Abstract
Serine/threonine (S/T) protein kinases are crucial components of diverse signaling pathways in eukaryotes, including the model filamentous fungus Neurospora crassa. In order to assess the importance of S/T kinases to Neurospora biology, we embarked on a global analysis of 86 S/T kinase genes in Neurospora. We were able to isolate viable mutants for 77 of the 86 kinase genes. Of these, 57% exhibited at least one growth or developmental phenotype, with a relatively large fraction (40%) possessing a defect in more than one trait. S/T kinase knockouts were subjected to chemical screening using a panel of eight chemical treatments, with 25 mutants exhibiting sensitivity or resistance to at least one chemical. This brought the total percentage of S/T mutants with phenotypes in our study to 71%. Mutants lacking apg-1, an S/T kinase required for autophagy in other organisms, possessed the greatest number of phenotypes, with defects in asexual and sexual growth and development and in altered sensitivity to five chemical treatments. We showed that NCU02245/stk-19 is required for chemotropic interactions between female and male cells during mating. Finally, we demonstrated allelism between the S/T kinase gene NCU00406 and velvet (vel), encoding a p21-activated protein kinase (PAK) gene important for asexual and sexual growth and development in Neurospora.
INTRODUCTION
Protein phosphorylation is a central component of numerous mechanisms that regulate critical cellular functions. In eukaryotic cells, signal transduction, metabolism, movement, the circadian rhythm, and many other processes are controlled through protein phosphorylation and dephosphorylation by protein kinases and phosphatases, respectively. Together with tyrosine kinases, serine-threonine (S/T) protein kinases comprise a large class in the eukaryotic protein kinase superfamily (29). S/T kinases have serine-threonine protein kinase catalytic domains and phosphorylate serine and threonine residues of target proteins. Important roles for S/T protein kinases in regulating cellular processes have been demonstrated in many eukaryotes (for a review, see references 19 and 81).
The number of S/T kinases in eukaryotic organisms is significant, with more than 100 genes in yeast, flies, and humans (57). Serine/threonine protein kinases (PKs) can be classified into groups based on their catalytic domains. The AGC (PKA, PKG, PKC) group includes PKC and the cyclic nucleotide-activated kinases PKA and PKG, the β-adrenergic receptor kinase (βARK) family, the ribosomal S6 kinase family, NDR (nuclear Dbf2-related) kinases, and other related families (29, 70). The CK1 (casein kinase 1, or cell kinase 1) group is a small but essential group of eukaryotic kinases that includes the CK1 protein family (29, 48). The CAMK (Ca2+/calmodulin-dependent protein kinase) group contains the CAMK1 and CAMK2 protein kinase families but also contains several families of non-calcium-regulated kinases (29). STE (homologs of yeast sterile 7, 11, and 20) group kinases have been implicated in regulation of numerous mitogen-activated protein kinase (MAPK) pathways in various organisms (29, 37). The CMGC (CDK, MAPK, GSK3, CLK) group includes CDK (cyclin-dependent kinase), MAPK, GSK3 (glycogen synthase kinase 3), and CLK (CDK-like kinase) family kinases (29). The “Other” group includes many families (e.g., aurora kinase family [AUR], never-in-mitosis [NIMA]-related kinase family [NEK], polo-like kinase family [PLK], halotolerance family [HAL], Wee1 kinase family [WEE]) that are clearly eukaryotic protein kinases but cannot be easily classified into the other groups (57, 58). The “Atypical” group contains kinases that display little or no sequence similarity to eukaryotic protein kinase domains. Kinase families within this group include the histidine kinase family (HisK), phosphatidyl inositol 3′-kinase-related family (PIKK), pyruvate dehydrogenase kinase family (PDHK), and many others (57, 58). Kinases belonging to the same catalytic group are often functionally related, and accumulating studies indicate that multiple kinases can be implicated in a given cellular process (57). Therefore, a full understanding of the roles of S/T kinases cannot be accomplished without large-scale analysis. Since genome sequences are now available for a large number of organisms, systematic examination of all protein kinases in a species is more feasible.
The original annotation of the genome of the filamentous fungus Neurospora crassa predicted 89 S/T kinases (25). The most recent annotation predicts an additional 18 genes (Table 1, bottom), for a total of 107 genes (Table 1). Of the 107 S/T kinase genes, 32 have been previously analyzed (Table 1). The list includes the nine components of the three MAPK cascades (24, 42, 49, 53, 56, 68), PKA (5, 32), PKC (23), checkpoint kinase 2 (73), ATM (Ataxia telangiectasia mutated; mus-21) (87), ATR (ATM and Rad3-related) homologs (mus-9) (87), the NDR (nuclear Dbf2-related) family of S/T kinases, cot-1 and dbf-2 (20, 95), and others (see Table 1 for a complete list). Characterized Neurospora kinases have been shown to regulate a diversity of cellular functions, including polarized growth, hyphal fusion, asexual sporulation (conidiation), female sexual development, stress regulation, DNA damage responses, and the circadian clock (4, 18, 20, 22, 60, 68, 73, 84, 87, 95).
Table 1.
Summary of Neurospora serine-threonine protein kinase groups, families, and phenotypes
| Groupa | Familyb | NCUno.c | Neurospora gened | S. cerevisiae homologe | Phenotype summary |
||||
|---|---|---|---|---|---|---|---|---|---|
| Inviable | Growth of basal hyphaef | Asexual developmentg | Sexual developmenth | Chemical sensitivityi | |||||
| AGC | AKT | 03200 | stk-10 | SCH9 | R | AH | SC | ||
| AGC | AKT | 07280 | stk-50 | YPK1/YPK2 | X | —j | — | — | — |
| AGC | NDR | 03242 | stk-21 | ||||||
| AGC | NDR | 07296 | cot-1 (95) | CBK1 | R | ||||
| AGC | NDR | 07378 | stk-12 | RIM15 | AH | ||||
| AGC | NDR | 09071 | dbf-2 (20) | DBF2/DBF20 | R | C, AH | PP, P, A | NSk | |
| AGC | PDK1/PKA | 03571 | stk-23 | PKH1/PKH2 | R | T | |||
| AGC | PKA | 00682 | pkac-2 (5) | AH | |||||
| AGC | PKA | 06240 | pkac-1 (5) | TPK1/TPK2/TPK3 | R | C, AH | PP, P | NS | |
| AGC | PKC | 06544 | pkc (23) | PKC1 | X | — | — | — | — |
| AGC | RSK | 01797 | nrc-2 (49) | KIN82/FPK1 | R | C, AH | P, A | NS | |
| AGC | RSK/AKT | 03197 | stk-11 | YPK3 | |||||
| AGC | YANK | 07062 | stk-49 | ||||||
| CAMK | CAMK1 | 02283 | camk-2 | ||||||
| CAMK | CAMK1 | 09123 | camk-1 (94) | CMK1/CMK2 | R | C, AH | PP, P, A | NS | |
| CAMK | CAMK1 | 09212 | camk-4 | RCK1/RCK2 | B | ||||
| CAMK | CAMKL | 00914 | stk-16 | FRK1/KIN4 | R | PP, P, A | B, SC, FL | ||
| CAMK | CAMKL | 02245 | stk-19 | YPL150W | P, A | N/S | |||
| CAMK | CAMKL | 04566 | prk-10 | SNF1 | SC, T | ||||
| CAMK | CAMKL | 04747 | stk-31 | KIN1/KIN2 | R | C, AH | PP, P, A | NS | |
| CAMK | CAMKL | 06249 | stk-40 | PSK1/PSK2 | |||||
| CAMK | CAMKL | 08346 | mus-58 (88) | CHK1 | AH | SC | |||
| CAMK | CAMKL | 09064 | stk-53 | ||||||
| CAMK | CAMKL/CAMK unique | 04143 | stk-26 | PRR1 | |||||
| CAMK | RAD53 | 02751 | mus-59 (88) | ||||||
| CAMK | RAD53 | 02814 | prd-4 (73) | DUN1 | |||||
| CAMK | CAMK1/RAD53/CAMKL | 06486 | stk-43 | ||||||
| CK1 | CK1 | 00685 | ck-1a (27) | HRR25 | X | — | — | — | — |
| CK1 | CK1 | 04005 | ck-1b (93) | YCK1/YCK2/YCK3 | X | — | — | — | — |
| CMGC | CDK | 01435 | stk-1 | X | — | — | — | — | |
| CMGC | CDK | 03659 | prk-3 | KIN28 | T | ||||
| CMGC | CDK | 04426 | div-4 | CAK1 | R | PP, P, A | M, NS | ||
| CMGC | CDK | 06685 | stk-47 | CTK1 | R | PP, P, A | C | ||
| CMGC | CDK | 07172 | stk-8 | SSN3 | R | C, AH | PP, P, A | NS | |
| CMGC | CDK | 07580 | mdk-1 | PHO85 | X | — | — | — | — |
| CMGC | CDK | 07880 | prk-6 | ||||||
| CMGC | CK2 | 03124 | cka (60) | CKA1/CKA2 | AH | SC | |||
| CMGC | CLK | 00230 | prk-4 | KNS1 | X | — | — | — | |
| CMGC | DYRK | 06638 | stk-46 | R | PP, P, A | M | |||
| CMGC | DYRK | 07872 | prk-2 | YAK1 | R | C, AH | PP, P, A | NS | |
| CMGC | DYRK | 10853 | stk-57 | ||||||
| CMGC | GSK | 04185 | gsk-3 (20) | RIM11/MRK1 | B | ||||
| CMGC | MAPK | 02393 | mak-2 (53) | FUS3/KSS1 | R | C, AH | PP, P, A | NS | |
| CMGC | MAPK | 07024 | os-2 (92) | HOG1 | C, | PP, P, A | NS | ||
| CMGC | MAPK | 09842 | mak-1 (56, 68) | SLT2 | R | C, AH | PP, P, A | N/S | |
| CMGC | RCK | 01498 | ime-2 (38) | IME2 | R | AH | F | ||
| CMGC | SRPK | 09202 | mdk-2 | SKY1 | R | C, AH | PP, P, A | NS | |
| CMGC | CLK/SRPK | 05655 | stk-35 | R | AH | PP, P, A | NS | ||
