Abstract
Shigella flexneri is the major Shigella species that causes diarrheal disease in developing countries. It is further subdivided into 15 serotypes based on O-antigen structure. Serotyping of S. flexneri is important for epidemiological purposes. In this study, we developed a multiplex PCR assay targeting the O-antigen synthesis gene wzx and the O-antigen modification genes gtrI, gtrIC, gtrII, oac, gtrIV, gtrV, and gtrX for molecular serotyping of S. flexneri. The multiplex PCR assay contained eight sets of specific PCRs in a single tube and can identify 14 of the 15 serotypes (the exception being serotype Xv) of S. flexneri recognized thus far. A nearly perfect concordance (97.8%) between multiplex PCR assay and slide agglutination was observed when 358 S. flexneri strains of various serotypes were analyzed, except that 8 strains were carrying additional cryptic and/or defective serotype-specific genes. The multiplex PCR assay provides a rapid and specific method for the serotype identification of S. flexneri.
INTRODUCTION
Shigella is the major pathogen causing bacterial dysentery in developing countries. There are about 164.7 million cases of shigellosis annually worldwide, resulting in 1,100,000 deaths, most of which are children under 5 years of age (10). Of the four species of Shigella, S. flexneri is the predominant species affecting economically poor populations.
S. flexneri is further divided into different serotypes based on the structure of the O antigen. At least 15 serotypes have been recognized: 1a, 1b, 1c, 2a, 2b, 3a, 3b, 4a, 4b, 5a, 5b, X, Xv, Y, and F6 (15, 17, 21). Serological analysis of S. flexneri has long been used to characterize isolates for epidemiological and bacteriological purposes.
A slide agglutination method using rabbit antiserum raised against specific type and group factors has been the only means to routinely identify the serotypes of S. flexneri. Commercially available diagnostic antisera have been used in microbiology laboratories worldwide. However, there exist some deficiencies in this scheme. First, it is time-consuming since up to 10 separate agglutination tests using antisera for type antigens I, II, III, IV, V, and VI and for group antigens 3;4, 7;8, and 6 and the monoclonal antibody MASF1c for 1c are required to serotype an isolate (16, 18, 21). Second, an incorrect reading can occur as a result of visual assessment of the agglutination reactions. Finally, expensive antiserum kits limit its application in the laboratories of developing countries.
All serotypes except serotype 6 of S. flexneri share a polysaccharide backbone comprising repeating units of a tetrasaccharide in the polysaccharide (15). The basic O antigen is referred to as serotype Y, and the addition of glucosyl and/or O-acetyl residues to the tetrasaccharide unit results in serological type (i.e., I, II, III, IV, and V)-, group (i.e., 3;4, 7;8, and 6)-, and 1c-specific antigenic determinants (17).
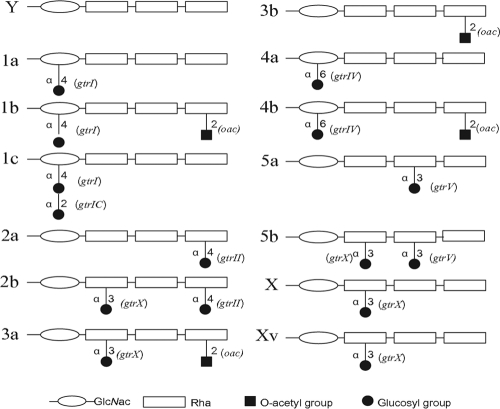
Three genes—gtrA, gtrB, and gtr (type)—are responsible for the glucosylation of S. flexneri. The first two genes are highly homologous and interchangeable, whereas the third gene, gtr (type), is unique and encodes the serotype-specific glucosyltransferase (3, 17). The serotype-specific gtr genes for types I, II, IV, and V, group 7;8, and 1c are gtrI, gtrII, gtrIV, gtrV, gtrX, and gtrIC, respectively (1, 2, 8, 9, 12, 17). The gtr genes are encoded by different prophage genomes within the host chromosome. O-acetylation, which results in group 6 and/or type III O-antigen modification in serotype 1b, 3a, 3b, and 4b strains, is mediated by the oac gene, which is also carried by prophage (6, 19). Strains of different serotypes characteristically carry one or more of the appropriate prophage genomes encoding specific O-antigen modification genes (Fig. 1).
