Abstract
In response to harsh environmental conditions, ascomycetes produce stress-resistant spores to promote survival. As sporulation requires a diploid DNA content, species with a haploid lifestyle, such as Kluyveromyces lactis, first induce mating in response to stress. In K. lactis, mating and mating-type switching are induced by the DNA-binding protein Mts1. Mts1 expression is known to be upregulated by nutrient limitation, but the mechanism is unknown. We show that a ras2 mutation results in a hyperswitching phenotype. In contrast, strains lacking the phosphodiesterase Pde2 had lower switching rates compared to that of the wild type (WT). As Ras2 promotes cyclic AMP (cAMP) production and Pde2 degrades cAMP, these data suggest that low cAMP levels induce switching. Because the MTS1 regulatory region contains several Msn2 binding sites and Msn2 is a transcription factor that is activated by low cAMP levels, we investigated if Msn2 regulates MTS1 transcription. Consistently with this idea, an msn2 mutant strain displayed lower switching rates than the WT strain. The transcription of MTS1 is highly induced in the ras2 mutant strain. In contrast, an msn2 ras2 double mutant strain displays WT levels of the MTS1 transcript, showing that Msn2 is a critical inducer of MTS1 transcription. Strains lacking Msn2 and Pde2 also exhibit mating defects that can be complemented by the ectopic expression of Mts1. Finally, we show that MTS1 is subjected to negative autoregulation, presumably adding robustness to the mating and switching responses. We suggest a model in which Ras2/cAMP/Msn2 mediates the stress-induced mating and mating-type switching responses in K. lactis.
INTRODUCTION
The three cell types in S. cerevisiae are the a- and α-haploid cell types and the a/α-diploid cell type. Diploids arise from a mating reaction, i.e., the cellular and nuclear fusion of haploids with opposite mating types (28). A single locus (MAT) determines the mating type, which encodes either the a (MATa) or α gene (MATα). Each haploid genome also contains two additional loci that encode mating-type information, and they are known as the cryptic mating-type loci (HMLα and HMRa) because they are not expressed. Mating-type switching in S. cerevisiae is induced by an endonuclease called HO (homothallism), which cuts the MAT locus in a position that shares homology with the cryptic mating-type loci (29, 38). A gene conversion then exchanges the genes present in the MAT locus for the genes present in one of the cryptic mating-type loci.
In S. cerevisiae, mating-type switching is a thoroughly characterized process which has contributed significantly to basic knowledge of gene conversion (26) and transcriptional regulation (17, 37). Mating-type switching in S. cerevisiae is regulated by controlling the transcription of the HO gene. HO transcription is repressed in MATa/MATα diploids (by the a1/α2 repressor) and induced in the G1 phase of the cell cycle (by SBF) (2). In addition, a lineage-specific regulation was described in which only mother cells express HO, since daughter cells contain a repressor of HO transcription (Ash1) (16). These regulatory mechanisms lead to a switching rate of ∼0.5/generation in haploid S. cerevisiae strains, resulting in a rapid return to the diplophase. Consistently, S. cerevisiae strains isolated from natural sources are diploid.
The function of mating-type switching in S. cerevisiae is to facilitate a rapid return to the diploid state. An obvious advantage of being diploid is that survival in harsh environmental conditions is enhanced through the formation of spores. Spores form after meiosis, and this developmentally distinct form of yeasts is resistant to many types of stresses, including heat, drought, and toxic chemicals (15). However, other ascomycetes grow predominantly as haploids in nature. One example of this is the distantly related ascomycetes Kluyveromyces lactis (42). Hence, in yeasts where the haplophase dominates there are additional mechanisms that ensure survival in a harsh environment.
Mating-type switching also has been observed in K. lactis (6, 27). K. lactis relies on gene conversion and cryptic mating-type loci for completing switching, but the inducer of switching is different from that of S. cerevisiae. In K. lactis, mating-type switching is induced by a DNA-binding protein called Mts1 (mating-type switch 1). MTS1 is orthologous to the S. cerevisiae RME1 gene, which has a role in repressing meiosis-specific genes during vegetative growth. Mts1 binds to several sites in both the MATa and MATα loci. Mts1 itself shares no homology with DNA nucleases but appears to stimulate the excision of a mobile DNA element present in the MATα locus. The current model posits that the mobilization of the MATα3 mutator-like element (MULE) induces switching. Consistently with this idea, the transposase-like protein encoded by the α3 MULE is essential for switching from MATα to MATa. Moreover, the mutational inactivation of the catalytic DDE motif of α3 abolishes switching. The mechanism underlying MATa to MATα switching remains unknown, but it probably involves a hairpin-generating nuclease. Hairpin intermediates can be observed in the MATa locus in genetic backgrounds that are compromised for hairpin resolution (6).
Switching rates in K. lactis have never been accurately determined, but it is clear that the rates are lower than those in S. cerevisiae (27). The regulation of switching also seems different between yeasts, as switching in K. lactis is induced in nutrient-limited conditions. Interestingly, MTS1 transcription is induced in nutrient-limited conditions as well (6, 9), leading to a model in which switching is controlled through the regulation of MTS1 expression. Apart from stimulating switching, Mts1 also induces mating through the direct activation of genes essential for mating (9). Hence, nutrient limitation stimulates both mating and switching. Mating-type switching followed by mating then can promote spore formation upon nutrient limitation, albeit with some delay compared to that of diploid S. cerevisiae.