| CMGC | CLK/SRPK | 05658 | stk-36 | R | C, AH | PP, P, A | NS | ||
| CMGC | CLK/SRPK/DYRK | 09189 | stk-54 | C | |||||
| CMGC | CLK/SRPK | 10004 | stk-56 | ||||||
| Other | AUR | 00108 | stk-13 | IPL1 | X | — | — | — | — |
| Other | CAMKK | 03523 | stk-22 | TOS3/SAK1 | R | AH | C, F | ||
| Other | CAMKK | 06177 | camk-3 | ||||||
| Other | CDC7 | 11410 | cdc7 | CDC7 | NAl | NA | NA | NA | NA |
| Other | HAL | 01940 | ptk-2 (52) | PTK1/PTK2 | R | F | |||
| Other | HAL | 04335 | stk-30 | NPR1/PRR2 | SC | ||||
| Other | HAL | 06179 | stk-5 | SAT4 | SC | ||||
| Other | IRE | 02202 | stk-14 | IRE1 | |||||
| Other | NAK | 06202 | stk-38 | ARK1/PRK1 | R | C, AH | PP, P | C, B, M | |
| Other | NAK | 07399 | stk-9 | YPL236CP | C, AH | PP, P, A | F, T, M | ||
| Other | NEK | 03187 | nim-1 (75) | KIN3 | A | B | |||
| Other | PEK | 01187 | cpc-3 (78) | GCN2 | R | ||||
| Other | PLK | 09258 | cdc5 | CDC5 | X | — | — | — | — |
| Other | RAN | 04990 | stk-17/fi (59) | SKS1/VHS1 | R | C, AH | PP, P, A | T | |
| Other | RAN | 06230 | stk-39 | ||||||
| Other | SCYI | 04755 | stk-32 | SCY1 | |||||
| Other | ULK | 00188 | apg-1 | ATG1 | R | AH | PP, P, A | F, S, SC, T, M | |
| Other | VPS15 | 06626 | stk-45 | VPS15 | |||||
| Other | WEE | 04326 | stk-29 | SWE1 | B | ||||
| Other | IKS | 08177 | stk-51 | IKS1 | |||||
| STE | STE11 | 01335 | cdc15 | PP, P, A | |||||
| STE | STE11 | 02234 | mik-1 (68) | BCK1 | R | C, AH | PP, P, A | NS | |
| STE | STE11 | 03071 | os-4 (24, 42) | SSK2/SSK22 | C, | PP, P, A | NS | ||
| STE | STE11 | 06182 | nrc-1 (49) | STE11 | R | C, AH | PP, P, A | NS | |
| STE | STE20 | 00406 | vel (this study) | CLA4/SKM1 | R | C, AH | PP, A | NS | |
| STE | STE20 | 00772 | mst-1 (20) | A | |||||
| STE | STE20 | 03894 | stk-4 | STE20 | R | T, M | |||
| STE | STE20 | 04096 | prk-9 | R | C, AH | PP, P, A | NS | ||
| STE | STE20 | 11235 | pod-6 (79) | NA | NA | NA | NA | NA | |
| STE | STE7 | 00587 | os-5 (24) | PBS2 | C, | PP, P, A | NS | ||
| STE | STE7 | 04612 | mek-2 (56) | STE7 | R | C, AH | PP, P, A | NS | |
| STE | STE7 | 06419 | mek-1 (68) | MKK1/MKK2 | NA | NA | NA | NA | NA |
| Unclassified | 02885 | stk-20 | |||||||
| Unclassified | 05638 | stk-34 | |||||||
| Unclassified | 06006 | stk-37 | |||||||
| Unclassified | 06421 | stk-41 | |||||||
| Unclassified | 06422 | stk-42 | |||||||
| Unclassified | 06583 | stk-44 | |||||||
| CMGC | CDK | 09778 | cdc28 | CDC28 | |||||
| Other | BUD32 | 04595 | prk-11 | BUD32 | |||||
| Other | SCY1 | 04279 | stk-28 | CEX1 | |||||
| Atypical | ABC1 | 03823 | stk-25 | ABC1 | |||||
| Atypical | ABC1 | 04259 | stk-27 | YLR253W | |||||
| Atypical | ABC1 | 05600 | stk-33 | YPL109C | |||||
| Atypical | BRD | 09595 | stk-55 | ||||||
| Atypical | PDHK | 03796 | stk-24 | YGL059W | |||||
| Atypical | PDHK | 06760 | stk-48 | ||||||
| Atypical | PDHK | 11744 | stk-58 | PKP1 | |||||
| Atypical | PIKK | 00274 | mus-21 (87) | TEL1 | |||||
| Atypical | PIKK | 01379 | stk-18 | TRA1 | |||||
| Atypical | PIKK | 05608 | div-18 | TOR1/TOR2 | |||||
| Atypical | PIKK | 11188 | mus-9 (87) | MEC1 | |||||
| Atypical | RIO | 07722 | rgb-40 | RIO2 | |||||
| Atypical | RIO | 08767 | stk-52 | RIO1 | |||||
| Atypical | TAF1 | 02556 | hat-2 | TAF1 | |||||
| TKL | LRRK | 05808 | tkl-1 | ||||||
Group abbreviations: AGC, PKA (protein kinase A/cyclic AMP-dependent protein kinase), PKG (protein kinase G/cGMP-dependent protein kinase), PKC (protein Kinase C); CK1 (casein kinase 1 or cell kinase 1); CAMK (Ca2+/calmodulin-dependent protein kinase); CMGC, cyclin-dependent, mitogen-activated, glycogen synthase and cyclin-dependent protein kinase-like kinases; STE, sterile; TKL, tyrosine kinase-like kinase.
Family abbreviations: AKT, oncogene protein of v-akt; NDR, nuclear Dbf2 related; PDK1, phosphoinoside-dependent protein kinase; PKA, protein kinase A or cyclic AMP-dependent protein kinase; PKC, protein kinase C; RSK, ribosomal s6 kinase; YANK, yet another novel kinase; CAMK, Ca2+/calmodulin-dependent protein kinase; CAMKL, Ca2+/calmodulin-dependent protein kinase-like kinase; RAD, radiation sensitive; CK, casein kinase or cell kinase; CDK, cyclin-dependent protein kinase; CLK, cyclin-dependent protein kinase-like kinase; DYRK, dual-specificity tyrosine phosphorylation-regulated kinase; GSK, glycogen synthase kinase; MAPK, mitogen-activated protein kinase; RCK, related to murine RCK (ros cross-hybridizing kinase); SRPK, serine-rich protein kinase; AUR, Aurora; CAMKK, Ca2+/calmodulin-dependent protein kinase kinase; CDC, cell division cycle; HAL, halotolerance; IRE, inositol-requiring protein; NAK, NF-κB-activating kinase; NEK, NIMA-related kinase; PEK, pancreatic alpha-subunit of eukaryotic initiation factor kinase; PLK, polo kinase; RAN, Ran GTPase kinase; SCY1, related to S. cerevisiae Scy1 kinase; ULK, Unc-51-like kinase; VPS, vacuolar protein sorting; WEE, small; IKS, Ira1 kinase suppressor; STE, sterile; ABC1, related to S. cerevisiae Abc1 kinase; BRD, bromodomain; PDHK, pyruvate dehydrogenase kinase; PIKK, phosphatidyl inositol 3′-kinase-related kinase; RIO, right open reading frame; TAF1, TATA-binding protein-associated factor 1; LRRK, leucine-rich repeat kinase.
Based on version 5 annotation of the Broad Institute's Neurospora crassa database (www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html).
References for previously studied genes are in parentheses. Kinase names are consistent with the Neurospora e-Compendium Project at Leeds University (http://bmbpcu36.leeds.ac.uk/∼gen6ar/newgenelist/genes/gene_list.htm). stk-18 through stk-58 were named during this study, in accordance with the e-Compendium system.
Yeast orthologs were obtained from the literature or assigned during this study based upon analyses of phylogenetic trees available at www.phylomedb.org (34, 35).
R, reduced growth.
Asexual phenotypes are represented by phenotypes in aerial hyphae (AH) or conidial development (C).
The sexual phenotypes are represented by their occurrence during protoperithecial (PP), perithecial (P), or ascospore (A) development.
Chemical phenotypes are summarized based on sensitivity or resistance to cytochalasin A (C), benomyl (B), FK-506 (F), sorbitol (S), sodium chloride (SC), fludioxonil (FL), tert-butyl hydroperoxide (T), and menadione (M).
—, phenotypic assay could not be performed due to inviability of knockout mutant.
NS, mutant was not analyzed by chemical screening due to poor growth.
NA, mutant was not analyzed for phenotypes due to simultaneous mutation of adjacent gene.
Chemical sensitivity screening has proven a robust method for identifying phenotypes for all gene deletion mutants in Saccharomyces cerevisiae, including those lacking S/T kinases (31). A proof-of-principle approach involved subjecting viable knockout mutants to sublethal concentrations of 12 chemicals, chosen to perturb a wide variety of cellular functions (69). Relative growth of mutants in the presence and absence of the chemicals was scored and compared relative to growth of the wild type. The data were then subjected to clustering analysis to allow grouping of chemicals with hypersensitive mutants. The results demonstrated known associations between gene products/pathways and chemicals, thus illustrating the viability of the approach. More importantly, this work revealed novel chemical interactions and pathways, including those with proteins of unknown function.
The Neurospora genome sequence is publicly available (25), and a large-scale gene knockout project for the ∼10,000 predicted Neurospora genes is nearing completion (12, 13). We previously investigated the impact of transcriptional regulation on vegetative growth and asexual and sexual differentiation in Neurospora by examining 103 mutants lacking transcription factor genes (13). Here, we further exploit the gene knockout collection by investigating mutants lacking each of 86 S/T protein kinase genes in the Neurospora genome. We were unable to analyze three genes due to misannotation, and another 18 were predicted after the inception of this study. Morphological and growth phenotypes were analyzed (86), with 44 out of 77 viable knockout mutants displaying defects. Chemical sensitivity screens using a panel of eight treatments were introduced to augment identification of phenotypes. Using the latter approach, we were able to identify phenotypes for 25 viable mutants, bringing the total number of kinase genes with obvious functions to 61 in Neurospora. Finally, we demonstrate allelism between one kinase gene and a mapped morphological mutation.