Fig. 1.
Chemical composition and specific O-antigen modification genes of different serotypes of S. flexneri. The basic O antigen consists of repeating units of tetrasaccharide. Serotypes differ by the addition of either glucosyl or O-acetyl groups to different sugars within the tetrasaccharide repeat unit via the linkages indicated. Specific O-antigen modification genes for different serotypes are indicated in parentheses.
In the present study, we developed a multiplex PCR assay targeting O-antigen modification genes to serotype S. flexneri. Except for the serotype X variant (Xv), which cannot be differentiated from serotype X, all other serotypes recognized up to now can be successfully serotyped using this approach. This multiplex PCR assay provides a rapid and specific method for molecular serotyping of S. flexneri.
MATERIALS AND METHODS
Bacterial strains.
A total of 14 S. flexneri strains (Table 1) representing 14 serotypes recognized (except for serotype 5b, of which we have no strains in our collection) are used to set up the conditions for a multiplex PCR using eight primer pairs (see below). A total of 358 S. flexneri strains (Table 2) were analyzed to evaluate the validation of the multiplex PCR assay. To test the cross-reaction of primers used in the present study, 50 strains of other species were tested (the number of strains of each species is given in parentheses): S. sonnei (n = 2), S. dysenteriae (n = 12, including all 12 serotypes), S. boydii (n = 18, including all 18 serotypes), enteroaggregative Escherichia coli (n = 2), enterohemorrhagic E. coli O157:H7 (n = 3), enteroinvasive E. coli (n = 1), enteropathogenic E. coli (n = 1), enterotoxigenic E. coli (n = 1), uropathogenic E. coli (n = 1), E. coli K-12 (n = 2), Listeria monocytogenes (n = 1), Vibrio cholerae (n = 1), Salmonella enterica serovar Paratyphi A (n = 1), S. enterica serovar Paratyphi B (n = 2), Yersinia enterocolitica (n = 1), and Salmonella enterica serovar Choleraesuis (n = 1). The serotypes of S. flexneri strains were identified using Shigella monovalent antisera (Denka Seiken, Japan) and monoclonal antibody (Reagensia AB, Sweden). The Chinese S. flexneri strains used here were isolated from diarrheal patients in China and preserved in our laboratory. Other strains were obtained from the National Collection of Type Cultures (NCTC), United Kingdom.
Table 1.
Serotype characteristics of S. flexneri reference strains by agglutination and multiplex PCR
| Strain | Serotype | Agglutination |
Multiplex PCR |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Type |
Group |
MASF1c | |||||||||||||||||
| I | II | III | IV | V | VI | 3;4 | 6 | 7;8 | wzx1-5 | gtrI | gtrIC | gtrII | oac | gtrIV | gtrV | gtrX | |||
| 2000019 | 1a | + | – | – | – | – | – | + | – | – | − | + | + | − | − | − | − | − | − |
| 1997020 | 1b | + | – | – | – | – | – | + | + | – | – | + | + | – | – | + | – | – | – |
| 06HN081a | – | – | – | – | – | – | – | + | – | + | + | + | + | – | + | – | – | – | |
| 301 | 2a | – | + | – | – | – | – | + | – | – | – | + | – | – | + | – | – | – | – |
| NCTC4 | 2b | – | + | – | – | – | – | – | – | + | – | + | – | – | + | – | – | – | + |
| 03HL12 | 3a | – | – | + | – | – | – | – | + | + | – | + | – | – | – | + | – | – | + |
| 2002110 | 3b | – | – | + | – | – | – | – | + | – | – | + | – | – | – | + | – | – | – |
| NCTC9725 | 4a | – | – | – | + | – | – | + | – | – | – | + | – | – | – | – | + | – | – |
| NCTC9726 | 4b | – | – | – | + | – | – | – | + | – | – | + | – | – | – | + | + | – | – |
| 51247 | 5a | – | – | – | – | + | – | + | – | – | – | + | – | – | – | – | – | + | – |
| 2003036 | Y | – | – | – | – | – | – | + | – | – | – | + | – | – | – | – | – | – | – |
| 2001014 | X | – | – | – | – | – | – | – | – | + | – | + | – | – | – | – | – | – | + |
| 2002017 | Xv | – | – | – | + | – | – | – | – | + | – | + | – | – | – | – | – | – | + |
| 2000007 | F6 | – | – | – | – | – | + | – | – | – | – | – | – | – | – | – | – | – | – |
06HN081 is an untypeable F1 strain that was positive both for MASF1c and group 6 antisera. Based on this serological reaction pattern, the strain belongs to the newly defined serotype 7b (see the text for details).
