Abstract
Viruses are the main etiological cause of central nervous system (CNS) infections. A rapid molecular diagnosis is recommended to improve the therapeutic management of patients. The aim of this study was to evaluate the performances of a DNA microarray, the Clart Entherpex kit (Genomica, Coslada, Spain), allowing the rapid and simultaneous detection of 9 DNA and RNA neurotropic viruses: herpes simplex virus 1 (HSV-1), HSV-2, varicella-zoster virus (VZV), cytomegalovirus (CMV), Epstein-Barr virus (EBV), human herpesvirus 6 (HHV-6), HHV-7, HHV-8, and the human enteroviruses (HEVs). This evaluation was performed with 28 samples from the European proficiency panels (Quality Control for Molecular Diagnostics [QCMD]; Glasgow, Scotland) and then with 78 cerebrospinal fluid (CSF) specimens. The majority of the QCMD results obtained by the DNA microarray were similar to those recorded by the overall QCMD participants. The main discrepant results were observed for low concentrations of HSV-2 and HEVs. From the clinical samples, the kit detected 27 of the 28 herpesvirus CNS infections and all of the 30 HEV-positive CSF samples. No false-positive result was observed among the 20 virus-negative CSF samples. The clinical sensitivity, specificity, and negative and positive predictive values of the assay were 98.3, 100, 95.2, and 100%, respectively, when the results were compared to those of commercially available PCR assays. Interestingly, HHV-7 was detected in 11 (37%) of the 30 HEV-positive CSF samples from children suffering from aseptic meningitis causing significantly longer lengths of stay at the hospital than infection with HEVs alone (2.4 versus 1.4 days; P = 0.038). In conclusion, this preliminary study showed that this DNA microarray could be a valuable molecular diagnostic tool for single and mixed DNA and RNA virus infections of the CNS.
INTRODUCTION
Viruses are the main etiological cause of central nervous system (CNS) infections, ahead of bacterial and fungal causes (14, 34). They are responsible for encephalitis and aseptic meningitis (3, 5, 35). Encephalitis is a rare but one of the most devastating neurological disorders. Encephalitis in humans is due mainly to herpesviruses, in particular herpes simplex (HSV) and varicella-zoster (VZV) viruses, but also to cytomegalovirus (CMV), Epstein-Barr virus (EBV), or human herpesvirus 6 (HHV-6) in the immunocompromised host (21, 35, 40). Human enteroviruses (HEVs) are the most common cause of aseptic meningitis outbreaks in children during the summer season (1, 23). In both these types of CNS infections, a rapid virological diagnosis is required to improve the therapeutic management by acyclovir therapy in cases of herpes simplex encephalitis or to increase cost savings in hospitalized cases of HEV-related aseptic meningitis during the epidemic season (20, 26, 30, 36). PCR has been recognized as the reference method for the diagnosis of viral central nervous system infections in cerebrospinal fluid (CSF) specimens (10, 30, 36, 38). However, the wide range of viruses potentially responsible for CNS infections as well as their genetic characteristics, both DNA and RNA viruses, rendered rapid and large virological diagnosis difficult by using monoplex reverse transcription (RT)-PCR and PCR assays (4, 27).
The aim of this preliminary study was to evaluate the analytical and clinical performances of a commercially available multiplex RT-PCR DNA microarray, the Clart Entherpex kit (Genomica, Coslada, Spain), allowing a rapid and simultaneous detection of 9 DNA and RNA neurotropic viruses, HSV-1, HSV-2, VZV, CMV, EBV, HHV-6, HHV-7, HHV-8, and HEVs in a single CSF sample. In the first phase, this multiplex DNA microarray assay was evaluated by testing proficiency samples of the 2008 and 2009 European proficiency panels (Quality Control for Molecular Diagnostics [QCMD]; Glasgow, Scotland). In the second phase, 78 CSF specimens from patients hospitalized for CNS infections that had been previously tested by standardized commercially available PCR and RT-PCR assays for neurotropic virus detection were retrospectively analyzed, assessing the application of this commercially available multiplex RT-PCR DNA microarray on a routine basis.
MATERIALS AND METHODS
European proficiency panels and clinical specimens.
The evaluation of the kit was first carried out with samples of the 2008 and 2009 European proficiency panels (QCMD; Glasgow, Scotland) stored at −80°C until processing with the multiplex RT-PCR DNA microarray. Four proficiency samples for each of HSV-1, HSV-2, VZV, CMV, EBV, HHV-6, and HEVs containing concentrations ranging from 102 to 105 copies/ml were selected and independently tested 4 times each in order to assess the specificity, the sensitivity, and the reproducibility of the DNA microarray.