MATERIALS AND METHODS
Strains and culture conditions.
Vogel's minimal medium (VM) was used for vegetative growth and synthetic crossing medium (SCM) for development of female sexual structures (17). Sorbose-containing FGS medium was used for isolation of colonies on plates (17). Hygromycin (Calbiochem, San Diego, CA) was used at 200 μg/ml in media where indicated. Wild-type strains ORS-SL6a (FGSC 4200; mat a) and 74-OR23-IVA (FGSC 2489; mat A) were obtained from the Fungal Genetics Stock Center (FGSC; Kansas City, MO). Gene replacement mutants for S/T kinase genes were generated using a previously described high-throughput method (see Table S1 in the supplemental material) (13).
Mutant constructions.
Deletion cassettes for kinase genes were constructed using yeast recombinational cloning in a 96-well plate system as described previously (12, 13). Cassettes were transformed into Neurospora mutants deficient in nonhomologous end-joining DNA repair (mus-51 or mus-52) by electroporation in 96-well plates, with selection on hygromycin-containing FGS medium. Transformants were picked onto VM agar slants containing hygromycin and then crossed to wild-type 74-OR23-IVA (mat A) on SCM. Ejected ascospores were harvested from each cross and plated on FGS agar plates containing hygromycin. Spot testing was used to identify progeny that lacked the mus-51 or mus-52 mutation (both marked with bar, conferring resistance to phosphinothricin [2]) and to determine mating type. Homokaryotic mutant strains were confirmed by Southern blot analysis as described previously (13, 67).
In cases where ascospores of viable knockout mutants could not be isolated, attempts were made to isolate homokaryotic mutants in the vegetative phase by isolation of uninucleate microconidia (21) or by serial plating of macroconidia. In both of these cases, the resulting homokaryotic mutant strains retained the mus-51 or mus-52 mutant background. For microconidia isolation, transformants were inoculated on 0.1× SCM agar slants containing 0.5% sucrose and 1.0 mM sodium iodoacetate and incubated at 25°C under constant light for at least 10 days (21). Microconidia were harvested using 2.5 ml sterile water and then filtered through a Millex Durapore filter unit (5 μm; Millipore, Bedford, MA; microconidia pass through, while larger macroconidia are retained on the filter). Microconidia were collected by centrifugation, resuspended in sterile water, and plated on FGS solid medium containing hygromycin. For single spore isolation, macroconidia were harvested and plated onto FGS plates containing hygromycin. After incubation of plates at 30°C in the dark, a colony was picked and transferred to a VM-hygromycin slant. After growth for 5 days, macroconidia were isolated and plated onto a new FGS hygromycin plate, and a resistant colony was picked onto a fresh VM-hygromycin slant. This process was repeated 2 to 3 times. The presence of the gene deletion mutation for each kinase was confirmed by Southern blot analysis as described above (13).
We were unable to isolate homokaryotic mutants for nine genes because ascospores, microconidia, or serially transferred single spores carrying the gene replacement were not viable. Knockout cassettes for NCU11235, NCU06419, and NCU11410 were designed based on incorrect gene models later found to include a second gene (Table 1). As a result, two genes were disrupted in these strains, so the corresponding knockout mutants were excluded from the phenotypic analysis. The NCU09071 mutant was generated in a his-3 background and was supplemented with histidine for phenotypic analysis.
Analysis of growth, morphological, and developmental phenotypes.
Phenotypic analysis of each knockout mutant was performed by students in the Neurospora Genetics and Genomics Summer Research Institute (UCLA) (13, 86). Analysis of colony growth and morphology was performed using two different media at two different temperatures: VM and VM containing 2% yeast extract (VM+YE) at 25°C and 37°C. After inoculation, plates were incubated under ambient light/dark conditions in the laboratory. Hyphae at the colony edge and the entire colony were then photographed. Growth rates of basal hyphae were measured in glass race tubes containing VM agar medium at 25°C under ambient light/dark conditions over a 72-h period (86).
Slant tubes containing VM medium were inoculated with strains and grown at room temperature for 6 to 8 days. Production of conidia and aerial hyphae and overall pigmentation were then scored. Aerial hyphal extension was measured in VM or VM+YE standing liquid cultures. Test tubes containing 2 ml of liquid medium were inoculated and incubated statically at 25°C for 24 h. The top edge of the mycelial mat was marked on the tubes, and then the cultures were incubated for an additional 72 h. Total height (in mm) was recorded.
For analysis of female sexual fertility, mutants were inoculated on SCM plates containing 1.5% sucrose and incubated under ambient light/dark conditions at room temperature for 7 to 8 days. Cultures were scored for the formation of protoperithecia and then fertilized using wild-type conidia of the opposite mating type. Perithecial formation and ascospore development were scored 2 weeks after fertilization.
Trichogyne assays.
Formation of trichogynes and chemotropic interactions between trichogynes and conidia of opposite mating type were analyzed as previously described (46). Neurospora wild-type (control) and mutant (ΔNCU02245) strains were grown on SCM agar for 6 days under constant light. A block of mycelia from the SCM plate was inoculated onto a 2% water-agar plate and incubated in a humid atmosphere at 25°C for 4 to 6 days under constant light. The plates were transferred to ambient humidity conditions and incubated 3 to 5 more days. A small square block (0.5 by 0.5 cm) of thin water agar was placed on the water agar plate in a region covering a few protoperithecia. A volume containing 1 μl of a wild-type microconidial suspension was placed on the water agar block. Migration of trichogynes into the block was examined after 16 h and then at 24-h intervals using an Olympus BX41 compound microscope (Olympus America, Lake Success, NY) with UM Plan fluorite objective lenses.
Chemical sensitivity screening.
All viable S/T kinase knockout mutants were screened for responses to the reactive oxygen species (ROS)-generating agent menadione. Race tubes containing 1× VM salts, 1.5% sucrose, 50 ng/ml biotin, and 1.5% agar with or without 100 μM menadione (M5750; Sigma) were inoculated with each mutant. Tubes were incubated in constant light at 25°C for 24 h before transfer to constant darkness at 25°C, after which time the growth front was marked every 24 h. Growth rates were measured and assigned to a growth rate range (±2.5 mm/day) estimated over 4 days. The growth rate in the presence of menadione was normalized to that on medium lacking menadione.
Cytochalasin A (40 ng/ml; Sigma, St. Louis, MO), benomyl (92 ng/ml; Fluka, St. Louis, MO), FK-506 (50 ng/ml; LC Laboratories, Woburn, MA), tert-butyl hydroperoxide (0.13 mM; Sigma, St. Louis, MO), sorbitol (0.8 M; Sigma), sodium chloride (0.35 M; EMD Chemicals, Gibbstown, NJ), and fludioxonil (2.75 ng/ml) were used for chemical screens of viable S/T kinase mutants with growth rates at least 50% of wild type on VM. The concentration of each chemical used was determined as the amount that inhibited wild-type growth by ∼70%. VM agar plates with or without chemical were inoculated with wild-type and knockout strains in quadruplicate. Plates were incubated in constant darkness at 30°C for 22 to 24 h. Colony radii were measured, and the percent growth in the presence versus absence of the chemical was calculated for each of the four measurements. Data from three independent experiments (four measurements/experiment) were subjected to a Q test to remove outliers. Data were then analyzed using a t test. Mutants were considered sensitive or resistant if they displayed results within at least 95% confidence in two out of three trials and at least 80% confidence in all three trials.
Complementation of ΔNCU00406 and velvet mutants.
To test for a possible allelic relationship between NCU00406 and the velvet (vel) mutation, a construct containing the NCU00406 open reading frame (ORF) and promoter region was generated and transformed into vel and ΔNCU00406 mutants. A fragment containing the NCU00406 ORF, as well as 3 kb upstream and 0.65 kb downstream (total size, ∼6 kb), was amplified by PCR using primers cla45F (5′-TTGAGAGCTCGCAGTTGGTAGGAACACA-3′) and cla43R (5′-CTTCTCTAGACTCCTCCTGATTCGTTGA-3′) and digested with SacI and XbaI. The fragment was then ligated into pTJK1 digested with SacI and XbaI to yield pGP7. pTJK1 contains the bar dominant selectable marker, conferring resistance to phosphinothricin (42). pGP7 was then transformed into conidia from the ΔNCU00406 and vel mutants by using electroporation as described previously (40). Transformants were selected on FGS solid medium containing phosphinothricin (66). Homokaryotic strains were isolated by serial plating of macroconidia as described above and then confirmed by Southern blot analysis (67).
RESULTS
Serine/threonine protein kinase genes in the Neurospora genome.
Currently, 107 serine/threonine protein kinase genes are predicted in N. crassa. At the inception of this study, 89 S/T kinases were annotated and selected for analysis. To classify each of the S/T kinases to a specific group (36, 58), protein sequences were submitted as a query for a BLAST algorithm at the Salk Institute's Protein Kinases, Kinomes and Evolution website, using a database of protein kinase genes. Among the 89 genes in our study, 6 appeared to be unique to filamentous fungi, while the remaining 83 could be categorized into the following groups: AGC, CAMK, CK1, CMGC, Other, and STE (Table 1).
To compare S/T kinases across diverse eukaryotic phyla, kinases from human, plant (Selaginella moellendorffii), yeast (S. cerevisiae), and Neurospora crassa were analyzed (Table 2). The distributions of kinases among the AGC, Atypical, CAMK, CK1, CMGC, Other, and STE groups were compared. While additional kinases belonging to other groups (e.g., tyrosine kinases and histidine kinases) are known for each of these organisms, the 83 Neurospora kinases with homology to proteins outside the filamentous fungi can be classified within six of the seven groups above. The Atypical kinases were annotated after the inception of this study. While specific kinases within the Atypical group are known to be S/T kinases (87), it is unclear whether all kinases in this group phosphorylate serine and threonine residues. We therefore focused our analyses on the AGC, CAMK, CK1, CMGC, Other, and STE groups. Neurospora has the fewest kinases among the organisms analyzed in Table 2. This is likely caused by the phenomenon of RIP (repeat-induced point mutation), where one copy of a duplicate sequence is detected and mutated (80). This has resulted in a streamlined genome containing fewer paralogs than most eukaryotic organisms (25, 26). This lack of redundancy makes Neurospora an ideal organism in which to study the function of kinases. Correcting for differences in the total number of kinases, relative kinase group sizes are essentially uniform across all of the organisms in our comparison. As expected, the relative group sizes of human, yeast, and Neurospora are slightly more similar to one another than they are to those in plants. Neurospora also contains members of protein kinase families and subfamilies that are known to be fungal specific (57). Neurospora S/T kinase gene names were taken from the literature, the e-compendium at Leeds University (http://bmbpcu36.leeds.ac.uk/∼gen6ar/newgenelist/genes/gene_list.htm), or assigned during this study (Table 1). S. cerevisiae orthologs were taken from the literature or assigned during this study based upon analyses of phylogenetic trees available at www.phylomedb.org (Table 1) (33, 34).