Table 2.
Correlation of multiplex PCR and conventional serotyping for 358 S. flexneri strains tested
| Serotype | No. of strains | No. of strains positive for target gene |
Multiplex PCR serotype classification (no. of strains) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| wzx1-5 | gtrI | gtrIC | gtrII | oac | gtrIV | gtrV | gtrX | |||
| F1a | 25 | 25 | 25 | 0 | 0 | 0 | 0 | 0 | 0 | 1a (25) |
| F1b | 14 | 14 | 14 | 0 | 0 | 14 | 0 | 0 | 0 | 1b (14) |
| F2a | 55 | 55 | 0 | 0 | 55 | 0 | 0 | 0 | 0 | 2a (55) |
| F2b | 50 | 50 | 0 | 0 | 50 | 0 | 0 | 0 | 50 | 2b (50) |
| F3a | 10 | 10 | 0 | 0 | 0 | 10 | 0 | 0 | 10 | 3a (10) |
| F3b | 2 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 3b (2) |
| F4a | 5 | 5 | 0 | 0 | 0 | 0 | 5 | 0 | 0 | 4a (5) |
| F4b | 5 | 5 | 0 | 0 | 0 | 5 | 5 | 0 | 0 | 4b (5) |
| F5a | 4 | 4 | 0 | 0 | 0 | 1 | 0 | 4 | 0 | 5a (3), untypeable (1) |
| Y | 36 | 36 | 0 | 0 | 5 | 0 | 0 | 0 | 0 | Y (31), 2a (5) |
| Xv | 78 | 78 | 0 | 0 | 0 | 0 | 0 | 0 | 78 | X or Xv (78) |
| X | 69 | 69 | 0 | 0 | 2 | 0 | 0 | 0 | 69 | X or Xv (67), 2b (2) |
| F6 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | F6 |
Preparation of DNA templates.
DNA templates were prepared directly from bacterial colonies by the boiling method. Briefly, a single colony from an overnight culture at 37°C on LB agar was suspended in 30 μl of distilled water and boiled at 100°C for 10 min. The sample was immediately cooled on ice for 5 min and centrifuged at 13,000 × g at 4°C for 10 min. The supernatant, containing DNA, was used as the template for PCR amplification.
PCR primers.
Primers used in the present study are listed in Table 3. Primers used for the amplification of the O-antigen flippase gene wzx were from Li et al. (11). The other primers were designed for our study based on the sequences of the S. flexneri serotype-specific genes gtrI, gtrIC, gtrII, oac, gtrIV, gtrV, and gtrX. The primers were synthesized by Sangon Biotech (Shanghai) and dissolved in TE buffer (10 mM Tris-Cl, 1 mM EDTA [pH 8.0]) to obtain a 50 μM stock solution.
Table 3.