Stress-induced gene transcription in S. cerevisiae is partly controlled by a set of redundant zinc finger-containing transcription factors called Msn2 and Msn4 (34, 44). These factors bind the stress response element (STRE) AG4, which is present in the regulatory regions of many stress-induced genes (11). In S. cerevisiae, large-scale analyses showed that Msn2/4 activate about 200 genes (22). The activation of several genes promotes a phenomenon known as cross-protection, i.e., that transient exposure to one type of stress will provide protection from future exposure to different stress conditions. There is also evidence from other ascomycetes that orthologs of Msn2/4 regulate stress-induced gene expression (39). Furthermore, environmental signals and the cyclic AMP (cAMP)-dependent protein kinase (PKA) (45) regulate the activity of the Msn2/4 proteins. Msn2/4 predominantly localize to the cytoplasm in nutrient-rich conditions (24) but relocalize to the nucleus after a variety of stress conditions (35), including calorie restriction (36), displaying an oscillatory behavior moving between the nuclear and cytoplasmic compartments (31). The nuclear localization sequences (NLS) of Msn2/4 are inhibited by PKA phosphorylation and activated by protein phosphatase 1 dephosphorylation (18). In S. cerevisiae, mutations of PKA-dependent phosphorylation sites render Msn2 constitutively nuclear, directly linking the Ras/cAMP/PKA pathway to the repression of Msn2 activity (18, 25).
In this study, we demonstrate that mating-type switching in K. lactis is regulated by the Ras/cAMP pathway and Msn2. This regulation is mediated by controlling the transcription of the MTS1 gene. The transcription of MTS1 links mating-type switching with mating, as both processes are directly induced by Mts1. We propose that this regulation of mating-type switching and mating serves as an adaptation to the mostly haploid lifestyle of K. lactis.
MATERIALS AND METHODS
Yeast strains.
The strains used in this study are listed in Table 1. Unless noted otherwise, all gene deletions/insertions were generated using a one-step gene disruption/insertion procedure (32) with a kanMX, NAT, or URA3 PCR fragment amplified from pFA6a-KanMX (5), pAG25 (23), or pRS306. Genetic manipulations were confirmed by DNA blots or locus-specific PCR. Sequences of oligonucleotides used are available on request. SAY724 (MATa hmlα1::kanMX nej1::LEU2) was generated through a cross between SAY130 (MATa hmlα1::kanMX) and SAY509 (MATα nej1::LEU2). SAY975 (mata2Δ::NAT hmlα1::kanMX nej1::LEU2) was generated in SAY724. SAY990 (mata2Δ::NAT hmlα1::kanMX) was obtained by crossing SAY724 with SAY119. The strains SAY1131 (pde2::kanMX), SAY1379 (msn2::kanMX), SAY1488 (MATa::URA3), and SAY1489 (HMRa::URA3) were generated in SAY572 (MATa nej1::LEU2). SAY1546 (MATa::URA3 pde2::NAT) was generated in SAY1488, whereas SAY1540 (MATa::URA3 msn2::kanMX) was generated in SAY1379. SAY1552 (MATa::URA3 lig4::kanMX) and SAY1554 (MATa::URA3 lig4::kanMX mts1::LEU2) were generated in SAY683 (lig4::kanMX) and SAY719 (lig4::kanMX mts1::LEU2), respectively. SAY988 (MTS1-TAP) was generated by amplifying a TAP-containing PCR fragment with 50 bp of flanking DNA corresponding to the MTS1 3′ end and introducing it into SAY572. SAY1069 (ras2::pPMB18; obtained by selection) was generated by the random transformation of linearized pPMB18 into SAY990. The genetic selection is described in more detail below. SAY1664 (ras2::pPMB18 msn2::kanMX) was generated by crossing strains SAY1069 and SAY1653, followed by random spore analysis.
Table 1.
Strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| WM52 | MATα ade1 adeX his7 uraA1 | 14 |
| SAY129 | MATα ade1 hmlα1Δ::kanMX leu2 metA1 trp1 uraA1 | 3 |
| SAY119 | MATα ade1 leu2 metA1 or META1 trp1 uraA1 | 3 |
| SAY130 | MATahmlα1Δ::kanMX leu2 lysA1 metA1 trp1 uraA1 | 3 |
| SAY509 | MATα ade1 leu2 metA1 nej1Δ::LEU2 trp1 uraA1 | 32 |
| SAY572 | MATaleu2 lysA1 metA1 nej1Δ::LEU2 trp1 uraA1 | 32 |
| SAY683 | MATaleu2 lig4Δ::kanMX lysA1 metA1 trp1 uraA1 | 32 |
| SAY719 | MATalysA1 lig4Δ::kanMX leu2 mts1Δ::LEU2 trp1 uraA1 | This study |
| SAY724 | MATahmlα1Δ::kanMX leu2 lysA1 metA1 nej1Δ::LEU2 trp1 uraA1 | This study |
| SAY975 | mata2Δ::NAT hmlα1Δ::kanMX nej1Δ::LEU2 uraA1 leu2 lysA1 trp1 metA1 | This study |
| SAY988 | MATaleu2 lysA1 metA1 MTS1-TAP::NAT nej1Δ::LEU2 trp1 uraA1 | This study |
| SAY990 | mata2Δ::NAT hmlα1Δ::kanMX leu2 lysA1 metA1 trp1 uraA1 | This study |
| SAY1069 | mata2Δ::NAT hmlα1Δ::kanMX leu2 lysA1 metA1 ras2::pPMB18(LEU2) trp1 uraA1 | This study |
| SAY1131 | MATaleu2 lysA1 metA1 nej1Δ::LEU2 pde2Δ::kanMX trp1 uraA1 | This study |
| SAY1379 | MATaleu2 lysA1 metA1 msn2Δ::kanMX nej1Δ::LEU2 trp1 uraA1 | This study |
| SAY1488 | MATa::URA3 leu2 lysA1 metA1 nej1Δ::LEU2 trp1 uraA1 | This study |
| SAY1489 | MATaHMRa::URA3 leu2 lysA1 metA1 nej1Δ::LEU2 trp1 uraA1 | This study |
| SAY1539 | MATa::URA3 leu2 lysA1 metA1 msn2Δ::kanMX nej1Δ::LEU2 trp1 uraA1 | This study |
| SAY1546 | MATa::URA3 leu2 lysA1 metA1 nej1::LEU2 pde2::NAT trp1 uraA1 | This study |
| SAY1552 | MATa::URA3 leu2 lig4Δ::kanMX lysA1 metA1 trp1 uraA1 | This study |
| SAY1554 | MATa::URA3 leu2 lig4Δ::kanMX lysA1 mts1Δ::LEU2 trp1 uraA1 | This study |
| SAY1664 | MATaMATα ade1 lysA1 leu2 metA1 or META1 msn2::kanMX nej1::LEU2 ras2::pPMB18 trp1 uraA1 | This study |
Plasmids.
