γH2AX in vitro (26). Recently, in mammalian cells, PP4C, the orthologue of S. cerevisiae Pph3, has been reported to dephosphorylate γH2AX (15, 32). Unexpectedly, we found that CaPph3 and CaPsy2 are dispensable for CaHta dephosphorylation during DNA damage recovery. In mammalian cells, PP2A isoforms, the orthologues of S. cerevisiae Pph21 and Pph22, have been reported to directly bind to and dephosphorylate γH2AX (14), and in some cases, other protein phosphatases have redundant functions with Pph3 (10). Therefore, we propose that CaHta dephosphorylation during DNA damage recovery might be dependent on other protein phosphatase(s) in C. albicans.
CaPPH3 and CaPSY2 play important roles in Rad53 dephosphorylation during DNA damage recovery.
Activation of the checkpoint kinase Rad53 is an essential event in activating cellular responses when DNA damage occurs (53). Checkpoint activation is crucial to the maintenance of genome integrity in the cell's response to genotoxic insults. Once the damage is repaired, checkpoint deactivation is necessary for the cell cycle to resume. Previous studies in S. cerevisiae found that the PP2C-type phosphatases Ptc2 and Ptc3 were necessary for Rad53 deactivation after the DNA double-strand break was repaired (28). More recently, the phosphatase Pph3 complex was shown to be required for Rad53 dephosphorylation after introduction of a repairable but long-lived double-strand break (35). Here, we demonstrate that CaPph3 and CaPsy2 are required for Rad53 dephosphorylation during DNA damage recovery. Deletion of CaPPH3 or CaPSY2 has little effect on the rate of Rad53 hyperphosphorylation upon exposure to MMS or cisplatin. However, after removing the genotoxins, Rad53 dephosphorylation was complete at 4 h in wild-type cells while Rad53 remained fully phosphorylated in pph3Δ or psy2Δ cells for at least 4 h. In S. cerevisiae, Rad53 plays a crucial role in the stabilization of replication forks in response to DNA damage and in the regulation of DNA replication restart during recovery (46). Our study shows that treatment of pph3Δ or psy2Δ mutants with cisplatin or MMS caused a significantly prolonged S-phase arrest compared with wild-type cells. In addition, the majority of mutant cells did not resume DNA synthesis and cell cycle progression even after the removal of the DNA-damaging agents, although they continued to elongate for hours. Taken together, our data indicate that CaPph3 and CaPsy2 are required for Rad53 dephosphorylation during DNA damage recovery, and Rad53 hyperphosphorylation in pph3Δ and psy2Δ mutants is most likely the reason behind the prolonged S-phase arrest after the removal of genotoxins.
DNA damage checkpoint and pathogen adaptation to host defense.
Cell cycle checkpoints are regulatory pathways that control the order and timing of cell cycle transitions (24, 34). When these events are impaired, the checkpoint pathways temporarily arrest cell cycle progression to provide time for cells to repair damage. In the present study, we showed that the Capph3Δ and Capsy2Δ mutants grew normally under unperturbed conditions but exhibited severe defects in their ability to adapt to genotoxic stresses, such as exposure to MMS or cisplatin. The mutant cells failed to resume cell cycle progression even after removing the genotoxins, and many cells eventually lost viability. The observed persistent hyperphosphorylation of Rad53 indicates that the pph3Δ and psy2Δ mutants cannot deactivate the DNA damage checkpoint.
When microbial pathogens are attacked by host defense mechanisms during infection, cell cycle checkpoints may prove to be important for pathogens to repair damage that threatens the integrity of the genome (6). For example, in the case of attacks by reactive oxygen or nitrogen species (33), the DNA damage checkpoint is expected to be activated. Because of the inability of pph3Δ and psy2Δ mutants to deactivate the DNA damage checkpoint, they may not be able to survive in the host. Thus, the Pph3 phosphatase might be targeted in developing antifungal drugs.
Roles of CaPph3 and CaPsy2 in the filamentous growth induced by genotoxic stress.
Several recent publications have reported that perturbation of cell cycle progression causes pseudohyphal growth in C. albicans and cell cycle regulatory factors and that checkpoint proteins can influence cell morphogenesis. Deleting RAD53 causes hypersensitivity to genotoxins, failure to arrest the cell cycle, and loss of filamentous growth induced by either DNA damage or DNA replication disruption (42). Furthermore, studies in S. cerevisiae clearly established an essential role for the Mec1/Rad53 checkpoint proteins in the induction of filamentous growth by slowed DNA synthesis (25). Our results showed that Capph3Δ and Capsy2Δ mutants grew highly elongated filaments when DNA damage occurred and failed to revert to yeast growth after the removal of the damaging agents. Flow cytometry data revealed that pph3Δ and psy2Δ mutants were mainly arrested in S phase under genotoxic stress, consistent with a similar result in S. cerevisiae showing cisplatin-induced intra-S-phase delay and constitutive filamentation in a PPH3 deletion mutant (48).