Primers used in this studya
| Target gene | Primer |
Amplicon size (bp) | Serotype specificity | Accession no. | |
|---|---|---|---|---|---|
| Orientationb | Sequence (5′−3′) | ||||
| gtrI | F | CTGTTAGGTGATGATGGCTTAG | 1,122 | 1a, 1b, 1c | AF139596 |
| R | ATTGAACGCCTCCTTGCTATGC | ||||
| gtrII | F | ATTTATTGTTATTGGGGGTGGTTG | 1,272 | 2a, 2b | AF021347 |
| R | ATTTGTTCTTTATTTGCTGGTT | ||||
| oac | F | CTGTTCGGCTTTGAAAGTGCTG | 604 | 1b, 3a, 3b, 4b | AF547987 |
| R | CGTAGGCGTACATAGCAAGCAAAGA | ||||
| gtrIV | F | ATGTTCCTCCTTCTTCCTTT | 378 | 4a, 4b | AF288197 |
| R | TCCTGATGCTACCTTATCCA | ||||
| gtrV | F | AATACGATTCTCCTGGTGCTAAAC | 905 | 5a, 5b | U82619 |
| R | TAGGGCATTGCTTGTATCTTTCAT | ||||
| gtrX | F | AATGCTGGATGGGATAATCACCTT | 425 | 2b, 3a, 5b, X, Xv | L05001 |
| R | GAGACGGCTTCTCCATGTTTTGCT | ||||
| wzx1-5d | F | CACTTGTTGGGTATGCTGG | 782 | 1-5, X, Xv, Y | AE005674 |
| R | CCGGCAAACAGATTAGAAA | ||||
| gtrIC | F | AGGGAATGGCATTAGGGATCGG | 518 | 1c | FJ905303 |
| R | GCTGCAAGTGGTTTTTGTTGGA | ||||
| wzx6c | F | TTAAGAGCGATCATTTC | 739 | F6 | EU294165 |
| R | CCATCCAAGCGGACATT | ||||
Data in the last three columns apply to both orientations of each primer.
F, forward; R, reverse.
The primer pair for wzx6 was used to confirm the F6 serotype.
The primer pair for wzx1-5 was used to amplify the wzx gene of all serotypes except for F6.
PCR amplification and detection.
Multiplex PCR were performed using Qiagen multiplex PCR kit (Qiagen). The reaction mixture for each PCR consisted of 1× PCR Master Mix (containing HotStar Taq DNA polymerase, multiplex PCR buffer, and deoxynucleoside triphosphate mix), 0.2 μM concentrations of each primer, and 3 μl of template DNA in a final reaction volume of 50 μl. PCR amplification was performed using a standard multiplex PCR cycling protocol according to the instruction for the kit: 95°C for 15 min, followed by 30 cycles of 94°C for 30 s, 55°C for 90 s, and 72°C for 60 s, with a final extension of 72°C for 10 min in a thermocycler (Senso, Germany). A portion (5 μl) of the reaction mixture was mixed with loading buffer, subjected to electrophoresis in 1.5% agarose gel, and visualized by ethidium bromide staining. If necessary, PCR products were either sequenced directly or cloned into the pMD20-T TA cloning vector (TaKaRa, Japan) for sequencing.
RESULTS AND DISCUSSION
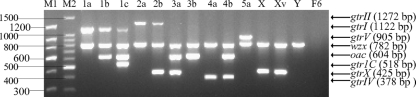
First, we performed singleplex PCR using template DNA extracted from reference strains. Each pair of primers obtained the predicted amplification product, which were 782 bp (wzx1-5), 1,122 bp (gtrI), 518 bp (gtrIC), 1,272 bp (gtrII), 604 bp (oac), 378 bp (gtrIV), 905 bp (gtrV), and 425 bp (gtrX). The identity of each product was further confirmed by DNA sequencing. We then performed multiplex PCR according to the protocols described in Materials and Methods. We tested annealing temperatures from 54 to 63°C, and the highest yield was obtained at 55°C (data not shown). Different serotypes show specific amplification patterns, as shown in Fig. 2 and Table 1. Each serotype gave the expected specific PCR products. The amplicon sizes showed good separation on 1.5% agarose gels and when two different molecular markers were used, and each PCR product could be unambiguously identified by size. The expected products for reference strains were as follows: 1a (wzx1-5 and gtrI), 1b (wzx1-5, gtrI, and oac), 1c (wzx1-5, gtrI, and gtrIC), 2a (wzx1-5 and gtrII), 2b (wzx1-5, gtrII, and gtrX), 3a (wzx1-5, oac, and gtrX), 3b (wzx1-5 and oac), 4a (wzx1-5 and gtrIV), 4b (wzx1-5, gtrIV, and oac), 5a (wzx1-5 and gtrV), X, Xv (wzx1-5 and gtrX), and Y (wzx1-5). wzx1-5 can also act as a control for PCR since it is expected to be positive for all S. flexneri serotypes except for serotype F6. There was no amplification for serotype F6, as expected, because the O-antigen synthesis genes of F6 are different from those of other S. flexneri serotypes and there are no modification genes (4, 15).