In a second phase, 78 CSF samples obtained from children or adults hospitalized at the Reims University Medical Centre (France) for suspected neurological virus infections from March 2002 to May 2009 were retrospectively analyzed. These samples had been routinely submitted to the virology laboratory for neurotropic virus detection and initially tested by both the multiplex herpes consensus generic endpoint PCR kit (Argène, Verniolle, France), screening for HSV-1, HSV-2, VZV, CMV, EBV, and HHV-6, and one of two HEV RT-PCR assays (either the enterovirus consensus endpoint PCR kit [Argène, Verniolle, France] or the enterovirus TaqMan one-step real-time RT-PCR kit [Andiatec, Kornwestheim, Germany]), depending on the year of CSF collection (6). They were then divided in aliquots and stored at −80°C until processing with the multiplex RT-PCR DNA microarray. Among the 78 clinical samples investigated, 28 had previously tested positive for herpesviruses (7 HSV-1, 2 HSV-2, 8 VZV, 1 CMV, 4 EBV, and 6 HHV-6) and negative by HEV RT-PCR assay; 30 samples obtained from children (sex ratio, 5 males to 1 female; mean age, 7 years) suffering from aseptic meningitis had previously tested positive for HEVs and negative by the herpes consensus generic kit; and 20 samples obtained from children (sex ratio, 1.5 males to 1 female; mean age, 6 years) suffering from aseptic meningitis had tested negative by both the herpes consensus generic kit and HEV RT-PCR assay. Since neither QCMD panels nor known clinical samples containing HHV-7 or HHV-8 were available, the performances of the DNA microarray regarding the detection of both these viruses have not been evaluated.
DNA/RNA extraction.
Total nucleic acid extraction was performed using a NucliSens easyMAG instrument (bioMérieux, Marcy l'Etoile, France) according to the manufacturer's instructions. Briefly, 200 μl of the QCMD specimen reconstituted according to the manufacturer's instructions or cerebrospinal fluid was added to 2 ml of lysis buffer in a plastic vessel and incubated for 10 min at room temperature. Fifty microliters of silica was then added to the mixture. This was followed by an automatic magnetic separation phase. Nucleic acids were recovered in 50 μl elution buffer and stored at −80°C until use.
Simultaneous detection of 9 viruses using a multiplex RT-PCR DNA microarray.
The Clart Entherpex kit (Genomica, Coslada, Spain) is based on viral genome-specific fragment amplification by multiplex PCR and its subsequent detection via hybridization with a microorganism-specific binding probe on low-density microarrays, allowing simultaneous detection and identification of the eight human herpesviruses (HSV-1, HSV-2, VZV, CMV, EBV, HHV-6, HHV-7, HHV-8) and HEVs (echovirus, poliovirus, and coxsackievirus) in clinical samples. For each sample, amplification reaction is performed in two tubes containing 5 μl of DNA/RNA extract and one of the two types of reaction mixtures, allowing the detection of either HSV-1, HSV-2, and VZV or CMV, EBV, HHV-6, HHV-7, HHV-8, and HEVs. The analyses were performed in 8-well strips according to the manufacturer's instructions. For each sample analyzed, the accuracy of the DNA microarray detection was controlled during extraction and amplification through an internal control and during hybridization, with at least 3 probes per amplified target detected. Following the phases of amplification, hybridization of the probes, and colorimetric detection of the hybridized probes, each microarray system was analyzed using a microarray reader piloted by specific software provided by the manufacturer, which allowed an automatic detection and interpretation of the results (33).
Viral load assessment in the CSF samples.
The viral load in the positive CSF samples selected was determined either by commercially available (EBV R-Gene, HSV1 HSV2 VZV R-Gene; Argene, Verniolle, France) or by previously described in-house real-time RT-PCR and PCR assays (12, 13, 17, 25). Viral load levels were then expressed as the number of genomic DNA or RNA copies per milliliter of CSF.
Statistical analyses.
Statistical comparison of demographic characteristics, laboratory findings, and clinical features between children demonstrating single infection by HEVs or mixed infection by HHV-7 and HEVs was conducted by utilizing the two-tailed Fisher exact test to compare qualitative variables and Student's t test for quantitative variables. These analyses were carried out with the SPSS 11.0 program (SPSS, Paris, France). A P value of <0.05 was considered statistically significant.
RESULTS
Analytical performances of the multiplex RT-PCR DNA microarray.