Table 2.
Classification of serine-threonine protein kinasesa in human, plant, yeast, and Neurospora crassa
| Group | No. of serine-threonine kinases from the group in: |
|||
|---|---|---|---|---|
| Human | Plantb | Yeastc | N. crassa | |
| AGC | 63 | 33 | 17 | 13 |
| Atypicald | 44 | 33 | 12 | 14 |
| CAMK | 74 | 139 | 22 | 14 |
| CK1 | 12 | 9 | 4 | 2 |
| CMGC | 64 | 93 | 23 | 23 |
| Other | 81 | 76 | 38 | 22 |
| STE | 47 | 36 | 14 | 12 |
Additional kinases and groups comprise the kinomes of each organism.
S. moellendorffii.
S. cerevisiae.
Histidine kinases were omitted from the Atypical group.
We attempted gene replacement of the 89 S/T protein kinase genes as part of the Neurospora Genome Project. In three cases (NCU06419, NCU11235, and NCU11410), the annotation suggests that adjacent genes were also mutated. Therefore, these three genes were excluded from further study (Table 1) (see Materials and Methods). We were able to generate homokaryotic knockout mutants for 49 genes as ascospore progeny from sexual crosses. We obtained homokaryotic mutants for another 28 kinase genes through isolation of uninucleate microconidia or after serial transfer of macroconidia. We were unable to isolate viable knockout mutants for the remaining nine kinase genes (Table 1). This left us with a total of 77 mutants for phenotypic analysis.
A majority of the viable S/T protein kinase mutants exhibit growth or developmental phenotypes.
We analyzed vegetative hyphal growth, asexual development, and sexual development in the 77 viable knockout mutants (13, 67). Neurospora grows vegetatively by apical extension, branching, and fusion of basal hyphae. Under the condition of nutrient deprivation or an air-water interface, Neurospora enters the developmental program for production of multinucleate asexual spores, macroconidia. Initially, aerial hyphae are differentiated from basal (vegetative) hyphae. These aerial hyphae then begin a budding routine at their tips, to form conidiophores which ultimately give rise to macroconidia (82). Sexual development is induced by nitrogen starvation in Neurospora, with differentiation of female reproductive structures (protoperithecia) (76). Fertilization is accomplished by chemotropic growth of a specialized female hypha (trichogyne) toward a male cell of opposite mating type, transport of the male nucleus into the protoperithecium, mitosis, and cell proliferation, followed by nuclear fusion. Meiosis ensues, and the protoperithecium develops into the mature fruiting body (perithecium) containing the meiotic progeny (ascospores) (76).
Our analysis revealed that 32 and 31 kinase mutants had abnormalities in basal hyphal growth and asexual development, respectively (Fig. 1 and Table 1; see also Table S1 in the supplemental material for detailed phenotypic data). Among these, 23 mutants were defective in both hyphal growth and asexual development (Fig. 1 and Table 1). Mutants lacking NDR family kinase cot-1, PDK1/PKA family member stk-23, HAL (halotolerance) family ptk-2, PEK (pancreatic eIF-2α kinase) family member cpc-3, and STE20 family member stk-4 were distinctive in that their only morphological phenotype was a defect in hyphal growth. Interestingly, the stk-4 ortholog ste20 is not only required for normal growth in S. cerevisiae but is also essential for mating and pseudohyphal differentiation (99). In contrast to the Neurospora genes specific for hyphal growth, NDR family mutant Δstk-12, PKA family mutant Δpkac-2, CAMKL (Ca2+/calmodulin-dependent protein kinase-like) family mutant Δmus-58, and CK2 (casein kinase 2) family mutant Δcka only possessed defects in asexual sporulation. S. cerevisiae does not possess an asexual sporulation pathway; however, loss of the stk-12 ortholog in S. cerevisiae leads to defects in sexual sporulation, along with other phenotypes (83).
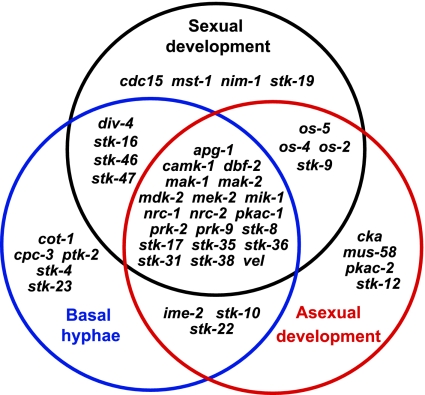
Fig. 1.
Venn diagram showing distribution of S/T protein kinase mutants with phenotypes. The 44 viable mutants exhibiting defects in at least one of the three major growth/developmental pathways are indicated as gene names for the deleted genes.
A large proportion of S/T kinase mutants displayed defects in sexual development. The genes replaced in STE20 family Δmst-1, STE11 family Δcdc15, CAMKL family Δstk-19, and NEK family Δnim-1 appear to be specific for sexual development, while the majority of genes implicated in sexual development also play roles during asexual growth and development (Fig. 1). Most of the sexual developmental mutants were not able to produce protoperithecia (Table 1; see also Table S1 in the supplemental material). The CAMKL family mutants Δstk-16 and Δstk-31 and the CDK family mutant Δdiv-4 formed protoperithecia but had defects in the timing or number of female structures produced. Three mutants formed normal perithecia but ejected no (Δnim-1), few (Δmst-1), or white ascospores (Δvel; 20% of ascospores were white.). One of the S. cerevisiae vel homologs, cla4, is required for normal pheromone sensitivity (10, 41). This suggests that Cla4p acts at an earlier point in sexual development in yeast than VEL in Neurospora.
Most of the 32 mutants that exhibited abnormalities in sexual development also possessed defects in basal hyphae growth and asexual differentiation (Table 1). Among these mutants are those lacking components of the MIK-1/MEK-1/MAK-1 and NRC-1/MEK-2/MAK-2 MAPK pathways. The MAK-1 pathway is required for normal growth, hyphal fusion, conidiation, differentiation of protoperithecia, and cell wall integrity (56, 68), while the MAK-2 cascade controls growth, hyphal fusion, conidiation, and protoperithecial development (49, 53, 56). Strains with mutations in the proteins of the osmotic stress resistance MAPK pathway (OS-4/OS-5/OS-2) have near-normal growth rates but exhibit defects in conidiation and sexual development while displaying sensitivity to osmotic stress and resistance to phenylpyrrole and dicarboximide fungicides (24, 42, 92). The roles for these MAPK pathways in Neurospora share many parallels with the corresponding pathways in S. cerevisiae and other fungi. For example, in S. cerevisiae, the MAK-2 homologous pathway is required for the pheromone response and filamentous growth, the MAK-1-related pathway is necessary for cell wall integrity and mitotic progression, and the OS-2 homologous MAPK pathway is required for resistance to osmotic stress (11).
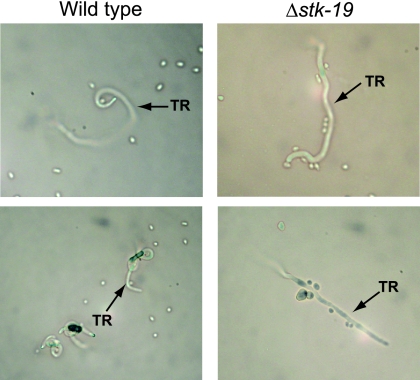
The Δstk-19 mutant was of particular interest, since it formed protoperithecia normally but produced very few perithecia after fertilization. We have previously observed such a phenotype in mutants with defects in chemotropic growth of female-specific hyphae, or trichogynes, toward male cells (conidia) (46). In order to determine whether Δstk-19 possessed such a chemotropism defect, we further investigated this mutant in a trichogyne chemotropism assay (46) (Fig. 2). The results showed that although many Δstk-19 protoperithecia form trichogynes, they are largely unable to recognize or encircle the male conidia (Fig. 2). This result suggests that the missing kinase is required for chemotropism of females toward males during fertilization. In S. cerevisiae, the stk-19 ortholog YPL150W is required for a normal growth rate and glycogen accumulation but has no known roles in mating or sexual development (91, 99).
Fig. 2.
Trichogyne chemotropism in the mutant lacking NCU02245/stk-19. The wild type and the ΔNCU02245 (Δstk-19) strain were cultured to produce female reproductive structures (protoperithecia). Sterile agar blocks were then placed on top of protoperithecia. Subsequently, a conidial suspension (males) from a wild-type strain of opposite mating type was applied to the top of the agar block. Trichogyne (TR) growth and migration through the agar blocks toward the males was monitored microscopically 16 to 24 h after application of the conidia.
Overall, our results showed that 44/77 mutants analyzed (57%) exhibit a defect in at least one of the three major phenotypes analyzed (Fig. 1; Table 1). Among these, 11 and 20 knockout mutants possess defects in two or three traits, respectively (Fig. 1; Table 1). The number of genes implicated in multiple traits (31) in combination with those for which no viable mutant could be isolated (9) suggest that a large proportion of Neurospora S/T kinases play crucial roles in important fungal life processes (Fig. 1).
Chemical sensitivity assays increase the number of S/T protein kinase mutants with phenotypes.
In S. cerevisiae, most genes that do not exhibit obvious functions during growth and development are essential for normal resistance to at least one chemical treatment (31). In order to better understand the functions of S/T kinases in Neurospora, we compared mutants to the wild type with regard to their sensitivity to a panel of chemical treatments (see Table 1 and the detailed chemical phenotypes in Table 3). The chemicals included those that induce oxidative (menadione and tert-butyl hydroperoxide) or osmotic (sorbitol and sodium chloride) stress, perturb the cytoskeleton (benomyl and cytochalasin A), act as a fungicide (fludioxonil), or inhibit the Ca2+-dependent phosphatase calcineurin (FK-506). In order to avoid issues with slow-growing strains that may have skewed our results, we included in our analysis only those viable mutants (56) that had growth rates of at least 50% of the wild type on normal medium.
