Abstract
We evaluated the performances of 4 commercial real-time PCR kits for Bordetella pertussis IS481 sequence detection in nasopharyngeal aspirates by comparison with an in-house real-time PCR assay. Among them, the Simplexa Bordetella pertussis/parapertussis assay (Focus Diagnostics), the SmartCycler Bordetella pertussis/parapertussis assay (Cepheid), and Bordetella R-gene (Argene) present sensitivities over 90%. One kit proved unsuitable for routine clinical use.
TEXT
Nucleic acid amplification tests (NAAT), including PCR and, more recently, real-time PCR, overcome some of the limitations of culture and serological methods for the diagnosis of Bordetella infection (1, 6, 12). NAAT targets include IS481 for Bordetella pertussis (this gene is also present in B. holmesii and sometimes in B. bronchiseptica) (12, 13), the pertussis toxin promoter or porin gene BP3385 for B. pertussis (10), and IS1001 for B. parapertussis (5, 13). NAAT methods are highly sensitive and may be either genus or species specific, depending on the choice of primers and targets. Real-time PCR accelerates the diagnostic process by combining amplification and detection (12, 13). These methods have proved more sensitive than the equivalent gel-based system (9, 12), and many medical laboratories have developed in-house tests. A voluntary external molecular quality control procedure for these in-house methods was set up in France in 2009 by the National Reference Centre for Whooping Cough and Other Bordetelloses (Institut Pasteur, Paris) (3), following a study performed in eight hospital laboratories throughout France to assess the performance of in-house methods and adaptations of the techniques developed by Reischl et al. (11), Kösters et al. (8), Caro et al. (3), and Templeton et al. (13). Commercial molecular diagnostic kits are now available but have never been comparatively tested.
The aim of this study was to compare the performances of 4 commercial real-time PCR assays for the detection of B. pertussis, using as a reference an in-house method evaluated during the French external molecular quality control (3).
Eighty-one nonredundant nasopharyngeal aspirates (NPA) from patients with suspected pertussis were tested in the bacteriology laboratories of three French teaching hospitals (Tours, Poitiers, and Limoges). Sixty-six samples tested positive with our in-house PCR, while 15 had tested negative but were from patients with a suspicion of pertussis. The samples and DNA extracts were stored at −20°C until use. DNA was extracted with the Invisorb spin cell minikit (Invitek, Berlin, Germany) in Tours and with the QIAamp DNA minikit (Qiagen) in Poitiers and Limoges by following the manufacturers' instructions. The principles and procedures of the two extraction methods are very similar. Tenfold serial dilutions of DNA from B. pertussis strain Tohama, containing 238 copies of IS481, were used to determine the analytical sensitivity of each method. DNA extracted from each of the 81 samples was tested with the in-house real-time PCR assay using primers Bp481F (5′-CCGAACCGGATTTGAGAAAC-3′) and Bp481R (5′-TAGGAAGGTCAATCGGGCAT-3′), which target a 100-bp fragment of IS481. Detection was based on hybridization of the Bp481S probe (5′-6-carboxyfluorescein [FAM]-CCGGCCGGATGAACACCCATAA-6-carboxytetramethylrhodamine [TAMRA]-3′). Primers were purchased from Eurogentec (Seraing, Belgium) and were used at a concentration of 10 μmol/liter. Amplification was performed with a SmartCycler device (Cepheid, Maurens-Scopont, France), with 5 μl of DNA, 12.5 μl of premix Ex Taq (TaKaRa, Foster City, CA), 4.2 μl of water, 0.4 μl of each primer, and 2.5 μl of Bp481S probe (1 μM). The thermal cycling conditions were as follows: 1 cycle of 15 s at 95°C, followed by 45 cycles of 5 s at 95°C and 10 s at 60°C. Each laboratory tested the specimens collected in its host hospital and used the same protocols as the other laboratories. The SmartCycler II apparatus (Cepheid) was used for all assays.
The four commercial PCR kits, all based on TaqMan technology, were Bordetella R-gene (Argene, Verniolle, France), the Simplexa Bordetella pertussis/parapertussis assay (Focus Diagnostics; distributed in France by Eurobio, Courtabeuf, France), the Bordetella pertussis real-time PCR kit (catalog no. RD-0061-02 [Shanghai ZJ Bio-Tech; distributed in France by BioAdvance, Bussy-Saint-Martin, France]), and the SmartCycler Bordetella pertussis/parapertussis assay (Cepheid). The different kits were used according to the recommendations of the manufacturers, including cycle thresholds (CT) of 7 for the Shanghai ZJ Bio-Tech kit, 15 and 30 for the Focus Diagnostics kit, and 30 for the other two methods. Differences between the methods are shown in Table 1.
Table 1.