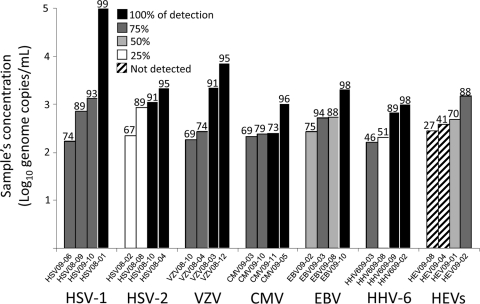
The European proficiency panels were processed as described in Materials and Methods, resulting in 4 replicates for each proficiency sample. Figure 1 depicted the results obtained from the analysis of the QCMD external quality assessment panels for HSV-1, HSV-2, VZV, CMV, EBV, HHV-6, and HEVs with the multiplex RT-PCR DNA microarray and comparison to the overall data sets submitted by the participants to the same QCMD programs. In summary, the lowest viral load detected at least once for the 6 herpesviruses tested ranged from 166 copies/ml for HHV-6 to 266 copies/ml for EBV. Moreover, the reproducibility tests indicated that viral loads of 166, 182, 211, 171, and 524 copies/ml were detected in 3 of the 4 analyses performed (75%) for HHV-6, VZV, CMV, HSV-1, and EBV, respectively. Concerning HSV-2, 861 copies were detected only once, whereas 1,099 copies were constantly detected during this evaluation. Overall, quality controls (QCs) of >250 copies/ml for the CMV, >500 copies/ml for HHV-6, and >2,000 copies/ml for HSV-1, HSV-2, VZV, and EBV were systematically detected, reflecting the minimal concentrations at which 4 of the 4 replicates performed were positive (Fig. 1). For HEVs, the kit failed to detect the QCs corresponding to 280 and 390 genome copies/ml. The viral loads at 480 and 1,480 copies/ml were detected in 50% and 75% of the analyses, respectively (Fig. 1). Neither cross-reactivity nor misidentification between herpesviruses was observed during the proficiency panel analyses with the multiplex RT-PCR DNA microarray.
Fig. 1.
Analysis of the QCMD external quality assessment panels with the multiplex RT-PCR DNA microarray and comparison to the overall data sets submitted by the participants to the QCMD. The quality controls were independently tested 4 times each. Rates of detection obtained with the microarray ranging from 0 to 100% are depicted with the shade of gray or the pattern used for the histograms, while the numbers at the top of the bars indicate the results obtained by the participants in the QCMD panel. The performances of the DNA microarray regarding the detection of HHV-7 and HHV-8 have not been evaluated, since no QCMD panels containing these viruses are available.
Finally, the rates of detection obtained for each virus with the DNA microarray were compared to the overall data sets for the corresponding external quality assessment (EQA) program submitted to QCMD by the participants, whatever the molecular technique used by the participants. Discrepant results were observed mainly for HSV-2 and HEVs and concerned samples containing low viral concentrations: 224 and 861 copies/ml of HSV-2 and 280 and 390 copies/ml of HEVs. These samples were detected by the microarray in 25% of the analyses for HSV-2 but never for HEVs, whereas the same specimens were, respectively, detected in 67 and 89% and in 27 and 41% of the overall participants in the QCMD panel. Besides, discrepancies were also observed for the lowest concentrations of the EBV (266 and 541 copies/ml) and HHV-6 (209 copies/ml) panel samples. These samples were detected by the DNA microarray both at 50% for EBV and at 25% for HHV-6, in comparison to the respective detection rates of 75, 88, and 51% obtained for the overall participants in the QCMD panel (Fig. 1).
Clinical evaluation from CSF samples.
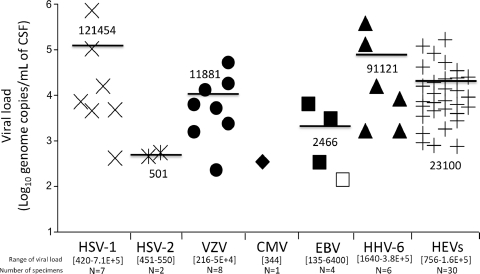
In order to perform a clinical evaluation of the multiplex RT-PCR DNA microarray, 78 CSF samples that had been previously analyzed by standardized commercially available RT-PCR and PCR assays were retrospectively tested. For each of the 58 CSF samples that had previously tested positive for herpesviruses or HEVs, the viral load was determined. Figure 2 shows for each virus the values of the viral load assessed per milliliter of CSF samples. Interestingly, HSV-1 and HHV-6 demonstrated the highest levels of viral loads in the CSF samples selected for this study, whereas the lowest viral concentrations were observed in CMV and HSV-2 infections (Fig. 2).
Fig. 2.
Viral load levels determined in CSF samples, initially positive for either herpesvirus (HSV-1 to HHV-6) or human enteroviruses, retrospectively tested with the DNA microarray. Copy numbers were plotted on a logarithmic scale. The mean value of the viral loads is indicated for each virus within the graph and depicted by a black line. The numbers indicated within the square brackets on the x axis correspond to the range of values of the viral loads determined for each virus in the CSF samples. The number of CSF specimens analyzed for each virus is indicated below. The sample that initially tested positive for EBV with EBV R-Gene and missed by Clart Entherpex is depicted by an empty square. No known CSF sample positive for HHV-7 or HHV-8 has been tested.