Fig. 2.
Multiplex PCR products of S. flexneri reference strains representing all 15 serotypes. PCR products were electrophoresed on a 1.5% (wt/vol) agarose gel, stained with ethidium bromide, and photographed under UV light. Serotypes are indicated above the lanes. M1 and M2, 150- and 100-bp DNA ladder markers (TaKaRa, Japan).
We attempted adding a pair of primers specific for serotype F6 to the reaction. However, since the reaction already contained eight primer pairs, adding an additional pair of primers significantly increases the difficulty of optimizing the multiplex PCR. After testing three pairs of primers based on the F6-specific O-antigen wzx6 gene sequence, we failed to optimize the multiplex PCR. Without a positive means for the identification of F6 in the multiplex PCR, caution should be exercised in the interpretation of negative PCR results as the F6 serotype. We strongly recommend confirmation of F6 serotype using singleplex PCR with the F6-specific primer pair listed in Table 3, which amplifies a 739-bp product. Since isolates would have been identified as S. flexneri using Shigella and subgroup B polyvalent sera prior to carrying out the multiplex PCR assay, the likelihood of an F6 false positive (multiplex PCR negative) is expected to be low. Serotype F6 prevalence was 6% on average, ranging from 0% (China) to 15% (Pakistan) in Asian countries in a study by von Seidlein et al. (20). Simultaneous singleplex F6 specific PCR may be conducted in countries where the prevalence of F6 is high.
Note that the primers used to amplify oac were designed based on the conserved region of oac and oac1b, which share only 88% identity (unpublished data). Note also that a reference strain for 5b was not available for testing; we expect amplification patterns of 905 bp (gtrV) and 425 bp (gtrX) for serotype 5b, based on the known O-antigen modification of 5b. Due to the unavailability of a typical serotype 1c strain, we used an F1 (untypeable) strain, 06HN081, which reacted with 1c-specific monoclonal antibody MASF1c, as a control for detecting the serotype 1c-specific modification gene gtrIC. In contrast to typical 1c strains, which reacted with MASF1c only, the F1 (untypeable) strain was positive both for MASF1c and for group 6 antisera. Correspondingly, the gtrIC and oac genes responsible for MASF1c and group 6 antigen were identified from the F1 (untypeable) strain by multiplex PCR and confirmed by sequencing. These results suggest that the O-antigen of the F1 (untypeable) strain was a modification of 1b by GtrIC and is a new serotype (Table 1). This new serotype has the same serological reaction pattern as the newly proposed serotype 7b by Forster et al. (7), suggesting that the serotype of 06HN081 is 7b. It should be noted that Forster et al. redefined 1c as 7a since both 7a and 7b share a positive reaction with MASF1c (7). Thus, our multiplex PCR can identify these two newly defined serotypes.
Strains of serotype Xv and serotype X presented the same amplification pattern (wzx1-5 and gtrX) by multiplex PCR. Therefore, they are classified into the same serotype Xv or X. Serotype Xv has been one of the most predominant serotypes in China since 2002 (21). It was initially named as S. flexneri serotype 4c due to reaction with both monovalent anti-type IV serum and anti-group 7;8 serum (13). However, factors responsible for type IV antigenic determinant (v factor) had not been identified (21), and it is not yet feasible to develop a PCR method for typing Xv. An additional slide agglutination method using monovalent anti-type IV serum is necessary to differentiate serotype Xv from serotype X (21).