Cloning was performed using standard methods (41). Plasmids pPMB35 (6) and pCXJ18 (13) are described elsewhere. pPMB18 (pRS405-pADH1-pTEF2) was generated in two steps. First, AKP183 was generated by a three-factor cloning using an XbaI-BamHI PCR fragment containing the S. cerevisiae ADH1 promoter (bp −492 to −13 relative to the start codon), an XhoI-BamHI PCR fragment containing the S. cerevisiae TEF2 promoter (bp −1000 to −0 relative to the start codon), and SpeI-XhoI-digested pRS404 (TRP1). Second, the ADH1-TEF2 promoters were released from pRS404 using a NotI-XhoI digest and ligated into NotI-XhoI-cut pRS405 (LEU2), generating pPMB18.
Media and standard methods.
Protocols for DNA and Western blotting, DNA/RNA preparations, and the transformation of yeast and bacteria, as well as the composition of growth media for yeast and bacteria, were published previously (4, 41, 43). For protein blots, the primary anti-TAP antiserum (Open Biosystems) and the anti-Pgk1 antiserum (a gift from Per Ljungdahl) both were diluted 1:1,000. The PCR-based determination of the MAT genotype was described before (6).
RT-qPCR.
Reverse transcriptase quantitative PCR (RT-qPCR) procedures and primers used for amplifying the ACT1 control cDNA were described before (7). The primers used for amplifying the MTS1 cDNA were 5′ TCCACAAAAACCCAAAAAGC and 5′ TTCTTTGGCAACGAGGTCTT.
Analysis of chromatin immunoprecipitation (ChIP) results.
Primary data published previously (9) were used in combination with the MochiView software (30) to scan the K. lactis genome for Mts1-Myc binding sites.
Genetic selection.
pPMB18 (330 μg) was linearized with BamHI and introduced into SAY990. Eleven independent transformations with 30 μg of linearized plasmid resulted in approximately 1.3 × 10−4 colonies on SC-Leu plates. The colonies were replica printed to SC-Leu plates containing 150 μg/ml G418 (designated SC-Leu + 150 μg/ml G418). Colonies growing on SC-Leu + 150 μg/m G418 plates were transferred to fresh plates (containing SC-Leu + 150 μg/ml G418) and then patched on SC-Leu and replica printed on a yeast extract-peptone-dextrose (YEPD) plate containing a lawn of the test mater SAY44 (MATα). The mating plates were further replica printed on SD+Ura to score mating-proficient colonies. Inverse PCR (32) followed by DNA sequencing were performed to identify the genomic position of pPMB18 in the mating-proficient strains. The pPMB18 insertion in the RAS2 gene (C13387g) corresponded to amino acid 73 (out of 284).
RESULTS
K. lactis switches mating type with a rate of 6 × 10−4 events/generation in rich medium.
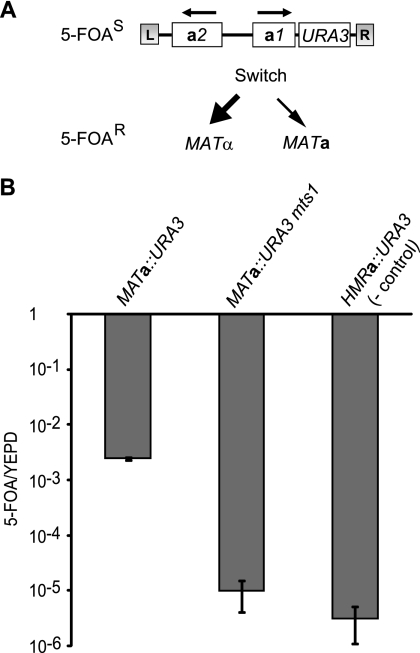
K. lactis strains grown under laboratory conditions exhibit stable mating types and hence switch mating type infrequently (3). Previously, we have observed low-level switching when strains were grown in nutrient-limited conditions (6), but we lacked a reliable assay for measuring switching rates. Measuring switching rates using quantitative PCR is unfeasible, as MAT, HMLα, and HMRa all are flanked by identical repetitive sequences (L and R), such that long amplicons would be necessary to distinguish between MATα and MATa. To establish a non-PCR-based assay measuring switching rates, we generated a strain that contained an insertion of the URA3 gene in the MATa locus (Fig. 1 A). The URA3 gene was inserted downstream of the MATa1 gene without disrupting the open reading frame (ORF). We predicted that mating-type switching would lead to the elimination of the URA3 gene. Because ura3 strains are resistant to 5-fluoro-orotic acid (5-FOA), mating-type switching rates could be determined by measuring the fraction of cells that became 5-FOAr. As a control, we also generated a strain with an insertion of the URA3 gene downstream of the HMRa1 gene. Since HMRa acts as a donor during switching rather than as a recipient, the HMRa::URA3 locus should be lost at a very low rate. The strains were inoculated into rich medium from a plate selecting for the insertion (SC-Ura), grown for six to eight generations, and then plated on 5-FOA and YEPD plates. The average 5-FOA/YEPD ratio was 4.6 × 10−3 (standard errors from the means [SEM], 1.2 × 10−3) for the MATa::URA3 strain in 11 independent experiments (Fig. 1B). For the HMRa::URA3 strain the average 5-FOA/YEPD ratio was only 3.0 × 10−6, which is 1,000-fold lower than that for the MATa::URA3 strain. We tested the mating type of eight 5-FOA-resistant segregants obtained from the MATa::URA3 strain by MAT-specific PCR and found that they had the MATα genotype. Hence, the 5-FOAr strains had switched their mating type and the MATa::URA3 strain preferentially used the HMLα locus as the donor.