Table 3.
Chemical sensitivity summary
| Deleted gene | Sensitivitya |
|||||||
|---|---|---|---|---|---|---|---|---|
| Cytochalasin A | Benomyl | FK506 | Sorbitol | Sodium chloride | Fludioxonil | tert-Butyl hydroperoxide | Menadione | |
| stk-10 | S | |||||||
| stk-23 | S | |||||||
| camk-4 | R | |||||||
| stk-16 | S | S | S | |||||
| prk-10 | R | S | ||||||
| mus-58 | S | |||||||
| prk-3 | S | |||||||
| div-4 | S | |||||||
| stk-47 | S | |||||||
| cka | S | |||||||
| stk-46 | S | |||||||
| gsk-3 | R | |||||||
| ime-2 | R | |||||||
| stk-54 | R | |||||||
| stk-22 | R | R | ||||||
| ptk-2 | S | |||||||
| stk-30 | S | |||||||
| stk-5 | S | |||||||
| stk-38 | S | S | S | |||||
| stk-9 | S | S | S | |||||
| nim-1 | R | |||||||
| stk-17/fi | S | |||||||
| apg-1 | S | S | S | S | S | |||
| stk-29 | S | |||||||
| stk-4 | S | S | ||||||
Mutants were classified as sensitive (S) or resistant (R) relative to the wild type based on the percentage of growth in the presence of the chemical compared to growth in the absence of the chemical, as follows: percent growth = (colony radius with chemical)/(colony radius without chemical) × 100. See Materials and Methods for details. For all compounds, sensitivity was scored directly from the average growth rate, except for menadione, for which sensitivity was scored after the average growth rates were binned (see Table S1 in the supplemental material). Corroboration of findings was based on the version 5 annotation of the Broad Institute's Neurospora crassa database (www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html).
Menadione generates a hyperoxidative condition by producing ROS, while tert-butyl hydrogen peroxide is a source of the oxidant peroxide (6, 44). Three mutants, ULK (Unc-51-like kinase) family Δapg-1, STE20 family Δstk-4, and NAK (NF-κB activating kinase) family Δstk-9, were hypersensitive to both menadione and tert-butyl hydrogen peroxide (Table 3), suggesting the missing kinases play general roles in resistance to oxidative stress. Another three and four mutants were more sensitive than the wild type to menadione or tert-butyl peroxide, respectively (Table 3). With a total of seven sensitive mutants, tert-butyl peroxide yielded the greatest number of phenotypes in our study. Reactive oxygen species have previously been implicated in control of conidiation in Neurospora (1). Therefore, it is of interest that four mutants (Δstk-38, Δstk-9, Δstk-17/fi, and Δapg-1) all possess defects in some aspect of conidiation, in addition to altered sensitivity to tert-butyl peroxide and/or menadione.
We assayed the response of kinase mutants to two agents that induce osmotic stress in fungi, sorbitol and sodium chloride. One mutant (Δapg-1) that was sensitive to menadione and peroxide also displayed decreased growth in the presence of sorbitol and sodium chloride, consistent with a general sensitivity to environmental stress. In S. cerevisiae, the apg-1 homologous gene, atg1, is required for autophagy, regulation of cellular ROS levels, and sensitivity to the complex chemical propolis but has no reported roles in resistance to osmotic stress (101). In total, seven Neurospora mutants exhibited significant sensitivity to sodium chloride, while one displayed increased resistance (Table 3). Of the sensitive mutants, two (Δstk-5 and Δstk-30) lack genes of the HAL (halotolerance) family. This result is a significant proof of principle, as the HAL family is known to function in salt tolerance in S. cerevisiae (63).
To explore the role of S/T kinases in cytoskeletal maintenance, the mutants were screened using two chemicals known to alter two major cytoskeletal components. Cytochalasin A interacts with actin filaments and inhibits their polymerization and elongation (14). Benomyl binds to microtubules, resulting in the inhibition of crucial cellular processes such as mitosis, meiosis, and cellular transport (90). The growth of two kinase mutants, Δstk-38 (NAK family) and Δstk-47 (CDK family), was decreased more than the wild type in the presence of cytochalasin A. Two mutants displayed increased resistance relative to the wild type, Δstk-22 (CAMKK [Ca2+/calmodulin-dependent protein kinase kinase] family) and Δstk-54 (CLK/SRPK [serine-rich protein kinase]/DYRK [dual-specificity tyrosine-regulated kinase] family) (Table 3). Three kinase mutants (Δnim-1 and Δgsk-3 [GSK family] and Δcamk-4 [CAMK1 family]) were more resistant to benomyl, while three mutants (Δstk-16 [CAMKL family], Δstk-29 [WEE family], and Δstk-38 [NAK family]) exhibited greater sensitivity (Table 3). Notably, mutant Δstk-38 exhibited significant sensitivity to both cytochalasin A and benomyl (Table 3). The observation of altered sensitivity of a mutant to cytochalasin A and/or benomyl is consistent with a defect in the actin and/or β-tubulin cytoskeleton, respectively. Four mutants, Δstk-16, Δstk-22, Δstk-38, and Δstk-47, exhibited defects during vegetative or sexual development in the absence of chemicals, possibly resulting from disturbance of the cytoskeleton. Interestingly, one of the S. cerevisiae stk-16 orthologs, KIN4, inhibits the mitotic exit network and is localized to the mother cell cortex, spindle pole bodies, and bud neck (16, 71). Additionally, the S. cerevisiae orthologs of stk-38, ARK1 and PRK1, are important for regulation of the cortical actin cytoskeleton (15). Deletion of ARK-1 results in formation of actin clumps and a growth defect in S. cerevisiae. Of interest, the Neurospora Δstk-38 mutant exhibits defects in hyphal growth and asexual and sexual development. Finally, while the sensitivity of the Neurospora Δstk-47 mutant to cytochalasin A suggests an influence on actin organization, the S. cerevisiae ortholog CTK1 may play a role in microtubule maintenance, given the sensitivity of the yeast ctk1 mutant to benomyl (69).
FK-506 is an immunosuppressant drug that inhibits calcineurin, an S/T protein phosphatase in the calcium signaling pathway (74). Three S/T kinase mutants, Δapg-1, Δptk-2, and Δstk-9 (NAK family), were more sensitive to FK-506 than the wild type, while two mutants, Δime-2 (RCK [ros cross-hybridizing kinase] family) and Δstk-22 (CAMKK family) were more resistant to FK-506 (Table 3). Altered sensitivity to FK-506 implicates these kinases in calcineurin function in Neurospora. The finding that the Δime-2 mutant has altered sensitivity to FK-506 is a novel finding in any fungal species. Work in the Glass laboratory has shown that ime-2 is not required for meiosis, as in S. cerevisiae, but instead negatively regulates formation of female reproductive structures in response to adequate nitrogen in Neurospora (38). This function is similar to that described for the filamentous fungus Cryptococcus neoformans (39, 55). In our study, we did not screen for protoperithecial formation under nitrogen-rich conditions. However, we did note that ime-2 mutants had defects in hyphal growth and aerial hyphae formation, in addition to altered sensitivity to FK-506.
Fludioxonil is a member of the phenylpyrrole class of fungicides (64). Fludioxonil inappropriately stimulates the osmotic sensitive-2 (OS-2) MAPK pathway, leading to excessive glycerol production and subsequent cell death in filamentous fungi (100). All Neurospora os mutants, including those with defects in the three genes of the os MAPK pathway, are resistant to fludioxonil (24, 100). In accordance with previous results, we observed that the three os MAPK pathway mutants were resistant to fludioxonil but sensitive to sorbitol and sodium chloride (data not shown). In addition, we noted another previously uncharacterized S/T kinase mutant (Δstk-16; CAMKL family) that was sensitive to both fludioxonil and sodium chloride, as well as benomyl (Table 3). This mutant is of interest, as it appears to have uncoupled the previously observed association between fungicide resistance and osmotic sensitivity and may participate in a novel pathway that modulates fungicide resistance in Neurospora. Δstk-16 also possesses defects in hyphal growth and protoperithecial development (Table 1). One of the homologous genes in S. cerevisiae, KIN4, modulates mitotic exit but has not been implicated in osmotolerance (16).
Taken together, the results revealed 25 mutants with chemical sensitivity phenotypes. Accounting for mutants that exhibited a significant sensitivity or resistance to more than one chemical, a total of 38 chemical phenotypes were observed. Importantly, these screens uncovered unique phenotypes for eight mutants that did not exhibit any morphological or developmental defects. This corresponds to more than 10% of the viable S/T kinase mutants and brings the total number of viable mutants with phenotypes to 52.
NCU00406 is allelic with VELVET, encoding a PAK with homology to Cla4p and Skm1p from S. cerevisiae.
While performing phenotypic analysis, we observed that the ΔNCU00406 mutant possessed severe defects in extension of basal and aerial hyphae and a distinctive colony morphology. The mutant was yellow, with aggregated hyphae and little production of conidia. We noted that the phenotype of ΔNCU00406 resembled that of the morphological mutant velvet (vel), isolated more than 50 years ago (72). Furthermore, the vel mutation maps near cloned genetic markers in the vicinity of NCU00406.
Based on the above information, we investigated whether vel is allelic with NCU00406. We transformed vel and the ΔNCU00406 mutants with a construct containing the NCU00406 open reading frame and promoter region. In both cases, introduction of a wild-type allele of NCU00406 complemented growth and morphological phenotypes (Fig. 3 A). In order to determine the position of the mutation(s) in vel, we amplified regions of the NCU00406 gene from the vel and wild-type strains and submitted them for sequencing. The results revealed two single base pair substitutions, a C-to-T change at nucleotide position 765 and another C-to-T change at position 1144 in the 2,502-bp ORF of the vel allele. The substitution at position 765 is synonymous, retaining the codon for phenylalanine. However, the second substitution produces a nonsense mutation at nucleotide position 1144, predicted to result in a truncated protein lacking the protein kinase domain (Fig. 3B). Taken together, these results demonstrate that vel is allelic with NCU00406, and they highlight the importance of this PAK-encoding gene to Neurospora biology. In S. cerevisiae, the VEL-related protein Cla4p is equally important to yeast biology, being required for normal bud morphology, pheromone responsiveness, filamentation, and invasive growth (10, 41, 89). In contrast, although S. cerevisiae Skm1p is necessary for normal resistance to hyperosmotic and other stresses (45, 98), this protein has not been implicated in the wide array of functions ascribed to Cla4p.