Of the 58 virus-positive clinical samples investigated, the Clart Entherpex kit detected 27 of the 28 (96%) CNS infections due to herpesviruses and all (100%) the 30 HEV-related aseptic meningitis infections. No misidentification among herpesviruses was observed. Only one EBV infection was missed, demonstrating a 98.3% agreement (57/58) with the results initially obtained with the Argène PCR and RT-PCR assays (Table 1). Concerning this specific case, the EBV load assessed by quantitative real-time PCR assay was 135 copies/ml, corresponding to the lowest viral load identified in CSF samples, regardless of the virus considered (Fig. 2). Of the 20 virus-negative clinical samples investigated, no discrepant result was observed between the DNA microarray and the Argène PCR and RT-PCR assays. In summary, the DNA microarray demonstrated, by comparison to the routinely used PCR and RT-PCR assays considered the gold standard, a sensitivity of 98.3%, a specificity of 100%, a positive predictive value of 100%, and a negative predictive value of 95.2%.
Table 1.
Summary of the CSF specimen testing results obtained with the multiplex RT-PCR DNA microarray by comparison to the standardized commercially available PCR and RT-PCR assays for neurotropic virus detection
| Multiplex RT-PCR DNA microarray result | No. of specimens |
|||
|---|---|---|---|---|
| Herpes consensus generic kit |
HEVs RT-PCR assays (enterovirus consensus or enterovirus TaqMan RT-PCR) |
|||
| Positive | Negative | Positive | Negative | |
| Positive | 27 | 0 | 30 | 0 |
| Negative | 1 | 50 | 0 | 48 |
Detection of HHV-7 and HEV mixed infections.
Among the 78 CSF samples tested, 12 (15%) HHV-7 infections were detected by the DNA microarray. HHV-7 was detected in 11 (37%) of the 30 HEV-infected CSF samples and as the sole virus in one of the 48 remaining CSF samples (2%), leading HHV-7 infection to be significantly associated with the HEV infection in children suffering from aseptic meningitis (37% versus 2%; P = 0.008). Detection of HHV-7 in the 12 positive CSF samples was confirmed by quantitative real-time PCR assay, demonstrating viral loads ranging from 60 to 300 genome copies per milliliter of CSF (mean value ± standard deviation, 163 ± 96 copies/ml) (12).
Table 2 summarizes the demographic, clinical, and therapeutic characteristics of the 11 patients coinfected by HHV-7 and HEVs by comparison to the 19 children presenting with single HEV infection. Univariate statistical analyses revealed that HHV-7 and HEV mixed infection caused significantly longer lengths of stay at the hospital than infection with HEVs alone (2.4 versus 1.4 days; P = 0.038) (Table 2). No other parameters, such as demographic characteristics, biological findings, clinical data, or treatments, appeared to be statistically associated with HHV-7 and HEV mixed infections (Table 2). Furthermore, no difference between the mean values of HEV load levels in CSF samples of children presenting with mixed HHV-7 and HEV (1.8 × 104 ± 2.0 × 104 copies/ml) or single HEV infection (2.0 × 104 ± 4.2 × 104 copies/ml) was observed (P = 0.212) (Table 2).
Table 2.
Comparison of demographic, clinical, and biological parameters and of the patient's management between children coinfected by HHV-7 and HEV and children presenting with single HEV infection
| Characteristic | Resulta |
P value | |
|---|---|---|---|
| HEV/HHV-1 mixed infection (n = 11 children) | HEV single infection (n = 19 children) | ||
| Median age (yr) | 5.73 ± 3.79 | 8.44 ± 6.66 | 0.19 |
| No. (%) of males | 9 (81.8) | 13 (81.3) | 0.97 |
| Fever (°C) | 38.3 ± 0.87 | 38.1 ± 0.39 | 0.64 |
| CSF analysis | |||
| Alpha interferon (IU/ml) | 7.60 ± 7.36 | 3.36 ± 3.32 | 0.12 |
| Protein (mg/dl) | 0.76 ± 0.88 | 0.39 ± 0.12 | 0.20 |
| No. of leukocytes/μl | 49 ± 34 | 91 ± 148 | 0.29 |
| Enterovirus load (copies/ml) | 1.8 × 104 ± 2.0 × 104 | 2.0 × 104 ± 4.2 × 104 | 0.21 |
| Blood analysis | |||
| Leukocytes (109 cells/liter) | 12.0 ± 2.6 | 12.5 ± 4.5 | 0.74 |
| Aspartate transaminase (IU/liter) | 37.0 ± 7.6 | 32.5 ± 17.2 | 0.56 |
| Alanine aminotransferase (IU/liter) | 42.0 ± 32.4 | 43.9 ± 50.4 | 0.94 |
| C-reactive protein (mg/liter) | 10.55 ± 14.6 | 25.1 ± 30.3 | 0.12 |
| Procalcitonin (ng/ml) | 0.82 ± 1.11 | 0.26 ± 0.18 | 0.28 |
| Patient's management | |||
| % Antibiotic use | 27.3 | 14.3 | 0.42 |
| % Antiviral use | 0 | 14.3 | 0.19 |
| No. of days of length of stay | 2.40 ± 1.26 | 1.38 ± 0.65 | 0.038 |
| % Deceased | 10 | 7.1 | 0.80 |
Values are presented as the means ± standard deviations.