Fig. 1.
Genetic assay for measuring switching rates. (A) Schematic of the tester strain used for measuring switching rates. The S. cerevisiae URA3 gene was inserted downstream of the MATa1 gene. Upon mating-type switching, the MATa::URA3 locus is replaced by the sequences present at HMLα (most frequently) or HMRa, resulting in 5-FOA resistance. (B) The ratios of 5-FOA/YEPD plating efficiencies were determined in strains SAY1488 (MATa::URA3), SAY1554 (MATa::URA3 mts1Δ::LEU2), and SAY1489 (HMRa::URA3). Error bars represent the standard errors from the means calculated from 11 (SAY1488), 3 (SAY1554), and 3 (SAY1489) independent experiments.
If this assay accurately measured switching, then it should be possible to block switching using the appropriate mutations. To test this idea, we determined switching in a MATa::URA3 mts1Δ mutant strain. Since Mts1 is critical for the induction of switching, we expected this strain to have a drastically reduced switching rate. In the mts1Δ mutant strain, the average 5-FOA/YEPD ratio was 9.7 × 10−6, which is comparable to the ratio in the HMRa::URA3 strain. We conclude that the assay accurately measured switching frequencies, with a mating-type switching frequency of cells grown in rich medium of approximately 6 × 10−4 events/generation.
Identification of Ras2 as a regulator of mating-type switching.
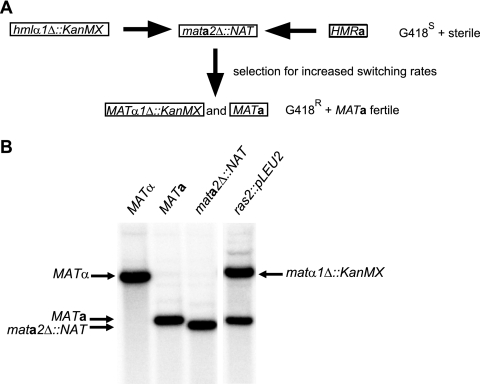
To identify genes that regulate switching in K. lactis, we performed a genetic selection for mutations that increased switching rates. A previous selection (6) showed that a physical translocation of a kanMX gene, encoding G418 resistance, from a silent locus into the expressed MAT locus would lead to its expression. We used a strain in which the HMLα1 gene was exchanged for the kanMX gene. In addition, the strain contained a deletion of the MATa2 gene, and since the MATa2 gene is necessary for mating, the resulting strain was sterile (Fig. 2 A). In K. lactis, illegitimate recombination frequencies are high, making it possible to use exogenously added DNA as a mutagen. Hence, the mata2Δ::NAT hmlα1Δ::kanMX strain was mutagenized using the random insertion of a plasmid containing a LEU2 selectable marker into the genome. We next selected for G418-resistant colonies and tested if such isolates could mate with a MATα tester strain. The goal of this procedure was to obtain mutations that resulted in constitutive switching, first switching to matα1Δ::kanMX, becoming G418 resistant, and then switching to MATa, becoming fertile. Among the isolated mutants was a strain that showed poor growth, and DNA blotting using a MAT-specific probe revealed that the strain formed a mixture of MATa and matα1Δ::kanMX genotypes (Fig. 2B). An inverse PCR procedure showed that the plasmid insertion was in the 5′ end of the RAS2 gene (C13387g), resulting in a disruption of the RAS2 ORF. RAS2 encodes a small GTPase with a central role in growth regulation in all eukaryotes. In fungi, Ras proteins are known to stimulate adenylate cyclase and hence cyclic AMP (cAMP) production (12, 46). K. lactis contains only one RAS ortholog, and in directed experiments (data not shown) we were unable to generate a haploid ras2 deletion, indicating that the RAS2 gene was essential in K. lactis. Hence, the allele we obtained most likely was a partial loss-of-function mutation. As the ras2::pLEU2 mutation imparted a serious germination defect (data not shown), we were unable to demonstrate that this mutation was responsible for the hyperswitching phenotype using an allelism test that involved tetrad analysis. To make sure that the ras2 mutation caused the hyperswitching phenotype, we introduced a plasmid-borne wild-type RAS2 gene into the ras2::pLEU2 strain. The introduction of RAS2 resulted in the restoration of the hyperswitching phenotype (data not shown). We concluded that Ras2 regulated mating-type switching in K. lactis.