Fig. 3.
Allelism between the S/T kinase gene NCU00406 and velvet (vel). (A) Colony morphology and vegetative growth of ΔNCU00406 and vel mutants complemented with a construct containing the wild-type NCU00406 gene. C00406-7-1 and Cvel-1-1 are ΔNCU00406 and vel mutants, respectively, transformed with the complementation construct for NCU00406. Strains were grown in VM agar plates (top) and flasks (bottom) for 24 h and 14 days, respectively. (B) Sequence analysis of the NCU00406 ORF region in the vel mutant. After amplification and cloning of the NCU00406 ORF region from the vel mutant, the 2,502-bp sequence was compared with the corresponding sequence from the wild type. The synonymous mutation at position 765 that does not alter the amino acid sequence is shown, along with the premature stop codon at 1144.
DISCUSSION
We identified 107 genes with the S/T protein kinase domain in the Neurospora genome. This number is similar to that for the yeast genome (∼130), but smaller than that in humans (∼380) and Drosophila melanogaster (∼185) (58). Our comparative sequence analysis demonstrates that most Neurospora S/T kinases are well-conserved among organisms such as yeasts, animals, and plants. Our classification of S/T protein kinases is largely in agreement with a previous assessment of eukaryotic S/T kinases that included N. crassa (61). Six kinases showed no or low homology with those in yeast, animals, and plants, indicating that they may be specific to filamentous fungi. All of these were dispensable for the three major developmental processes analyzed (vegetative growth and asexual and sexual development), and none displayed chemical sensitivities, suggesting that these kinases may be involved in specific functions for optimized growth and development.
Our phenotypic analysis of kinase mutants provides important information about functions of S/T protein kinases during Neurospora growth and differentiation. The analysis was performed on only 86 kinase genes, because in three cases (NCU06419, NCU11235, and NCU11410), annotation errors resulted in removal of additional genes along with the kinase, and in 18 cases (cdc28, div-18, hat-2, mus-9, mus-21, prk-11, rgb-40, stk-18, stk-24, stk-25, stk-27, stk-28, stk-33, stk-48, stk-52, stk-55, stk-58, and tkl-1) the annotation was made after the start of this study. A total of 42/86 (49%) Neurospora kinase mutants did not exhibit any obvious phenotype during hyphal growth or asexual or sexual development, compared to 59/103 (57%) of transcription factor mutants studied in identical assays (13). In contrast, nine kinase genes (cdc5, ck-1a, ck-1b, mdk-1, pkc, prk-4, stk-1, stk-13, and stk-50) were observed to be essential for fungal survival. In nearly all cases, these essential kinases were also found to be essential in S. cerevisiae, suggesting that these kinases are universal regulators of cell growth in fungi. The number of apparent essential S/T kinase genes (9/86) is approximately double that observed previously for transcription factor genes (4/103) (13), suggesting less redundancy in the S/T kinase gene class in Neurospora.
Our results revealed important differences between S/T kinases and transcription factors in the regulation of growth and development. A greater number of kinase mutants (44/77 viable mutants) were defective in at least one phenotype compared to transcription factor mutants (40/99 viable mutants). The difference is even greater when the number of genes involved in two or more growth/developmental functions is considered. Significantly more kinase genes (40%; 31/77 viable mutants) are involved in the regulation of two or more functions, compared to transcription factors (18%; 18/99 viable mutants). More than twice as many kinases as transcription factors (20 versus 9) are indispensable for all three functions. Thus, the data demonstrate that the impacts of kinases on fungal growth and differentiation are more dramatic than that on transcription factors, again, likely due to less functional redundancy in the kinases (7).
It is notable that 43% of the viable S/T kinases have no obvious roles during vegetative growth or asexual or sexual development. However, our chemical screening analysis revealed phenotypes for a total of 25 mutants, with 8 that did not exhibit defects in the growth and developmental assays. Of these eight, only one had been previously characterized, gsk-3 (20). That earlier study linked gsk-3 to glycogen metabolism and mitosis in Neurospora, but most of the phenotypes were either not analyzed in our study (arthro and microconidiation) or were relatively subtle. However, we did note altered resistance of the Δgsk-3 mutant to the microtubule-destabilizing chemical benomyl, perhaps related to its role in mitotic regulation. The remaining seven genes included CAMK1 kinase camk-4, CAMKL kinase prk-10, CDK kinase prk-3, CLK/SRPK/DYRK kinase stk-54, HAL family kinases stk-5 and stk-30, and WEE kinase stk-29. The discovery of chemical phenotypes for these morphologically normal mutants should inform future functional studies of these genes. Our observation of novel phenotypes using chemical screens is consistent with the results of a large-scale chemical genomic screen using S. cerevisiae. In that study, genes having no obvious phenotypic consequences when deleted were essential for optimal growth under various chemical and environmental stress conditions (31).
Of all the mutants analyzed, Δapg-1 possessed the greatest number of chemical phenotypes (sensitivity to the four osmotic/oxidative stresses and FK-506), as well as defects in asexual and sexual growth and development. It has been demonstrated that the homolog of this kinase in S. cerevisiae, ATG1, is required for normal growth, sexual sporulation, autophagy, and accumulation of reactive oxygen species (85, 101), but apg-1 had not been previously studied in Neurospora. Our results underscore the importance of this kinase to Neurospora environmental stress resistance, growth, and development. More work is needed to determine whether these functions are linked to the regulation of autophagy in this filamentous fungus.
Our study uncovered several S/T kinases that regulate different stages of sexual differentiation. A majority of the mutants that did not produce protoperithecia also possessed defects in hyphal growth and asexual development, indicating that abolishment of protoperithecial formation may often be linked to poor growth or lack of conidial differentiation. However, cdc15 seems to be a unique kinase that is required only for protoperithecial formation. Our analysis also demonstrated that stk-19 is primarily involved in the process by which the female trichogyne recognizes and fuses with the male cell. Since a G protein-coupled receptor, PRE-1, and the heterotrimeric Gα, Gβ, and Gγ proteins, GNA-1, GNB-1, and GNG-1, have been shown to regulate trichogyne recognition and fusion with male cells in previous studies (46, 51), it would be interesting to further investigate a possible interaction between STK-19 and PRE-1, GNA-1, GNB-1, or GNG-1.
We observed functional conservation for a number of S/T kinases when comparing Neurospora to other filamentous fungi. For example, PAKs and GCKs are known to regulate polarized hyphal growth in many filamentous and dimorphic fungi (for a review, see reference 8), and our analysis showed that three Neurospora PAKs/GCKs (vel, stk-4, and prk-9) are required for normal hyphal growth. In particular, the Δvel PAK mutant had a colony morphology (reduced growth and shortened aerial hyphae) that has also been observed in corresponding mutants in the filamentous fungi Magnaporthe grisea and Claviceps purpurea (54, 77).
We noted examples of interesting variations in the functions of S/T kinases implicated in mitotic regulation in Neurospora and the filamentous fungus Aspergillus nidulans. S/T kinases homologous to Cdc2 are at the top of the phosphorylation cascade that regulates mitosis in numerous eukaryotes (reviewed in reference 65). Cdc2 phosphorylates S/T kinases in three different families: Aurora, Polo-like, and NEK. The Neurospora CDC2 homolog (cdc28) was not annotated at the initiation of our study, but the A. nidulans homolog (nimX) is an essential gene (97). The downstream Aurora and the Polo-like kinase genes are essential in Neurospora (stk-13 and cdc5 [this study]), A. nidulans (Aurora and plkA [3, 28]) and S. cerevisiae (47, 62). However, although the NEK nimA is required for mitotic entry and is thus essential in A. nidulans (96), it is dispensable for viability in Neurospora (this study), as well as the yeasts Schizosaccharomyces pombe (50) and S. cerevisiae (43). The different requirement for the NEK in the two filamentous fungi is perhaps surprising, in light of the ability of the Neurospora gene to complement the A. nidulans nimA mutation (75). However, accumulating evidence suggests that the functions of NEKs are not restricted to mitotic entry, as first demonstrated in A. nidulans, but also influence other aspects of mitotic control, in roles that are shared with other kinases (65).
Another example of a difference between Neurospora and A. nidulans concerns the role of the STE11 family kinase cdc15 (in Neurospora) and sepH (in A. nidulans). sepH regulates septum formation and hyphal differentiation in A. nidulans (9, 30). In contrast, mutation of cdc15 did not lead to defects in growth of vegetative hyphae in Neurospora, but it blocked the formation of protoperithecia during the sexual cycle. Since sepH regulates septum formation through control of cytoskeletal structure in A. nidulans, it is possible that a role for cdc15 in reorganizing the cytoskeleton is only observed during protoperithecia formation in Neurospora.
The observation that 71% of the S/T kinases mutated in our study were either essential or necessary for normal growth, development, or chemical resistance underscores the central importance of S/T protein kinases to Neurospora biology. Further investigations will illuminate specific roles for individual kinases, reveal those that operate in shared pathways, and determine the extent of functional conservation between S/T kinases from Neurospora and other organisms.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to the undergraduates in the Neurospora Genetics and Genomics Summer Research Institute at UCLA who performed the morphological and growth assays presented in this study: Charity Achara, Diana Almanza, Shannon Aulakh, Jeffery Buenaflor, Gun Woo Byeon, Shuk Han Chan, Aaron Craddolph, Eugenia del Valle, Isais Deresa, Melinda Hernandez, Jessica Ji, David Jiang, Anqi Li, Alanna Malicdem, Heydi Nicanor-Lewis, Refugio Parra, Diana Perez, Yessenia Quintero, Tamara Restrepo, and Daizy Villalvazo. We acknowledge undergraduates in the Borkovich laboratory for development of chemical screening procedures: Antonia Adeniji, Suzanne Barrios, and Roxanne Sebeny.