In order to evaluate the specificity of the primers used here, a multiplex PCR assay was also performed against 50 strains of other Shigella species and bacterial pathogens commonly present in stool samples. No amplification was detected from any of these strains. Thus, the specificity of our multiplex PCR is 100%.
To determine whether this multiplex PCR assay was generally applicable to all S. flexneri strains and to evaluate its validity, 358 S. flexneri strains of various serotypes were analyzed by this method (Table 2). All except eight isolates determined by multiplex PCR were the same as that determined by slide agglutination, with a concordance rate of 97.8%. For the eight exceptions, five, one, and two isolates were serotypes Y, 5a, and X, respectively (Table 2). A multiplex PCR showed that the serotype Y and X isolates were originally 2a and 2b, respectively, while the serotype 5a strain (NCTC8523) was originally a novel F5 serotype. Sequencing revealed that for the five Y isolates, four were due to frameshift mutations in gtrII with one having a four-base deletion (bases 1197 to 1282) and three having a single base deletion (base 1031). One has an intact gtrII, but the defect may lie in elsewhere. For the two serotype X strains, both had a single base deletion (base 1024) in gtrII. The anomalous serotype 5a isolate possesses unexpectedly an oac gene. DNA sequencing found that there is a two-base deletion (bases 345 to 346) in the oac gene, rendering it defective in this isolate. Serotype Y strains carrying a defective gtrII gene have been reported previously. One case was an insertion sequence disruption of gtrII, while another was a single amino acid residual change leading to an inactive GtrII (5, 14). The higher number of gtrII mutations found in serotype 2a and 2b S. flexneri strains may be due to a higher proportion of these serotypes in nature.
This study developed a multiplex PCR method for molecular serotyping of S. flexneri. With a single reaction, most serotypes (14 of 15) recognized up to now can be easily and specifically identified. In contrast to the conventional slide agglutination method, which needs up to 10 separate reactions to identify the serotype of an isolate, the multiple PCR assay saves time and does not require expensive antisera, especially for high-throughput serotype identification. PCR results can be obtained in less than 3.5 h, and isolates in batches of 96 (depending on the capacity of the thermocycler) can be processed simultaneously. The cost of PCR typing is only 25% of the cost of agglutination based serotyping in our experience, providing a substantial saving. The disadvantage of molecular serotyping is the presence of cryptic/defective O-antigen modification genes that can give rise to discordant results with serology-based typing. However, the proportion of such strains is small. On the other hand, this variation offers further differentiation than conventional agglutination-based serotyping. This method is available for high-throughput detection in any laboratories equipped with PCR facilities.
ACKNOWLEDGMENTS
This study was supported by grants 2011CB504901, 2008ZX10004-008, 2008ZX10004-009, 2009ZX10004-203, 2011SKLID203, 2008SKLID106, and YB20098450101 from the Ministry of Science and Technology and the State Key Laboratory for Infectious Disease Prevention and Control, People's Republic of China.
We thank Naresh Verma for providing the gtrIC sequence.