Rad53 activation correlates with its hyperphosphorylation, as does its deactivation with its dephosphorylation (36). The failure of pph3Δ and psy2Δ cells to dephosphorylate Rad53 suggests continuous activation of the DNA damage checkpoint. Rad53 may promote cell elongation in two ways. One is by activating cell cycle arrest, which has been shown to lead to cell elongation in many previous reports (1, 3, 4, 6, 8, 41, 42). Rad53 has also been suggested to have a direct role in C. albicans true hyphal growth (42) and bud morphogenesis in S. cerevisiae (18, 43). Recently, two studies provided evidence suggesting that Rad53 controls cell morphogenesis through interaction with septins in S. cerevisiae (18, 43). Taken together, our data provide further evidence that Rad53 hyperphosphorylation plays an important role in DNA damage-induced filamentous growth.
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (no. 2007CB914401) and the Fundamental Research Funds for the Central Universities (no. 105566GK).
Footnotes
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Andaluz E., Ciudad T., Gomez-Raja J., Calderone R., Larriba G. 2006. Rad52 depletion in Candida albicans triggers both the DNA-damage checkpoint and filamentation accompanied by but independent of expression of hypha-specific genes. Mol. Microbiol. 59:1452–1472 [DOI] [PubMed] [Google Scholar]
- 2. Atir-Lande A., Gildor T., Kornitzer D. 2005. Role for the SCFCDC4 ubiquitin ligase in Candida albicans morphogenesis. Mol. Biol. Cell 16:2772–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bachewich C., Nantel A., Whiteway M. 2005. Cell cycle arrest during S or M phase generates polarized growth via distinct signals in Candida albicans. Mol. Microbiol. 57:942–959 [DOI] [PubMed] [Google Scholar]
- 4. Bachewich C., Thomas D. Y., Whiteway M. 2003. Depletion of a polo-like kinase in Candida albicans activates cyclase-dependent hyphal-like growth. Mol. Biol. Cell 14:2163–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bachewich C., Whiteway M. 2005. Cyclin Cln3p links G1 progression to hyphal and pseudohyphal development in Candida albicans. Eukaryot. Cell 4:95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bai C., Ramanan N., Wang Y. M., Wang Y. 2002. Spindle assembly checkpoint component CaMad2p is indispensable for Candida albicans survival and virulence in mice. Mol. Microbiol. 45:31–44 [DOI] [PubMed] [Google Scholar]
- 7. Bambach A., et al. 2009. Goa1p of Candida albicans localizes to the mitochondria during stress and is required for mitochondrial function and virulence. Eukaryot. Cell 8:1706–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bensen E. S., Clemente-Blanco A., Finley K. R., Correa-Bordes J., Berman J. 2005. The mitotic cyclins Clb2p and Clb4p affect morphogenesis in Candida albicans. Mol. Biol. Cell 16:3387–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berman J., Sudbery P. E. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918–930 [DOI] [PubMed] [Google Scholar]
- 10. Bertram P. G., et al. 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275:35727–35733 [DOI] [PubMed] [Google Scholar]
- 11. Booher R. N., Deshaies R. J., Kirschner M. W. 1993. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 12:3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calderone R. A., Fonzi W. A. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327–335 [DOI] [PubMed] [Google Scholar]
- 13. Chapa y Lazo B., Bates S., Sudbery P. 2005. The G1 cyclin Cln3 regulates morphogenesis in Candida albicans. Eukaryot. Cell 4:90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chowdhury D., et al. 2005. gamma-H2AX dephosphorylation by protein phosphatase 2A facilitates DNA double-strand break repair. Mol. Cell 20:801–809 [DOI] [PubMed] [Google Scholar]
- 15. Chowdhury D., et al. 2008. A PP4-phosphatase complex dephosphorylates gamma-H2AX generated during DNA replication. Mol. Cell 31:33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cohen P. T., Philp A., Vazquez-Martin C. 2005. Protein phosphatase 4—from obscurity to vital functions. FEBS Lett. 579:3278–3286 [DOI] [PubMed] [Google Scholar]
- 17. Enloe B., Diamond A., Mitchell A. P. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Enserink J. M., Smolka M. B., Zhou H., Kolodner R. D. 2006. Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae. J. Cell Biol. 175:729–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ernst J. F. 2000. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology 146:1763–1774 [DOI] [PubMed] [Google Scholar]
- 20. Fernandez-Capetillo O., Lee A., Nussenzweig M., Nussenzweig A. 2004. H2AX: the histone guardian of the genome. DNA Repair 3:959–967 [DOI] [PubMed] [Google Scholar]
- 21. Fonzi W. A., Irwin M. Y. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gow N. A., Brown A. J., Odds F. C. 2002. Fungal morphogenesis and host invasion. Curr. Opin. Microbiol. 5:366–371 [DOI] [PubMed] [Google Scholar]