Fludioxonil was a gift from Frank Wong and Allison Tally. We thank Michael Plamann, Kevin McCluskey, and Aric Wiest at the Fungal Genetics Stock Center for maintenance of Neurospora knockout mutants.
This work was supported by P01 GM068087 to J.C.D. and K.A.B., by R01 GM086565 to K.A.B., and by R01 GM34985 and R01 GM08336 to J.C.D. Undergraduate Fitz-Gerald Diala is a MARC U Star trainee at UCR (5T34GM062756). Antonia Adeniji was supported by MARC U Star grant 5T34GM062756, Suzanne Barrios by a CCRAA/HSI grant from the U.S. Department of Education, and Roxanne Sebeny by NSF REU DBI1004793.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 30 September 2011.
REFERENCES
- 1. Aguirre J., Rios-Momberg M., Hewitt D., Hansberg W. 2005. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 13:111–118 [DOI] [PubMed] [Google Scholar]
- 2. Avalos J., Geever R. F., Case M. E. 1989. Bialaphos resistance as a dominant selectable marker in Neurospora crassa. Curr. Genet. 16:369–372 [DOI] [PubMed] [Google Scholar]
- 3. Bachewich C., Masker K., Osmani S. 2005. The polo-like kinase PLKA is required for initiation and progression through mitosis in the filamentous fungus Aspergillus nidulans. Mol. Microbiol. 55:572–587 [DOI] [PubMed] [Google Scholar]
- 4. Baker C. L., Kettenbach A. N., Loros J. J., Gerber S. A., Dunlap J. C. 2009. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol. Cell 34:354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Banno S., et al. 2005. A catalytic subunit of cyclic AMP-dependent protein kinase, PKAC-1, regulates asexual differentiation in Neurospora crassa. Genes Genet. Syst. 80:25–34 [DOI] [PubMed] [Google Scholar]
- 6. Belden W. J., et al. 2007. The band mutation in Neurospora crassa is a dominant allele of ras-1 implicating RAS signaling in circadian output. Genes Dev. 21:1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Borkovich K. A., et al. 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68:1–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boyce K. J., Andrianopoulos A. 2011. Ste20-related kinases: effectors of signaling and morphogenesis in fungi. Trends Microbiol. 19:400–410 [DOI] [PubMed] [Google Scholar]
- 9. Bruno K. S., Morrell J. L., Hamer J. E., Staiger C. J. 2001. SEPH, a Cdc7p orthologue from Aspergillus nidulans, functions upstream of actin ring formation during cytokinesis. Mol. Microbiol. 42:3–12 [DOI] [PubMed] [Google Scholar]
- 10. Chasse S. A., et al. 2006. Genome-scale analysis reveals Sst2 as the principal regulator of mating pheromone signaling in the yeast Saccharomyces cerevisiae. Eukaryot. Cell 5:330–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen R. E., Thorner J. 2007. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773:1311–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collopy P. D., et al. 2010. High-throughput construction of gene deletion cassettes for generation of Neurospora crassa knockout strains. Methods Mol. Biol. 638:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Colot H. V., et al. 2006. A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc. Natl. Acad. Sci. U. S. A. 103:10352–10357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cooper J. A. 1987. Effects of cytochalasin and phalloidin on actin. J. Cell Biol. 105:1473–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cope M. J., Yang S., Shang C., Drubin D. G. 1999. Novel protein kinases Ark1p and Prk1p associate with and regulate the cortical actin cytoskeleton in budding yeast. J. Cell Biol. 144:1203–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. D'Aquino K. E., et al. 2005. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects. Mol. Cell 19:223–234 [DOI] [PubMed] [Google Scholar]
- 17. Davis R. H., J. deSerres F. 1970. Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol. 71A:79–143 [Google Scholar]
- 18. de Paula R. M., Lamb T. M., Bennett L., Bell-Pedersen D. 2008. A connection between MAPK pathways and circadian clocks. Cell Cycle 7:2630–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dickman M. B., Yarden O. 1999. Serine/threonine protein kinases and phosphatases in filamentious fungi. Fungal Genet. Biol. 26:99–117 [DOI] [PubMed] [Google Scholar]
- 20. Dvash E., Kra-Oz G., Ziv C., Carmeli S., Yarden O. 2010. The NDR kinase DBF-2 is involved in regulation of mitosis, conidial development, and glycogen metabolism in Neurospora crassa. Eukaryot. Cell 9:502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ebbole D., Sachs M. S. 1990. A rapid and simple method for isolation of Neurospora crassa homokaryons using microconidia. Fungal Genet. Newslett. 37:17–18 [Google Scholar]
- 22. Fleissner A., Leeder A. C., Roca M. G., Read N. D., Glass N. L. 2009. Oscillatory recruitment of signaling proteins to cell tips promotes coordinated behavior during cell fusion. Proc. Natl. Acad. Sci. U. S. A. 106:19387–19392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Franchi L., Fulci V., Macino G. 2005. Protein kinase C modulates light responses in Neurospora by regulating the blue light photoreceptor WC-1. Mol. Microbiol. 56:334–345 [DOI] [PubMed] [Google Scholar]
- 24. Fujimura M., et al. 2003. Putative homologs of SSK22 MAPKK kinase and PBS2 MAPK kinase of Saccharomyces cerevisiae encoded by os-4 and os-5 genes for osmotic sensitivity and fungicide resistance in Neurospora crassa. Biosci. Biotechnol. Biochem. 67:186–191 [DOI] [PubMed] [Google Scholar]
- 25. Galagan J. E., et al. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868 [DOI] [PubMed] [Google Scholar]
- 26. Galagan J. E., Selker E. U. 2004. RIP: the evolutionary cost of genome defense. Trends Genet. 20:417–423 [DOI] [PubMed] [Google Scholar]
- 27. Gorl M., et al. 2001. A PEST-like element in Frequency determines the length of the circadian period in Neurospora crassa. EMBO J. 20:7074–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hage N. 2010. Senior thesis. The Ohio State University, Columbus, OH [Google Scholar]
- 29. Hanks S. K., Hunter T. 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9:576–596 [PubMed] [Google Scholar]
- 30. Harris S. D. 2001. Septum formation in Aspergillus nidulans. Curr. Opin. Microbiol. 4:736–739 [DOI] [PubMed] [Google Scholar]
- 31. Hillenmeyer M. E., et al. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Huang G., et al. 2007. Protein kinase A and casein kinases mediate sequential phosphorylation events in the circadian negative feedback loop. Genes Dev. 21:3283–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huerta-Cepas J., Bueno A., Dopazo J., Gabaldon T. 2008. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 36:D491–D496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huerta-Cepas J., et al. 2011. PhylomeDB v3.0: an expanding repository of genome-wide collections of trees, alignments and phylogeny-based orthology and paralogy predictions. Nucleic Acids Res. 39:D556–D560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huerta-Cepas J., Dopazo H., Dopazo J., Gabaldon T. 2007. The human phylome. Genome Biol. 8:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hunter T. 1995. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80:225–236 [DOI] [PubMed] [Google Scholar]
- 37. Hunter T., Plowman G. D. 1997. The protein kinases of budding yeast: six score and more. Trends Biochem. Sci. 22:18–22 [DOI] [PubMed] [Google Scholar]
- 38. Hutchison E. A., Glass N. L. 2010. Meiotic regulators Ndt80 and ime2 have different roles in Saccharomyces and Neurospora. Genetics 185:1271–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Irniger S. 2011. The Ime2 protein kinase family in fungi: more duties than just meiosis. Mol. Microbiol. 80:1–13 [DOI] [PubMed] [Google Scholar]
- 40. Ivey F. D., Hodge P. N., Turner G. E., Borkovich K. A. 1996. The G alpha i homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol. Biol. Cell 7:1283–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin R., Dobry C. J., McCown P. J., Kumar A. 2008. Large-scale analysis of yeast filamentous growth by systematic gene disruption and overexpression. Mol. Biol. Cell 19:284–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones C. A., Greer-Phillips S. E., Borkovich K. A. 2007. The response regulator RRG-1 functions upstream of a mitogen-activated protein kinase pathway impacting asexual development, female fertility, osmotic stress, and fungicide resistance in Neurospora crassa. Mol. Biol. Cell 18:2123–2136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jones D. G., Rosamond J. 1990. Isolation of a novel protein kinase-encoding gene from yeast by oligodeoxyribonucleotide probing. Gene 90:87–92 [DOI] [PubMed] [Google Scholar]
- 44. Kato A., Akamatsu Y., Sakuraba Y., Inoue H. 2004. The Neurospora crassa mus-19 gene is identical to the qde-3 gene, which encodes a RecQ homologue and is involved in recombination repair and postreplication repair. Curr. Genet. 45:37–44 [DOI] [PubMed] [Google Scholar]
- 45. Kawahata M., Masaki K., Fujii T., Iefuji H. 2006. Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res. 6:924–936 [DOI] [PubMed] [Google Scholar]
- 46. Kim H., Borkovich K. A. 2004. A pheromone receptor gene, pre-1, is essential for mating type-specific directional growth and fusion of trichogynes and female fertility in Neurospora crassa. Mol. Microbiol. 52:1781–1798 [DOI] [PubMed] [Google Scholar]
- 47. Kitada K., Johnson A. L., Johnston L. H., Sugino A. 1993. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol. Cell. Biol. 13:4445–4457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Knippschild U., et al. 2005. The casein kinase 1 family: participation in multiple cellular processes in eukaryotes. Cell Signal. 17:675–689 [DOI] [PubMed] [Google Scholar]
- 49. Kothe G. O., Free S. J. 1998. The isolation and characterization of nrc-1 and nrc-2, two genes encoding protein kinases that control growth and development in Neurospora crassa. Genetics 149:117–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Krien M. J., et al. 1998. A NIMA homologue promotes chromatin condensation in fission yeast. J. Cell Sci. 111:967–976 [DOI] [PubMed] [Google Scholar]