Footnotes
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Adams M. M., Allison G. E., Verma N. K. 2001. Type IV O antigen modification genes in the genome of Shigella flexneri NCTC 8296. Microbiology 147:851–860 [DOI] [PubMed] [Google Scholar]
- 2. Adhikari P., Allison G., Whittle B., Verma N. K. 1999. Serotype 1a O-antigen modification: molecular characterization of the genes involved and their novel organization in the Shigella flexneri chromosome. J. Bacteriol. 181:4711–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Allison G. E., Verma N. K. 2000. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 8:17–23 [DOI] [PubMed] [Google Scholar]
- 4. Cheah K. C., Beger D. W., Manning P. A. 1991. Molecular cloning and genetic analysis of the rfb region from Shigella flexneri type 6 in Escherichia coli K-12. FEMS Microbiol. Lett. 67:213–218 [DOI] [PubMed] [Google Scholar]
- 5. Chen J. H., Hsu W. B., Chiou C. S., Chen C. M. 2003. Conversion of Shigella flexneri serotype 2a to serotype Y in a shigellosis patient due to a single amino acid substitution in the protein product of the bacterial glucosyltransferase gtrII gene. FEMS Microbiol. Lett. 224:277–283 [DOI] [PubMed] [Google Scholar]
- 6. Clark C. A., Beltrame J., Manning P. A. 1991. The oac gene encoding a lipopolysaccharide O-antigen acetylase maps adjacent to the integrase-encoding gene on the genome of Shigella flexneri bacteriophage Sf6. Gene 107:43–52 [DOI] [PubMed] [Google Scholar]
- 7. Forster R. A. Structural elucidation of the O-antigen of the Shigella flexneri provisional serotype 88-893: structural and serological similarities with S. flexneri provisional serotype Y394 (1c). Carbohydr. Res. 346:872–876 [DOI] [PubMed] [Google Scholar]
- 8. Guan S., Bastin D. A., Verma N. K. 1999. Functional analysis of the O antigen glucosylation gene cluster of Shigella flexneri bacteriophage SfX. Microbiology 145:1263–1273 [DOI] [PubMed] [Google Scholar]
- 9. Huan P. T., Bastin D. A., Whittle B. L., Lindberg A. A., Verma N. K. 1997. Molecular characterization of the genes involved in O-antigen modification, attachment, integration and excision in Shigella flexneri bacteriophage SfV. Gene 195:217–227 [DOI] [PubMed] [Google Scholar]
- 10. Kotloff K. L., et al. 1999. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bull. World Health Organ. 77:651–666 [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y., et al. 2009. Molecular detection of all 34 distinct O-antigen forms of Shigella. J. Med. Microbiol. 58:69–81 [DOI] [PubMed] [Google Scholar]
- 12. Mavris M., Manning P. A., Morona R. 1997. Mechanism of bacteriophage SfII-mediated serotype conversion in Shigella flexneri. Mol. Microbiol. 26:939–950 [DOI] [PubMed] [Google Scholar]
- 13. Pryamukhina N. S., Khomenko N. A. 1988. Suggestion to supplement Shigella flexneri classification scheme with the subserovar Shigella flexneri 4c: phenotypic characteristics of strains. J. Clin. Microbiol. 26:1147–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roberts F., Jennison A. V., Verma N. K. 2005. The Shigella flexneri serotype Y vaccine candidate SFL124 originated from a serotype 2a background. FEMS Immunol. Med. Microbiol. 45:285–289 [DOI] [PubMed] [Google Scholar]
- 15. Simmons D. A., Romanowska E. 1987. Structure and biology of Shigella flexneri O antigens. J. Med. Microbiol. 23:289–302 [DOI] [PubMed] [Google Scholar]
- 16. Stagg R. M., Cam P. D., Verma N. K. 2008. Identification of newly recognized serotype 1c as the most prevalent Shigella flexneri serotype in northern rural Vietnam. Epidemiol. Infect. 136:1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stagg R. M., et al. 2009. A novel glucosyltransferase involved in O-antigen modification of Shigella flexneri serotype 1c. J. Bacteriol. 191:6612–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Talukder K. A., et al. 2003. Phenotypic and genotypic characterization of provisional serotype Shigella flexneri 1c and clonal relationships with 1a and 1b strains isolated in Bangladesh. J. Clin. Microbiol. 41:110–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Verma N. K., Brandt J. M., Verma D. J., Lindberg A. A. 1991. Molecular characterization of the O-acetyl transferase gene of converting bacteriophage SF6 that adds group antigen 6 to Shigella flexneri. Mol. Microbiol. 5:71–75 [DOI] [PubMed] [Google Scholar]
- 20. von Seidlein L., et al. 2006. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 3:e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye C., et al. 2010. Emergence of a new multidrug-resistant serotype X variant in an epidemic clone of Shigella flexneri. J. Clin. Microbiol. 48:419–426 [DOI] [PMC free article] [PubMed] [Google Scholar]