DISCUSSION
Molecular techniques are now considered the gold standard for the detection in CSF samples of the viruses responsible for CNS infections (11, 19). The molecular tests used in routine diagnosis have to be specific and highly sensitive, allowing rapid and valuable detection of RNA and DNA viruses. At this time, the diagnosis of viral CNS infections is usually obtained through the combination of multiple PCR and RT-PCR assays, resulting in laboratory confirmation of approximately 45% of physician-diagnosed cases (4, 27). This failure can be explained by the inconsistency between the small volume of CSF available and the large number of viruses potentially responsible for meningitis or encephalitis (15). In the first step, the use of a multiplex PCR approach allowed us to broaden the detection of neurotropic viruses (2, 6, 7, 8, 22, 29, 31, 32, 41, 44). Nevertheless, some multiplex PCR requires confirmation by hybridization of the amplicons to specific probes fixed onto microwell plates, which have limited capacities of probe-binding sites (6). The hybridization of hundreds of probes on a DNA microarray has risen above this constraint, increasing the diagnostic output of the multiplex PCR assay. The previously published multiplex PCR followed by microarray-based detection showed concordant results, with single-endpoint PCR tests assessing the reliability of the method (4, 16). The present study aimed at the preliminary evaluation of the analytical and clinical performances of the first European Community (CE)- and in vitro diagnosis (IVD)-marked commercially available multiplex RT-PCR DNA microarray allowing the simultaneous detection of nine neurotropic DNA and RNA viruses in CSF samples.
The first phase of the evaluation was carried out using 28 samples of the 2008 and 2009 QCMD panels to assess the sensitivity and the reproducibility of the DNA microarray for herpesvirus and HEV detection. Through the 4 independent analyses performed, the kit demonstrated a limit of detection of less than 500 copies/ml for all of the 6 herpesviruses tested (Fig. 1). However, the analytical sensitivities of the test corresponding to the copy number of each one of the viruses that can be detected in 100% of the analyses performed were between 500 to 1,000 copies/ml for HHV-6 and greater than 2,000 copies/ml for HSV-1, HSV-2, VZV, and EBV. Regarding HEV detection, the low viral loads corresponding to 280 and 390 genome copies/ml were not detected, whereas a viral load of 1,480 copies/ml tested positive in 3 of the 4 analyses performed, therefore suggesting a lower sensitivity of the microarray regarding these viruses. By comparison, the manufacturer's claims regarding the analytical sensitivities of the assay are 10 copies for VZV, HHV-7, and HSV-1, 100 copies for HSV-2, CMV, EBV, HHV-6, HHV-8, and the coxsackieviruses, and 1,000 copies for the echoviruses and the polioviruses per PCR mixture containing 5 μl of DNA/RNA extract. Finally, the comparison of these data to those recorded by the overall QCMD participants, whatever the molecular technique used, showed discrepant results for QCMD samples containing low concentrations of HSV-2 and HEVs (Fig. 1).
The second phase of the evaluation consisted of the retrospective analysis of 78 CSF samples initially tested negative or positive for either herpesviruses or HEVs by standardized commercially available RT-PCR and PCR assays. This clinical evaluation confirmed the reliable diagnosis of the herpesvirus CNS infections by the DNA microarray since 27 of the 28 herpesvirus-positive CSF samples were detected, demonstrating high concordance with the results initially obtained using the herpes consensus generic multiplex endpoint PCR kit (Argène, Verniolle, France) (Table 1). Only one EBV infection characterized by a low viral load was not detected by the DNA microarray. Moreover, the Clart Entherpex kit did not reveal any sensitivity defect regarding HEV detection, as previously suggested by the QCMD sample analysis, since all (100%) the 30 HEV-positive CSF samples were detected. This could be explained by the viral load levels assessed in CSF samples, which ranged from 2.9 to 5.2 log10 copies/ml. The viral loads thus appeared to be high in the case of HEV-related meningitis and most often higher than the virus quantities contained in the lowest QCMD samples not detected by the microarray (Fig. 2). This finding was confirmed by previously published studies demonstrating that the viral loads ranged from 3.1 to 4.6 log10 copies/ml of CSF in HEV CNS infections (9, 18, 24).