- 51. Krystofova S., Borkovich K. A. 2005. The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gβγ dimer required for normal female fertility, asexual development, and gα protein levels in Neurospora crassa. Eukaryot. Cell 4:365–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lew R. R., Kapishon V. 2009. Ptk2 contributes to osmoadaptation in the filamentous fungus Neurospora crassa. Fungal Genet. Biol. 46:949–955 [DOI] [PubMed] [Google Scholar]
- 53. Li D., Bobrowicz P., Wilkinson H. H., Ebbole D. J. 2005. A mitogen-activated protein kinase pathway essential for mating and contributing to vegetative growth in Neurospora crassa. Genetics 170:1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li L., Xue C., Bruno K., Nishimura M., Xu J. R. 2004. Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol. Plant Microbe Interact. 17:547–556 [DOI] [PubMed] [Google Scholar]
- 55. Liu K. H., Shen W. C. 2011. Mating differentiation in Cryptococcus neoformans is negatively regulated by the Crk1 protein kinase. Fungal Genet. Biol. 48:225–240 [DOI] [PubMed] [Google Scholar]
- 56. Maerz S., et al. 2008. The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion and sexual development in Neurospora crassa. Genetics 179:1313–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Manning G., Plowman G. D., Hunter T., Sudarsanam S. 2002. Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27:514–520 [DOI] [PubMed] [Google Scholar]
- 58. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. 2002. The protein kinase complement of the human genome. Science 298:1912–1934 [DOI] [PubMed] [Google Scholar]
- 59. McCluskey K., et al. 2011. Rediscovery by whole genome sequencing: classical mutations and genome polymorphisms in Neurospora crassa. Genes Genomes Genet. 1:303–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mehra A., et al. 2009. A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell 137:749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miranda-Saavedra D., Barton G. J. 2007. Classification and functional annotation of eukaryotic protein kinases. Proteins 68:893–914 [DOI] [PubMed] [Google Scholar]
- 62. Monje-Casas F., Prabhu V. R., Lee B. H., Boselli M., Amon A. 2007. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell 128:477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mulet J. M., et al. 1999. A novel mechanism of ion homeostasis and salt tolerance in yeast: the Hal4 and Hal5 protein kinases modulate the Trk1-Trk2 potassium transporter. Mol. Cell. Biol. 19:3328–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ochiai N., et al. 2001. Characterization of mutations in the two-component histidine kinase gene that confer fludioxonil resistance and osmotic sensitivity in the os-1 mutants of Neurospora crassa. Pest. Manag. Sci. 57:437–442 [DOI] [PubMed] [Google Scholar]
- 65. O'Connell M. J., Krien M. J., Hunter T. 2003. Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 13:221–228 [DOI] [PubMed] [Google Scholar]
- 66. Pall M. 1993. The use of Ignite (basta; glufosinate; phosphinothricin) to select transformants of bar-containing plasmids in Neurospora crassa. Fungal Genet. Newslett. 40:57 [Google Scholar]
- 67. Park G., et al. 2011. High-throughput production of gene replacement mutants in Neurospora crassa. Methods Mol. Biol. 722:179–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Park G., Pan S., Borkovich K. A. 2008. Mitogen-activated protein kinase cascade required for regulation of development and secondary metabolism in Neurospora crassa. Eukaryot. Cell 7:2113–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Parsons A. B., et al. 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22:62–69 [DOI] [PubMed] [Google Scholar]
- 70. Pearce L. R., Komander D., Alessi D. R. 2010. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 11:9–22 [DOI] [PubMed] [Google Scholar]
- 71. Pereira G., Schiebel E. 2005. Kin4 kinase delays mitotic exit in response to spindle alignment defects. Mol. Cell 19:209–221 [DOI] [PubMed] [Google Scholar]
- 72. Perkins D. D. 1959. New markers and multiple point linkage data in Neurospora. Genetics 44:1185–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pregueiro A. M., Liu Q., Baker C. L., Dunlap J. C., Loros J. J. 2006. The Neurospora checkpoint kinase 2: a regulatory link between the circadian and cell cycles. Science 313:644–649 [DOI] [PubMed] [Google Scholar]
- 74. Prokisch H., Yarden O., Dieminger M., Tropschug M., Barthelmess I. B. 1997. Impairment of calcineurin function in Neurospora crassa reveals its essential role in hyphal growth, morphology and maintenance of the apical Ca2+ gradient. Mol. Gen. Genet. 256:104–114 [DOI] [PubMed] [Google Scholar]
- 75. Pu R. T., et al. 1995. Isolation of a functional homolog of the cell cycle-specific NIMA protein kinase of Aspergillus nidulans and functional analysis of conserved residues. J. Biol. Chem. 270:18110–18116 [DOI] [PubMed] [Google Scholar]
- 76. Raju N. B., Leslie J. F. 1992. Cytology of recessive sexual-phase mutants from wild strains of Neurospora crassa. Genome 35:815–826 [DOI] [PubMed] [Google Scholar]
- 77. Rolke Y., Tudzynski P. 2008. The small GTPase Rac and the p21-activated kinase Cla4 in Claviceps purpurea: interaction and impact on polarity, development and pathogenicity. Mol. Microbiol. 68:405–423 [DOI] [PubMed] [Google Scholar]
- 78. Sattlegger E., Hinnebusch A. G., Barthelmess I. B. 1998. cpc-3, the Neurospora crassa homologue of yeast GCN2, encodes a polypeptide with juxtaposed eIF2α kinase and histidyl-tRNA synthetase-related domains required for general amino acid control. J. Biol. Chem. 273:20404–20416 [DOI] [PubMed] [Google Scholar]
- 79. Seiler S., Vogt N., Ziv C., Gorovits R., Yarden O. 2006. The STE20/germinal center kinase POD6 interacts with the NDR kinase COT1 and is involved in polar tip extension in Neurospora crassa. Mol. Biol. Cell 17:4080–4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Selker E. U. 2002. Repeat-induced gene silencing in fungi. Adv. Genet. 46:439–450 [DOI] [PubMed] [Google Scholar]
- 81. Shchemelinin I., Sefc L., Necas E. 2006. Protein kinases, their function and implication in cancer and other diseases. Folia Biol. (Praha) 52:81–100 [PubMed] [Google Scholar]
- 82. Springer M. L. 1993. Genetic control of fungal differentiation: the three sporulation pathways in Neurospora crassa. Bioessays 15:365–374 [DOI] [PubMed] [Google Scholar]
- 83. Su S. S., Mitchell A. P. 1993. Identification of functionally related genes that stimulate early meiotic gene expression in yeast. Genetics 133:67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tang C. T., et al. 2009. Setting the pace of the Neurospora circadian clock by multiple independent FRQ phosphorylation events. Proc. Natl. Acad. Sci. U. S. A. 106:10722–10727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tsukada M., Ohsumi Y. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333:169–174 [DOI] [PubMed] [Google Scholar]
- 86. Turner G. E. 2011. Phenotypic analysis of Neurospora crassa gene deletion strains. Methods Mol. Biol. 722:191–198 [DOI] [PubMed] [Google Scholar]
- 87. Wakabayashi M., Ishii C., Hatakeyama S., Inoue H., Tanaka S. 2010. ATM and ATR homologes of Neurospora crassa are essential for normal cell growth and maintenance of chromosome integrity. Fungal Genet. Biol. 47:809–817 [DOI] [PubMed] [Google Scholar]
- 88. Wakabayashi M., Ishii C., Inoue H., Tanaka S. 2008. Genetic analysis of CHK1 and CHK2 homologues revealed a unique cross talk between ATM and ATR pathways in Neurospora crassa. DNA Repair (Amst.) 7:1951–1961 [DOI] [PubMed] [Google Scholar]
- 89. Watanabe M., Watanabe D., Nogami S., Morishita S., Ohya Y. 2009. Comprehensive and quantitative analysis of yeast deletion mutants defective in apical and isotropic bud growth. Curr. Genet. 55:365–380 [DOI] [PubMed] [Google Scholar]
- 90. Willhite C. C. 1983. Benomyl. J. Appl. Toxicol. 3:261–264 [DOI] [PubMed] [Google Scholar]
- 91. Wilson W. A., Wang Z., Roach P. J. 2002. Systematic identification of the genes affecting glycogen storage in the yeast Saccharomyces cerevisiae: implication of the vacuole as a determinant of glycogen level. Mol. Cell Proteomics 1:232–242 [DOI] [PubMed] [Google Scholar]
- 92. Yamashita K., et al. 2007. Involvement of OS-2 MAP kinase in regulation of the large-subunit catalases CAT-1 and CAT-3 in Neurospora crassa. Genes Genet. Syst. 82:301–310 [DOI] [PubMed] [Google Scholar]
- 93. Yang Y., Cheng P., He Q., Wang L., Liu Y. 2003. Phosphorylation of Frequency protein by casein kinase II is necessary for the function of the Neurospora circadian clock. Mol. Cell. Biol. 23:6221–6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Yang Y., Cheng P., Zhi G., Liu Y. 2001. Identification of a calcium/calmodulin-dependent protein kinase that phosphorylates the Neurospora circadian clock protein Frequency. J. Biol. Chem. 276:41064–41072 [DOI] [PubMed] [Google Scholar]
- 95. Yarden O., Plamann M., Ebbole D. J., Yanofsky C. 1992. cot-1, a gene required for hyphal elongation in Neurospora crassa, encodes a protein kinase. EMBO J. 11:2159–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Ye X. S., Fincher R. R., Tang A., Osmani A. H., Osmani S. A. 1998. Regulation of the anaphase-promoting complex/cyclosome by bimAAPC3 and proteolysis of NIMA. Mol. Biol. Cell 9:3019–3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ye X. S., et al. 1995. The NIMA protein kinase is hyperphosphorylated and activated downstream of p34cdc2/cyclin B: coordination of two mitosis promoting kinases. EMBO J. 14:986–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yoshikawa K., et al. 2009. Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae. FEMS Yeast Res. 9:32–44 [DOI] [PubMed] [Google Scholar]
- 99. Yoshikawa K., et al. 2011. Comprehensive phenotypic analysis of single-gene deletion and overexpression strains of Saccharomyces cerevisiae. Yeast 28:349–361 [DOI] [PubMed] [Google Scholar]
- 100. Zhang Y., Lamm R., Pillonel C., Lam S., Xu J. R. 2002. Osmoregulation and fungicide resistance: the Neurospora crassa os-2 gene encodes a HOG1 mitogen-activated protein kinase homologue. Appl. Environ. Microbiol. 68:532–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhang Y., et al. 2007. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy 3:337–346 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.