Fig. 2.
ras2 mutation resulted in increased switching rates. (A) Schematic drawing of the genetic selection used for isolating strains with increased switching rates. The starting strain (mata2Δ::NAT hmlα1::kanMX), which was G418s and sterile, was mutagenized, and revertant strains were isolated that were G418r (matα1Δ::kanMX) and fertile (MATa). (B) DNA blot analysis of BamHI-digested DNA from SAY129 (MATα), SAY130 (MATa), SAY990 (mata2Δ::NAT), and SAY1069 (isolated from the selection described for panel A, ras2::pLEU2) hybridized with a MAT-specific probe. The MATα, MATa, mata2Δ::NAT, and matα1Δ::kanMX bands are indicated.
PDE2 is required for normal switching rates.
Because compromised Ras2 activity will lower cAMP levels and result in hyperswitching, we tested if higher cAMP levels would inhibit switching. To obtain a strain with increased cAMP levels, we generated a strain with a deletion of the PDE2 gene (A3619g). Pde2 is a cAMP phosphodiesterase that degrades cAMP, hence limiting intracellular cAMP levels. We predicted that pde2 mutants would have lower switching rates, the phenotype opposite that of ras2.
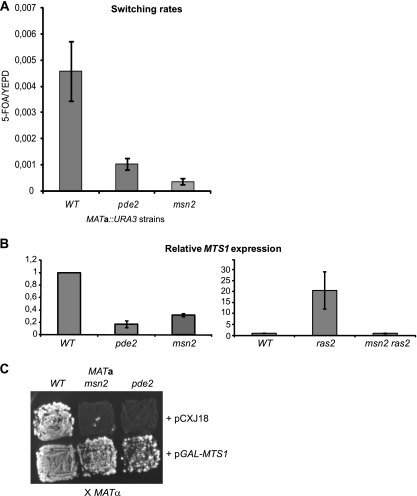
The average 5-FOA/YEPD ratio in the pde2 strain was 1.0 × 10−3 (SEM, 2.3 × 10−4), indicating a 5-fold reduced switching rate compared to that of the wild type (Fig. 3 A). These results are consistent with a role for cAMP in the regulation of switching.
Fig. 3.
ras2, pde2, and msn2 mutations affected MTS1 transcription. (A) The ratios of 5-FOA/YEPD plating efficiencies were determined in strains SAY1488 (MATa::URA3), SAY1546 (MATa::URA3 pde2Δ::NAT), and SAY1539 (MATa::URA3 msn2Δ::kanMX) to determine switching rates. Error bars represent the standard errors from the means calculated from 11 (SAY1488), 7 (SAY1546), and 6 (SAY1539) independent experiments, respectively. (B) RT-qPCR analysis using primers specifically amplifying the MTS1 cDNA. The y axes show relative expression levels of MTS1 mRNA. In the left panel, mRNA was prepared from strains SAY1488 (WT), SAY1546 (pde2Δ::NAT), and SAY1539 (msn2Δ::kanMX). In the right panel, mRNA was prepared from SAY990 (WT), SAY1069 (ras2::pLEU2), and SAY1664 (msn2Δ::kanMX ras2::pLEU2). The level of expression in the wild-type strains was set to 1.0. The data were normalized to the expression of the ACT1 gene using the comparative threshold cycle (CT) method. Error bars indicate the maximum and minimum values obtained in two separate experiments. (C) Mating tests of the MATa strains WT (SAY572), msn2Δ::kanMX (SAY1379), and pde2Δ::NAT (SAY1131) to the MATα tester strain (WM52). Diploids were selected on SC-Ade-Lys.
MSN2 also is required for normal switching rates.
We attempted to identify the transcription factor that is responsible for the cAMP-dependent regulation. Two key transcription factors responding to cAMP levels in S. cerevisiae are Msn2 and Msn4. The activity of the paralogous Msn2/4 proteins is inactivated by PKA-dependent phosphorylation (18, 25), leading to reduced transcription from Msn2/4-activated genes when levels of cAMP are high. Hence, a K. lactis MSN2/4 ortholog was an excellent candidate for mediating a cAMP-dependent regulation of switching. BLAST searches of the K. lactis genome using S. cerevisiae Msn2/4 as queries identified a single Msn2/4 ortholog (F26961g) which had the highest similarity to Msn2.
We deleted the MSN2 gene and measured switching rates in the resulting strain. In the msn2 strain the 5-FOA/YEPD ratio was only 3.5 × 10−4 (SEM, 1.2 × 10−4), demonstrating a 13-fold decreased switching rate compared to that of the WT. Hence, Msn2 regulates switching in K. lactis.
Transcription of MTS1 is regulated by Msn2, Ras2, and Pde2.
The Mts1 protein induces mating-type switching in K. lactis by binding to sites in both the MATa and MATα loci. Since we previously determined that the transcription of MTS1 was induced in nutrient-limited conditions (6), we hypothesized that Ras2/Pde2/Msn2 exert their effects on switching through regulating the transcription of the MTS1 gene. Consistently with this idea, Msn2 binds the stress response element (STRE) in S. cerevisiae, which has the sequence AGGGG. In the 3.2-kb intergenic region upstream of the MTS1 ORF there are five consensus STREs and six close matches to the STRE. We investigated the expression of the MTS1 gene normalized to the expression of ACT1 (Fig. 3B) by RT-qPCR. In the ras2 mutant strain, MTS1 transcription was upregulated 20-fold compared to that of the isogenic wild-type strain, explaining the hyperswitching phenotype. In contrast, MTS1 mRNA levels in the pde2Δ and msn2Δ mutant strains were reduced 5- and 3-fold, respectively, compared to that of their parental strain. To test if the increased MTS1 transcription in the ras2 mutant required Msn2, we measured MTS1 transcription in a ras2 msn2 double mutant strain. This strain expressed levels of MTS1 comparable to those of the wild type, indicating that Msn2 acts downstream of Ras2 in the regulation of switching. These data suggest that low levels of cAMP induce MTS1 transcription and switching, and that high levels of cAMP repress MTS1 transcription and switching.