Comparison of main parameters of 4 real-time PCR kits tested
| Manufacturer or source | Name of kit | Target pathogen(s) | Target gene(s) | Kit includes: |
Reagent storage temp (°C) | Final reaction volb (μl) | Vol (μl) of DNA from sample added | No. of cycles used for comparison (no. of cycles recommended by manufacturer) | No. of tests with kit | Apparatus for which kit is available | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ready-to-use mix containing primers and probe | ICa detection | Extraction controls | Positive control | ||||||||||
| In-house | B. pertussis | IS481 | No | No | No | −20 | 20 | 5 | 45 | ||||
| Argene | Bordetella R-gene | B. pertussis | IS481 | Yes | Yes (10 μl IC to add in sample before extraction) | Yes | Yes | −20 | 25 | 10 | 45 (45) | 60 | LightCycler (LC480/LC2.0), SmartCycler, ABI Prism, Rotor-Gene |
| Shanghai ZJ Bio-Tech (distributed in France by BioAdvance) | Bordetella pertussis real-time PCR kit | B. pertussis | IS481 | No | Yes (1 μl IC to add in mix before amplification) | No | Yes (107 copies/ml) | −20 | 25 | 2.5 | 45 (40) | 25 | LightCycler (LC480), iCycler (iQ4/iQ5), SmartCycler II, ABI 7000/7300/ 7500/7900, Rotor-Gene 6000, Mx3000P/ 3005P, MJ Option2/ Chromo4 |
| Cepheid | SmartCycler Bordetella pertussis/parapertussis assay | B. pertussis and B. parapertussis | IS481 (B. pertussis), IS1001 (B. parapertussis) | Yes | Yes (contained in mix [20 copies/μl]) | No | No | +4 (beads) | 25 | 5 | 45 (40) | 40 | SmartCycler II |
| Focus Diagnostics (distributed in France by Eurobio) | Simplexa Bordetella pertussis/parapertussis assay | B. pertussis and B. parapertussis | IS481 (B. pertussis), IS1001 (B. parapertussis) | No | Yes (5 μl IC to add in sample before extraction) | Yes | Yes | −20 | 25 | 5 | 45 (45) | 48 | LightCycler LC480, SmartCycler II, ABI 7500 |
IC, internal control.
As recommended by the manufacturer of the SmartCycler apparatus.
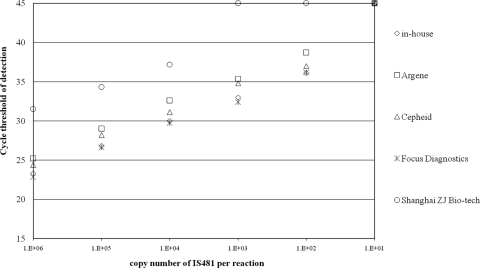
The analytical sensitivity of each method, tested in duplicate, is shown in Fig. 1. The CT values were similar for the different methods with up to 104 copies of IS481 per reaction, except that the CT of the Shanghai ZJ Bio-Tech test was about 7 cycles above that of the other methods whatever the number of copies. Only the in-house method and the Argene, Cepheid, and Focus Diagnostics kits were positive with the dilution containing 100 copies of IS481 per reaction.
Fig. 1.
Representation of the crossing threshold obtained for each commercial kit and the in-house real-time PCR for the detection of the Bordetella IS481 sequence.
Each clinical DNA specimen was tested with the in-house method and with the 4 commercial kits. All the DNA specimens were thawed and tested on the same day (no refreezing occurred between the different assays). The results are shown in Table 2. No false positives were noted for any assay. With the in-house method as the gold standard, clinical sensitivity was 97% with the Focus Diagnostics kit, 93.9% with the Cepheid kit, 90.9% with the Argene kit, and 51.5% with the Shanghai ZJ Bio-Tech kit. The Focus Diagnostics kit had the lowest mean CT (Table 2), which was not significantly different from that of the in-house assay. The Argene and Cepheid kits had slightly but not significantly higher mean CT values than the in-house assay. In contrast, the Shangai ZJ Bio-Tech kit had significantly higher mean CT values than the in-house assay. It is noteworthy that the manufacturer of the Shanghai ZJ Bio-Tech kit recommends using only half the amount of DNA used in the other kits. The performance of the Focus Diagnostics kit was unaffected when a CT of 15 rather than 30 was used (data not shown).
Table 2.
Clinical sensitivities and mean CT values for real-time PCR kitsa
| Kit | No. of positive specimens identified/total no. of positive specimens | Sensitivity (%) | Specificity (%) | Mean CT ± SD | P valued |
|---|---|---|---|---|---|
| In-house kit | 66/66 | 100 | 100 | 25.5 ± 8.6 | |
| Argene kit | 60/66 | 90.9 | 100 | 27.1 ± 9.5 | NS |
| Cepheid kit | 62/66 | 93.9 | 100 | 25.8 ± 9.2 | NS |
| Focus Diagnostics kit | 64/66 | 97 | 100 | 24.8 ± 8.8b | NS |
| Shanghai ZJ Bio-Tech kit | 34/66 | 51.5 | 100 | 37.3 ± 8c | <0.01 |
Data for the four real-time PCR kits tested are in comparison to results of the in-house real-time PCR assay for detection of the Bordetella IS481 sequence in 66 Bordetella-positive specimens.