Interestingly, the multiplex RT-PCR DNA microarray detected 11 (37%) HHV-7 and HEV mixed infections among the 30 pediatric aseptic meningitis cases initially related to HEVs. While HEVs are well-known neurotropic viruses, HHV-7 infection remains a neglected topic (42, 45). However, this CD4+ T-lymphotropic herpesvirus has been shown to contribute significantly to the burden of disease in young children with suspected encephalitis or severe convulsions with fever (37, 39, 46). In the present study, the presence of HHV-7 was confirmed by quantitative PCR (12). Moreover, the lack of correlation between HHV-7 detection and CSF leukocyte counts suggested that the HHV-7 DNA was from actively replicating virus and not just latent HHV-7 DNA carried in inflammatory cells (P = 0.29) (Table 2). Statistical analyses of the demographic, clinical, and therapeutic characteristics revealed that HHV-7 and HEV mixed infection resulted in significantly longer lengths of hospitalization for children suffering from aseptic meningitis than those with infection with HEVs alone (P = 0.038) (Table 2). Risk factors and consequences of this coinfection should be assessed in larger prospective studies. However, these preliminary data left open the question of the role of HHV-7 as the HEV meningitis cofactor associated with increased severity of the disease. Finally, this first description of combined HHV-7 and HEV infection highlighted the advantage of the DNA microarray technology for the detection of mixed viral CNS infections as well as for the investigation of the pathogenicity of neglected viruses (33).
Concerning the practical aspects of the kit, the CSF sample analysis with the DNA microarray was performed with 10 μl of total nucleic acid extract. The total time to complete the assay was approximately 8 h from specimen extraction to microarray detection, thereby allowing the laboratory to provide the answer to the clinician in a single work day. However, several limitations of the DNA microarray were observed during this study. Numerous handlings were required during the analysis, and opportunities of automation remained limited. Moreover, the kit cannot be used without a microarray reader piloted by specific software provided by the manufacturers, thus limiting the implementation of the kit in virology laboratories. By comparison to the one-step real-time PCR assays frequently used in current molecular virological diagnosis, the major drawback of the technique was the handling of amplicons. Although no contamination event was observed during this evaluation, a potential risk of contamination could not be ruled out. Finally, DNA microarray technology, which is a multiplex endpoint PCR system, did not allow the quantitation of the viral load in the clinical samples. However, it has been assumed that quantitation of viral nucleic acids may be useful in monitoring the effectiveness of antiviral therapy and for establishing the prognosis of the disease (9, 28, 43).
In conclusion, the evaluated multiplex RT-PCR DNA microarray appeared to be a sensitive and a specific test allowing rapid (in one work day) and simultaneous detection from a small volume of CSF sample of the main neurotropic DNA (herpesviruses) and RNA (enteroviruses) viruses. Moreover, our findings suggest that this molecular diagnostic tool could improve the routine virological diagnosis and the therapeutic management of CNS infections by detecting multiple viral infections potentially responsible for meningitis or encephalitis with increased severity. However, due to the small number of CSF specimens tested, further studies are needed to better characterize the performances of this test before its use in routine patient care.
ACKNOWLEDGMENTS
This study was supported by a grant for clinical and virological research (EA-4303/IFR53) from the Medical University and School of Medicine of Reims, France. Fanny Renois is supported by an official grant from the French Army Department (Bourse DGA: Délégation Générale de l'Armement, Ministère de la Défense; topic of microbiology and infectious diseases, 2009 to 2012).
None of us have a commercial or other association that might pose a conflict of interest (e.g., pharmaceutical stock ownership, consultancy).