Msn2 and Pde2 regulate mating.
Previously, an expression microarray experiment comparing a wild-type strain to an mts1Δ strain revealed that 20 genes were downregulated (>2-fold) and 52 genes were upregulated (>4-fold) in the mts1Δ strain (9). Specifically, Mts1 directly activated the transcription of a group of genes required for mating, which is consistent with the observation that mts1Δ strains are sterile. Because Mts1 directly activates genes required for mating, we anticipated that the Ras2/cAMP/Msn2 pathway would regulate mating in addition to its role in mating-type switching. We tested this notion by comparing the mating efficiency of MATa msn2 and MATa pde2 to that of an isogenic wild-type MATa strain. The strains were mixed with a MATα tester mater strain on medium that only allowed the growth of diploid cells (SC-Ade-Leu). Both the pde2 and msn2 genes were required for efficient mating, as the formation of diploids was severely reduced in the mutant strains (Fig. 3C). We predicted that the mating defect of msn2 and pde2 were the result of defective Mts1 induction. To test this idea, we introduced a pGAL-MTS1 plasmid into these strains, expecting that ectopic Mts1 expression would complement the mating defect. Ectopic Mts1 expression indeed improved mating in the msn2 and pde2 strains (Fig. 3C). Hence, Msn2/Pde2 regulates both mating and mating-type switching in K. lactis.
Autoregulation of MTS1 transcription.
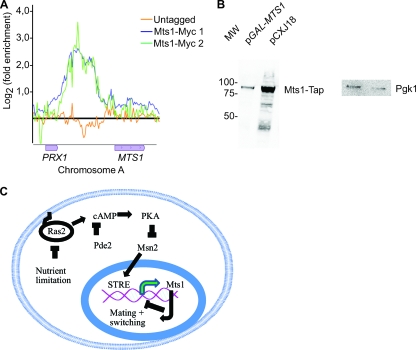
Others recently published a chromatin immunoprecipitation analyzed genome-wide by hybridization to a microarray (designated ChIP-on-chip) using epitope-tagged Mts1 (9). By further examining this data set, we discovered that a prominent Mts1 binding peak was present upstream of the MTS1 gene (Fig. 4 A). This indicated that Mts1 regulates the transcription of its own gene. To test this idea, we generated an MTS1-TAP allele regulated by the natural promoter at the endogenous chromosomal locus. We then introduced a plasmid into this strain in which Mts1 was overexpressed using an ectopic GAL1 promoter (pGAL-MTS1). We then investigated the steady-state levels of endogenous Mts1-Tap. Compared to plasmid alone, the endogenous Mts1-Tap protein was reduced 7-fold when Mts1 was overexpressed (Fig. 4B). Given that Mts1 binds the MTS1 regulatory region and that the overexpression of Mts1 represses MTS1 transcription, we conclude that MTS1 transcription is negatively autoregulated.
Fig. 4.
Negative autoregulation by Mts1 and preliminary model for regulation of switching. (A) An MTS1-Myc strain was subjected to ChIP-on-chip analysis (9) and analyzed using MochiView software (30). The x axis shows log2 (fold enrichment) ratios of two independent Mts1-Myc ChIPs to the input (green and blue lines) and an untagged strain (orange line). The genomic location is shown on the y axis. (B) Protein blot analysis of protein extracts from strain SAY988 (MTS1-TAP) containing plasmid alone (pCXJ18) or pGAL-MTS1 (pPMB35) using an anti-TAP antiserum. The right panel shows the same membrane using an anti-Pgk1 antiserum. (C) Schematic model for regulation of switching in K. lactis. PKA, protein kinase A; STRE, stress response element.
DISCUSSION
In this study, we demonstrate that mating-type switching is regulated through the RAS/cAMP pathway in K. lactis. We suggest a first model (Fig. 4C) in which Ras2 senses nutrient levels, thereby regulating cAMP production. High cAMP levels repress the activity of the Msn2 transcription factor. We assume that Msn2 activity is regulated by PKA in K. lactis similarly to Msn2 regulation by PKA in S. cerevisiae. An alignment of Saccharomyces and Kluyveromyces Msn2 reveals that residues in the NLS predicted to be targets of PKA in S. cerevisiae are partly conserved in K. lactis (data not shown). However, we have not been able to demonstrate that the subcellular localization of Msn2 changes in ras2 mutants compared to that in the WT.
When Ras2 is inactive or its function is compromised, Msn2 activates the transcription of Mts1, which in turn promotes mating-type switching. Given the presence of Msn2 binding sites (STREs) in the MTS1 regulatory region, we reason that the role of Msn2 in this aspect is direct. It turns out that the epitope tagging of Msn2 renders the protein inactive (data not shown), which has prevented us from demonstrating Msn2 binding to the MTS1 promoter by ChIP. We tested another cAMP-responsive candidate protein for the regulation of MTS1 transcription, called Gis1, but the deletion of GIS1 did not affect switching rates (data not shown). A prediction from this model is that MTS1 induction should rely on Msn2. This prediction was confirmed (Fig. 3B), as the absence of Msn2 suppressed the 20-fold overexpression of MTS1 observed in the ras2 mutant background. However, the ras2 msn2 double mutant expressed more MTS1 than the msn2 single mutant did (Fig. 3B). Hence, it is possible that Ras2 also affects Mts1 expression in an Msn2-independent manner.