As recommended by the manufacturer, the CT retained for the comparison was 15.
As recommended by the manufacturer, the CT retained for the comparison was 7.
Determined by paired t test (2). NS, not significant.
Internal controls are included to detect PCR inhibition. Beta-globin was used for this purpose in the in-house method in a separate vial. The Argene and Focus Diagnostics kits also include extraction controls (Table 1). The list of apparatus on which kits can be used is shown in Table 1. Two kits (the Cepheid and Focus Diagnostics assays) are claimed to detect B. pertussis and B. parapertussis in the same tube, while the other two kits and the in-house method target only IS481. B. holmesii, which also possesses IS481, cannot be distinguished from B. pertussis with these kits. This species rarely induces pertussis-like symptoms and can be identified by real-time PCR-based on the recA gene (7).
In conclusion, all four kits tested here were highly specific, whereas their sensitivities were highly variable. The Argene, Focus Diagnostics, and Cepheid kits performed as well as the in-house method in terms of both analytical and clinical sensitivities. These three kits include an internal control to detect inhibitors, which is important in routine practice (4), but only two of them, the Argene and Focus Diagnostics tests, validate the extraction step. The Shanghai ZJ Bio-Tech kit needs to be improved if it is to be used in routine clinical settings.
Acknowledgments
We thank Marie Odile Viaud, Lucie Yzon, and Catherine Louin for their technical assistance.
The different kits were supplied free of charge by the different corporations, which provided no other financial support for this study.
Footnotes
Published ahead of print on 14 September 2011.
REFERENCES
- 1. André P., et al. 2008. Comparison of serological and real-time PCR assays to diagnose Bordetella pertussis infection in 2007. J. Clin. Microbiol. 46:1672–1677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burucoa C., Garnier M., Silvain C., Fauchere J.-L. 2008. Quadruplex real-time PCR assay using allele-specific scorpion primers for detection of mutations conferring clarithromycin resistance to Helicobacter pylori. J. Clin. Microbiol. 46:2320–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caro V., et al. 2009. Proficiency program for real-time PCR diagnosis of Bordetella pertussis infections in French hospital laboratories and at the French national reference center for whooping cough and other bordetelloses. J. Clin. Microbiol. 47:3197–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cloud J. L., et al. 2003. Description of a multiplex Bordetella pertussis and Bordetella parapertussis LightCycler PCR assay with inhibition control. Diagn. Microbiol. Infect. Dis. 46:189–195 [DOI] [PubMed] [Google Scholar]
- 5. Farrell D. J., et al. 2000. Rapid-cycle PCR method to detect Bordetella pertussis that fulfills all consensus recommendations for use of PCR in diagnosis of pertussis. J. Clin. Microbiol. 38:4499–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guiso N., et al. 2011. What to do and what not to do in serological diagnosis of pertussis: recommendations from EU reference laboratories. Eur. J. Clin. Microbiol. Infect. Dis. 30:307–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guthrie J. L., Robertson A. V., Tang P., Jamieson F., Drews S. J. 2010. Novel duplex real-time PCR assay detects Bordetella holmesii in specimens from patients with pertussis-like symptoms in Ontario, Canada. J. Clin. Microbiol. 48:1435–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kösters K., Riffelmann M., Wirsing von König C. H. 2001. Evaluation of a real-time PCR assay for detection of Bordetella pertussis and B. parapertussis in clinical samples. J. Med. Microbiol. 50:436–440 [DOI] [PubMed] [Google Scholar]
- 9. Livak K. J., Flood S. J., Marmaro J., Giusti W., Deetz K. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4:357–362 [DOI] [PubMed] [Google Scholar]
- 10. Register K. B., Nicholson T. L., Guthrie J. L. 2010. Evaluation of specificity of BP3385 for Bordetella pertussis detection. J. Clin. Microbiol. 48:3334–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Reischl U., Lehn N., Sanden G. N., Loeffelholz M. J. 2001. Real-time PCR assay targeting IS481 of Bordetella pertussis and molecular basis for detecting Bordetella holmesii. J. Clin. Microbiol. 39:1963–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Riffelmann M., Wirsing von König C. H., Caro V., Guiso N. 2005. Nucleic acid amplification tests for diagnosis of Bordetella infections. J. Clin. Microbiol. 43:4925–4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Templeton K. E., et al. 2003. Evaluation of real-time PCR for detection of and discrimination between Bordetella pertussis, Bordetella parapertussis, and Bordetella holmesii for clinical diagnosis. J. Clin. Microbiol. 41:4121–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]