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Antona D., et al. 2007. Surveillance of enteroviruses in France, 2000–2004. Eur. J. Clin. Microbiol. Infect. Dis. 26:403–412 [DOI] [PubMed] [Google Scholar]
- 2. Bergallo M., et al. 2007. Development of a multiplex polymerase chain reaction for detection and typing of major human herpesviruses in cerebrospinal fluid. Can. J. Microbiol. 53:1117–1122 [DOI] [PubMed] [Google Scholar]
- 3. Big C., Reineck L. A., Aronoff D. M. 2009. Viral infections of the central nervous system: a case-based review. Clin. Med. Res. 7:142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Boriskin Y. S., et al. 2004. DNA microarrays for virus detection in cases of central nervous system infection. J. Clin. Microbiol. 42:5811–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brunel D., et al. 2008. Clinical and virological features of an aseptic meningitis outbreak in North-Eastern France, 2005. J. Clin. Virol. 42:225–228 [DOI] [PubMed] [Google Scholar]
- 6. Calvario A., et al. 2002. Herpes consensus PCR test: a useful diagnostic approach to the screening of viral diseases of the central nervous system. J. Clin. Virol. 25:S71–S78 [DOI] [PubMed] [Google Scholar]
- 7. Casas I., Tenorio A., Echevarrıa J. M., Klapper P. E., Cleator J. M. 1997. Detection of enteroviral RNA and specific DNA of herpesviruses by multiplex genome amplification. J. Virol. Methods 66:39–50 [DOI] [PubMed] [Google Scholar]
- 8. Cassinotti P., Mietz H., Siegl G. 1996. Suitability and clinical application of a multiplex nested PCR assay for the diagnosis of herpes simplex virus infections. J. Med. Virol. 50:75–81 [DOI] [PubMed] [Google Scholar]
- 9. Dalwai A., Ahmad S., Pacsa A., Al-Nakib W. 2009. Echovirus type 9 is an important cause of viral encephalitis among infants and young children in Kuwait. J. Clin. Virol. 44:48–51 [DOI] [PubMed] [Google Scholar]
- 10. DeBiasi R. L., Kleinschmidt-DeMasters B. K., Weinberg A., Tyler K. L. 2002. Use of PCR for the diagnosis of herpesvirus infections of the central nervous system. J. Clin. Virol. 25:S5–S11 [DOI] [PubMed] [Google Scholar]
- 11. DeBiasi R. L., Tyler K. L. 2004. Molecular methods for diagnosis of viral encephalitis. Clin. Microbiol. Rev. 17:903–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fernandez C., et al. 2002. Quantitation of HHV-7 genome by real-time polymerase chain reaction assay using MGB probe technology. J. Virol. Methods 106:11–16 [DOI] [PubMed] [Google Scholar]
- 13. Gautheret-Dejean A., et al. 2002. Development of a real-time polymerase chain reaction assay for the diagnosis of human herpesvirus-6 infection and application to bone marrow transplant patients. J. Virol. Methods 100:27–35 [DOI] [PubMed] [Google Scholar]
- 14. Glaser C. A., et al. 2006. Beyond viruses: clinical profiles and etiologies associated with encephalitis. Clin. Infect. Dis. 43:1565–1577 [DOI] [PubMed] [Google Scholar]
- 15. Huang C., et al. 2004. Multiple-year experience in the diagnosis of viral central nervous system infections with a panel of polymerase chain reaction assays for detection of 11 viruses. Clin. Infect. Dis. 39:630–635 [DOI] [PubMed] [Google Scholar]
- 16. Jääskeläinen A. J., Piiparinen H., Lappalainen M., Vaheri A. 2008. Improved multiplex-PCR and microarray for herpesvirus detection from CSF. J. Clin. Virol. 42:172–175 [DOI] [PubMed] [Google Scholar]
- 17. Kares S., et al. 2004. Real-time PCR for rapid diagnosis of entero- and rhinovirus infections using LightCycler. J. Clin. Virol. 29:99–104 [DOI] [PubMed] [Google Scholar]
- 18. Kawashima H., et al. 2008. Viral loads of cerebrospinal fluid in infants with enterovirus meningitis. J. Clin. Lab. Anal. 22:216–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kimberlin D. W. 2005. Diagnosis of herpes simplex virus infections of the CNS. Expert Rev. Mol. Diagn. 5:537–547 [DOI] [PubMed] [Google Scholar]
- 20. King R. L., et al. 2007. Routine cerebrospinal fluid enterovirus polymerase chain reaction testing reduces hospitalization and antibiotic use for infants 90 days of age or younger. Pediatrics 120:489–496 [DOI] [PubMed] [Google Scholar]
- 21. Kleinschmidt-DeMasters B. K., Gilden D. H. 2001. The expanding spectrum of herpesvirus infections of the nervous system. Brain Pathol. 11:440–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Markoulatos P., et al. 2001. Laboratory diagnosis of common herpesvirus infections of the central nervous system by a multiplex PCR assay. J. Clin. Microbiol. 39:4426–4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Michos A. G., et al. 2007. Aseptic meningitis in children: analysis of 506 cases. PLoS One 2:e674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Monpoeho S., et al. 2002. Application of a real-time polymerase chain reaction with internal positive control for detection and quantification of enterovirus in cerebrospinal fluid. Eur. J. Clin. Microbiol. Infect. Dis. 21:532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Najioullah F., Thouvenot D., Lina B. 2001. Development of a real-time PCR procedure, including an internal control for the measurement of HCMV viral load. J. Virol. Methods 92:55–64 [DOI] [PubMed] [Google Scholar]