The intergenic region representing the upstream region of MTS1 and the downstream region of the next ORF (PRX1) is unusually large (3.2 kb), considering that the average intergenic region in K. lactis is only about 500 bp (20). We speculate that this reflects a complex regulation of MTS1 transcription, giving room for binding sites for multiple regulatory proteins. Apart from the Msn2 binding sites presumably mediating a cAMP-dependent response, there is also a binding site for the a1/α2 heterodimer (9). The a1/α2 heterodimer represses the transcription of MTS1 in diploids or haploids with defects in the silencing of the cryptic mating-type loci. In S. cerevisiae, several genes required for mating are directly repressed by a1/α2 and are collectively known as haploid-specific genes. However, K. lactis haploid-specific genes are directly induced by Mts1 and not repressed by a1/α2. As MTS1 is repressed by a1/α2, the final output in S. cerevisiae and K. lactis is the same, but during the evolution of ascomycetes a rewiring of the regulation has occurred.
We also show that Mts1 is autoregulated, mediating the repression of its own transcript (Fig. 4). Negative autoregulation in other systems has been shown to reduce cell-to-cell fluctuations in steady-state levels of transcription factors (8) and also to speed up the response times of transcriptional networks (40). Hence, negative autoregulation by Mts1 is likely to add robustness to the mating and switching responses.
We have shown previously that Mts1 directly regulates switching through binding sites in the MATα locus. Mts1 also plays a pivotal role in regulating mating by inducing haploid-specific genes (9). Hence, Mts1 induces both mating and switching, but presumably an individual cell must do either. We hypothesize that this choice is regulated. It seems as if switching should be the secondary choice, as switching is costly and results in an inbreeding of the resulting offspring. A nutrient limitation-induced mating reaction, on the other hand, is more direct and also opens up the possibility for outcrossing. Hence, if there are cells of the opposite mating type in the vicinity, it would make sense to mate rather than switch. An attractive possibility is that mating pheromones inhibit switching (obviously pheromones induce mating), which is an idea that will be interesting to test in the future.
It is interesting that the regulation of switching in K. lactis is completely different from the regulation of switching in S. cerevisiae. In homothallic S. cerevisiae strains, switching occurs in mother cells in the G1 phase of the cell cycle, resulting in a very high switching rate. In contrast, in K. lactis switching rates are relatively low (6 × 10−4) in rich medium (Fig. 1B) and are regulated by nutrient availability (6, 27). Because the K. lactis genome contains an HO pseudogene in a syntenic position compared to the S. cerevisiae genome, the common ancestor probably used an HO-mediated switching mechanism. After their separation, K. lactis evolved a novel switching mechanism that acquired a different type of regulation. We argue that the differential regulation of switching in K. lactis has evolved as a consequence of the haplophase being the predominant life cycle. Natural isolates of K. lactis strains are haploid, in contrast to S. cerevisiae, where natural isolates are diploid. The diplophase is unstable in K. lactis, probably explaining the inclination for a haploid DNA content. Hence, K. lactis growing in nature cannot immediately sporulate in response to nitrogen and carbon starvation but must mate and form diploids first. Therefore, the Ras/cAMP regulation of mating and switching in K. lactis may promote survival during starvation by ensuring the formation of resilient spores.
Cyclic AMP signaling controls a wide range of responses in fungi (10, 19). A few examples are the pseudohyphal growth response in S. cerevisiae, which is controlled by intracellular cAMP concentrations (33). Morphological changes in fungi such as Candida albicans (21) and Cryptococcus neoformans (1) also are regulated by Ras/cAMP, and these morphological switches are critical for pathogenicity. Hence, our data add to an extensive list of responses to cAMP signaling in fungi, but to our knowledge this work is the first link between Ras/cAMP and mating-type switching.
ACKNOWLEDGMENTS
L. Booth and A. Johnson are acknowledged for sharing the Mts1-Myc ChIP-on-chip data and for assistance using the MochiView software. We thank P. Ljungdahl and Claes Andreásson for the critical reading of the manuscript and the gift of the anti-Pgk1 antiserum.
This work was supported by grants from the Swedish Cancer Society and the Carl Trygger Foundation to S.U.Å.