- 23. Hanway D., et al. 2002. Previously uncharacterized genes in the UV- and MMS-induced DNA damage response in yeast. Proc. Natl. Acad. Sci. U. S. A. 99:10605–10610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hartwell L. H., Weinert T. A. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science 246:629–634 [DOI] [PubMed] [Google Scholar]
- 25. Jiang Y. W., Kang C. M. 2003. Induction of S. cerevisiae filamentous differentiation by slowed DNA synthesis involves Mec1, Rad53 and Swe1 checkpoint proteins. Mol. Biol. Cell 14:5116–5124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Keogh M. C., et al. 2006. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 439:497–501 [DOI] [PubMed] [Google Scholar]
- 27. Lane S., Birse C., Zhou S., Matson R., Liu H. 2001. DNA array studies demonstrate convergent regulation of virulence factors by Cph1, Cph2, and Efg1 in Candida albicans. J. Biol. Chem. 276:48988–48996 [DOI] [PubMed] [Google Scholar]
- 28. Leroy C., et al. 2003. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell 11:827–835 [DOI] [PubMed] [Google Scholar]
- 29. Liu H., Kohler J., Fink G. R. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723–1726 [DOI] [PubMed] [Google Scholar]
- 30. Lo H. J., et al. 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90:939–949 [DOI] [PubMed] [Google Scholar]
- 31. Morgan J. 2005. Global trends in candidemia: review of reports from 1995-2005. Curr. Infect. Dis. Rep. 7:429–439 [DOI] [PubMed] [Google Scholar]
- 32. Nakada S., Chen G. I., Gingras A. C., Durocher D. 2008. PP4 is a gamma H2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 9:1019–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nathan C., Shiloh M. U. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U. S. A. 97:8841–8848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nyberg K. A., Michelson R. J., Putnam C. W., Weinert T. A. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617–656 [DOI] [PubMed] [Google Scholar]
- 35. O'Neill B. M., et al. 2007. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc. Natl. Acad. Sci. U. S. A. 104:9290–9295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pellicioli A., et al. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petrini J. H., Stracker T. H. 2003. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 13:458–462 [DOI] [PubMed] [Google Scholar]
- 38. Richardson M. D. 2005. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 56(Suppl. 1):i5–i11 [DOI] [PubMed] [Google Scholar]
- 39. Rocha C. R., et al. 2001. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans. Mol. Biol. Cell 12:3631–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rogakou E. P., Boon C., Redon C., Bonner W. M. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanchez Y., et al. 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286:1166–1171 [DOI] [PubMed] [Google Scholar]
- 42. Shi Q. M., Wang Y. M., Zheng X. D., Lee R. T., Wang Y. 2007. Critical role of DNA checkpoints in mediating genotoxic-stress-induced filamentous growth in Candida albicans. Mol. Biol. Cell 18:815–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Smolka M. B., et al. 2006. An FHA domain-mediated protein interaction network of Rad53 reveals its role in polarized cell growth. J. Cell Biol. 175:743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stoldt V. R., Sonneborn A., Leuker C. E., Ernst J. F. 1997. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 16:1982–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sudbery P., Gow N., Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317–324 [DOI] [PubMed] [Google Scholar]
- 46. Szyjka S. J., et al. 2008. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev. 22:1906–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Travesa A., Duch A., Quintana D. G. 2008. Distinct phosphatases mediate the deactivation of the DNA damage checkpoint kinase Rad53. J. Biol. Chem. 283:17123–17130 [DOI] [PubMed] [Google Scholar]
- 48. Vázquez-Martin C., Rouse J., Cohen P. T. 2008. Characterization of the role of a trimeric protein phosphatase complex in recovery from cisplatin-induced versus noncrosslinking DNA damage. FEBS J. 275:4211–4221 [DOI] [PubMed] [Google Scholar]
- 49. Wang B., et al. 2008. Protein phosphatase PP4 is overexpressed in human breast and lung tumors. Cell Res. 18:974–977 [DOI] [PubMed] [Google Scholar]
- 50. Wittenberg C., La Valle R. 2003. Cell-cycle-regulatory elements and the control of cell differentiation in the budding yeast. Bioessays 25:856–867 [DOI] [PubMed] [Google Scholar]
- 51. Wu H. I., Brown J. A., Dorie M. J., Lazzeroni L., Brown J. M. 2004. Genome-wide identification of genes conferring resistance to the anticancer agents cisplatin, oxaliplatin, and mitomycin C. Cancer Res. 64:3940–3948 [DOI] [PubMed] [Google Scholar]
- 52. Zheng X., Wang Y. 2004. Hgc1, a novel hypha-specific G1 cyclin-related protein regulates Candida albicans hyphal morphogenesis. EMBO J. 23:1845–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou B. B., Elledge S. J. 2000. The DNA damage response: putting checkpoints in perspective. Nature 408:433–439 [DOI] [PubMed] [Google Scholar]