- 26. Nigrovic L. E., Chiang V. W. 2000. Cost analysis of enteroviral polymerase chain reaction in infants with fever and cerebrospinal fluid pleocytosis. Arch. Pediatr. Adolesc. Med. 154:817–821 [DOI] [PubMed] [Google Scholar]
- 27. Oostenbrink R., et al. 2001. Signs of meningeal irritation at the emergency department: how often bacterial meningitis? Pediatr. Emerg. Care 17:161–164 [DOI] [PubMed] [Google Scholar]
- 28. Persson A., Bergström T., Lindh M., Namvar L., Studahl M. 2009. Varicella-zoster virus CNS disease—viral load, clinical manifestations and sequels. J. Clin. Virol. 46:249–253 [DOI] [PubMed] [Google Scholar]
- 29. Quereda C., et al. 2000. Diagnostic utility of a multiplex herpesvirus PCR assay performed with cerebrospinal fluid from human immunodeficiency virus-infected patients with neurological disorders. J. Clin. Microbiol. 38:3061–3067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramers C., Billman G., Hartin M., Ho S., Sawyer M. H. 2000. Impact of a diagnostic cerebrospinal fluid enterovirus polymerase chain reaction test on patient management. JAMA 283:2680–2685 [DOI] [PubMed] [Google Scholar]
- 31. Read S. J., Mitchell J. L., Fink C. G. 2001. LightCycler multiplex PCR for the laboratory diagnosis of common viral infections of the central nervous system. J. Clin. Microbiol. 39:3056–3059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Read S. J., Kurtz J. B. 1999. Laboratory diagnosis of common viral infections of the central nervous system by using a single multiplex PCR screening assay. J. Clin. Microbiol. 37:1352–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Renois F., et al. 2010. Rapid detection of respiratory tract viral infections and coinfections in patients with influenza-like illnesses by use of RT-PCR DNA microarray systems. J. Clin. Microbiol. 48:3836–3842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Somand D., Meurer W. 2009. Central nervous system infections. Emerg. Med. Clin. North Am. 27:89–100 [DOI] [PubMed] [Google Scholar]
- 35. Steiner I. 2011. Herpes simplex virus encephalitis: new infection or reactivation? Curr. Opin. Neurol. 24:268–274 [DOI] [PubMed] [Google Scholar]
- 36. Stellrecht K. A., Harding I., Woron A. M., Lepow M. L., Venezia R. A. 2002. The impact of an enteroviral RT-PCR assay on the diagnosis of aseptic meningitis and patient management. J. Clin. Virol. 25:S19–S26 [DOI] [PubMed] [Google Scholar]
- 37. Tanaka K., et al. 1994. Human herpesvirus 7: another causal agent for roseola (exanthem subitum). J. Pediatr. 125:1. [DOI] [PubMed] [Google Scholar]
- 38. Tunkel A. R., et al. 2008. The management of encephalitis: clinical practice guidelines by the Infectious Diseases Society of America. Clin. Infect. Dis. 47:303–327 [DOI] [PubMed] [Google Scholar]
- 39. Van den Berg J. S., et al. 1999. Neuroinvasion by human herpesvirus type 7 in a case of exanthem subitum with severe neurologic manifestations. Neurology 52:1077. [DOI] [PubMed] [Google Scholar]
- 40. Volpi A. 2004. Epstein-Barr virus and human herpesvirus type 8 infections of the central nervous system. Herpes 11:120A–127A [PubMed] [Google Scholar]
- 41. Wada K., et al. 2009. Multiplex real-time PCR for the simultaneous detection of herpes simplex virus, human herpesvirus 6, and human herpesvirus 7. Microbiol. Immunol. 53:22–29 [DOI] [PubMed] [Google Scholar]
- 42. Ward K. N. 2005. Human herpesviruses-6 and -7 infections. Curr. Opin. Infect. Dis. 18:247–252 [DOI] [PubMed] [Google Scholar]
- 43. Weinberg A., Li S., Palmer M., Tyler K. L. 2002. Quantitative CSF PCR in Epstein-Barr virus infections of the central nervous system. Ann. Neurol. 52:543–548 [DOI] [PubMed] [Google Scholar]
- 44. Wolffs P. F., et al. 2009. Evaluation of MeningoFinder, a novel multiplex ligation-dependent probe amplification assay for simultaneous detection of six virus species causing central nervous system infections. J. Clin. Microbiol. 47:2620–2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wyatt L. S., Rodriguez W. J., Balachandran N., Frenkel N. 1991. Human herpesvirus 7: antigenic properties and prevalence in children and adults. J. Virol. 65:6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoshikawa T., et al. 2000. Invasion by human herpesvirus 6 and human herpesvirus 7 of the central nervous system in patients with neurological signs and symptoms. Arch. Dis. Child. 83:170–171 [DOI] [PMC free article] [PubMed] [Google Scholar]