Footnotes
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Alspaugh J. A., Cavallo L. M., Perfect J. R., Heitman J. 2000. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans. Mol. Microbiol. 36:352–365 [DOI] [PubMed] [Google Scholar]
- 2. Andrews B. J., Herskowitz I. 1989. Identification of a DNA binding factor involved in cell-cycle control of the yeast HO gene. Cell 57:21–29 [DOI] [PubMed] [Google Scholar]
- 3. Åström S. U., Kegel A., Sjöstrand J. O., Rine J. 2000. Kluyveromyces lactis Sir2p regulates cation sensitivity and maintains a specialized chromatin structure at the cryptic alpha-locus. Genetics 156:81–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ausubel F. M. 1999. Short protocols in molecular biology: a compendium of methods from current protocols in molecular biology. Wiley, New York, NY [Google Scholar]
- 5. Bähler J., et al. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943–951 [DOI] [PubMed] [Google Scholar]
- 6. Barsoum E., Martinez P., Åström S. U. 2010. Alpha3, a transposable element that promotes host sexual reproduction. Genes Dev. 24:33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Barsoum E., Sjöstrand J. O., U. Åström S. 2010. Ume6 is required for the MATa/MATalpha cellular identity and transcriptional silencing in Kluyveromyces lactis. Genetics 184:999–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Becskei A., Serrano L. 2000. Engineering stability in gene networks by autoregulation. Nature 405:590–593 [DOI] [PubMed] [Google Scholar]
- 9. Booth L. N., Tuch B. B., Johnson A. D. 2010. Intercalation of a new tier of transcription regulation into an ancient circuit. Nature 468:959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Borges-Walmsley M. I., Walmsley A. R. 2000. cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 8:133–141 [DOI] [PubMed] [Google Scholar]
- 11. Boy-Marcotte E., Perrot M., Bussereau F., Boucherie H., Jacquet M. 1998. Msn2p and Msn4p control a large number of genes induced at the diauxic transition which are repressed by cyclic AMP in Saccharomyces cerevisiae. J. Bacteriol. 180:1044–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Broek D., et al. 1985. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell 41:763–769 [DOI] [PubMed] [Google Scholar]
- 13. Chen X. J. 1996. Low- and high-copy-number shuttle vectors for replication in the budding yeast Kluyveromyces lactis. Gene 172:131–136 [DOI] [PubMed] [Google Scholar]
- 14. Chen X. J., Clark-Walker G. D. 1994. sir2 mutants of Kluyveromyces lactis are hypersensitive to DNA-targeting drugs. Mol. Cell. Biol. 14:4501–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coluccio A. E., Rodriguez R. K., Kernan M. J., Neiman A. M. 2008. The yeast spore wall enables spores to survive passage through the digestive tract of Drosophila. PLoS One 3:e2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cosma M. P. 2004. Daughter-specific repression of Saccharomyces cerevisiae HO: Ash1 is the commander. EMBO Rep. 5:953–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cosma M. P., Tanaka T., Nasmyth K. 1999. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell 97:299–311 [DOI] [PubMed] [Google Scholar]
- 18. De Wever V., Reiter W., Ballarini A., Ammerer G., Brocard C. 2005. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 24:4115–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. D'Souza C. A., Heitman J. 2001. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25:349–364 [DOI] [PubMed] [Google Scholar]
- 20. Dujon B., et al. 2004. Genome evolution in yeasts. Nature 430:35–44 [DOI] [PubMed] [Google Scholar]
- 21. Feng Q., Summers E., Guo B., Fink G. 1999. Ras signaling is required for serum-induced hyphal differentiation in Candida albicans. J. Bacteriol. 181:6339–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gasch A. P., et al. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goldstein A. L., McCusker J. H. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- 24. Görner W., et al. 1998. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 12:586–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Görner W., et al. 2002. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 21:135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haber J. E. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561–599 [DOI] [PubMed] [Google Scholar]
- 27. Herman A., Roman H. 1966. Allele specific determinants of homothallism in Saccharomyces lactis. Genetics 53:727–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Herskowitz I. 1988. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol. Rev. 52:536–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Herskowitz I., Rine J., Strathern J. N. 1992. Mating type determination and mating-type interconversion in Saccharomyces cerevisiae, p. 583–656 In Jones E. W., Pringle J. R., Broach J. R. (ed.), The molecular and cellular biology of the yeast Saccharomyces, vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. Homann O. R., Johnson A. D. 2010. MochiView: versatile software for genome browsing and DNA motif analysis. BMC Biol. 8:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jacquet M., Renault G., Lallet S., De Mey J., Goldbeter A. 2003. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J. Cell Biol. 161:497–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kegel A., Martinez P., Carter S. D., Åström S. U. 2006. Genome wide distribution of illegitimate recombination events in Kluyveromyces lactis. Nucleic Acids Res. 34:1633–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lorenz M. C., Heitman J. 1997. Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 16:7008–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martínez-Pastor M. T., et al. 1996. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J. 15:2227–2235 [PMC free article] [PubMed] [Google Scholar]
- 35. Mayordomo I., Estruch F., Sanz P. 2002. Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (stress response element)-regulated genes. J. Biol. Chem. 277:35650–35656 [DOI] [PubMed] [Google Scholar]
- 36. Medvedik O., Lamming D. W., Kim K. D., Sinclair D. A. 2007. MSN2 and MSN4 link calorie restriction and TOR to sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. PLoS Biol. 5:e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nasmyth K. 1993. Regulating the HO endonuclease in yeast. Curr. Opin. Genet. Dev. 3:286–294 [DOI] [PubMed] [Google Scholar]
- 38. Nickoloff J. A., Chen E. Y., Heffron F. 1986. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc. Natl. Acad. Sci. U. S. A. 83:7831–7835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roetzer A., et al. 2008. Candida glabrata environmental stress response involves Saccharomyces cerevisiae Msn2/4 orthologous transcription factors. Mol. Microbiol. 69:603–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rosenfeld N., Elowitz M. B., Alon U. 2002. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 323:785–793 [DOI] [PubMed] [Google Scholar]
- 41. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 42. Schaffrath R., Breunig K. D. 2000. Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet. Biol. 30:173–190 [DOI] [PubMed] [Google Scholar]
- 43. Schiestl R. H., Gietz R. D. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339–346 [DOI] [PubMed] [Google Scholar]
- 44. Schmitt A. P., McEntee K. 1996. Msn2p, a zinc finger DNA-binding protein, is the transcriptional activator of the multistress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 93:5777–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith A., Ward M. P., Garrett S. 1998. Yeast PKA represses Msn2p/Msn4p-dependent gene expression to regulate growth, stress response and glycogen accumulation. EMBO J. 17:3556–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Toda T., et al. 1985. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell 40:27–36 [DOI] [PubMed] [Google Scholar]