Abstract
The majority of Staphylococcus aureus infections from Isfahan, Iran, were caused by epidemic methicillin-susceptible S. aureus (MSSA) lineages, sequence type 8 (ST8), ST22, ST30, and ST6. The predominant methicillin-resistant S. aureus strain was ST239. We observed a high prevalence of Panton-Valentine leukocidin-positive MSSA strains (19.7%), which is a matter of considerable concern, since these strains have the ability to cause severe infections.
TEXT
Staphylococcus aureus is a human pathogen of increasing public health concern. The introduction of methicillin during the late 1950s to treat infections caused by penicillin-resistant S. aureus soon resulted in the emergence of methicillin-resistant S. aureus (MRSA) (6), a serious clinical problem worldwide (7, 11). Enright and colleagues, performing a population genetic study on an international collection of 912 MRSA and methicillin-susceptible S. aureus (MSSA) isolates, identified the major epidemic MRSA (EMRSA) lineages, including clonal complex 8 (CC8), CC22, CC30, CC45, and CC5 (2). Further, the same authors have shown that the major EMRSA lineages have arisen on multiple occasions from the same epidemic MSSA strains by acquisition of the staphylococcal cassette chromosome mec (SCCmec), which encodes methicillin resistance (5). Therefore, the genetic characteristics of S. aureus strains that are well adapted to the hospital or community environment, and which pose major public health concerns, should be investigated to further an appreciation of the global breadth of epidemic S. aureus.
In the present study, we characterized a population of 83 consecutive S. aureus isolates collected between January 2010 and May 2010 at the Alzahra Hospital in Isfahan, Iran. Our objectives were to determine the composition of clinical S. aureus genotypes from Isfahan, Iran, and to compare them to worldwide data. Each of these 83 S. aureus isolates represents a single initial isolate from different patients. The collection of S. aureus isolates comprised (i) 17 community-acquired S. aureus (CA-SA) isolates and (ii) 66 hospital-acquired S. aureus (HA-SA) isolates. Hospital-acquired infections were classified according to Garner et al. (4). The methicillin antimicrobial susceptibility of S. aureus isolates was assessed by the agar dilution method, while vancomycin susceptibility was performed using the disk dilution method; both procedures and interpretative criteria were those of the Clinical and Laboratory Standards Institute (13, 14). Genomic DNA was extracted using the Qiagen DNeasy tissue kit (Qiagen Inc., Valencia, CA) according to the manufacturer's instruction. DNA was shipped to the Vaccine Infectious Disease Organization, Saskatoon, Canada. The presence of two virulence genes encoding γ-hemolysin and Panton-Valentine leukocidin (PVL) synergohymenotropic toxin was determined by PCR assays as previously described (10). Multilocus sequence typing (MLST) was performed on the 83 isolates as described previously (1). Amplicons were subjected to DNA sequence analysis, and their nucleotide sequences on both strands were determined using an Applied Biosystems 3730xl DNA analyzer (Plant Biotechnology Institute, National Research Council of Canada, Saskatoon, Saskatchewan, Canada). Sequence editing and alignments were done using ClustalW (17). All sequences were compared, and every unique sequence was assigned as a distinct allele using the MLST Web site hosted at Imperial College London (www.mlst.net). MEGA4 (16) was used to construct phylogenetic trees from concatenated sequences of the housekeeping genes by a combination of several methods, including the unweighted-pair group method using arithmetic means (UPGMA), with p-distance and Kimura 2-parameter (8) substitution models. We used a comparative eBURST analysis (3) of already deposited worldwide data and our data in order to predict the ancestral profile and clonal complex of each genotype identified in this study. For identification of clonal complexes, we used the most stringent default group definition, where all sequence types (STs) have to be single-locus variants of at least one ST in the clonal complex.
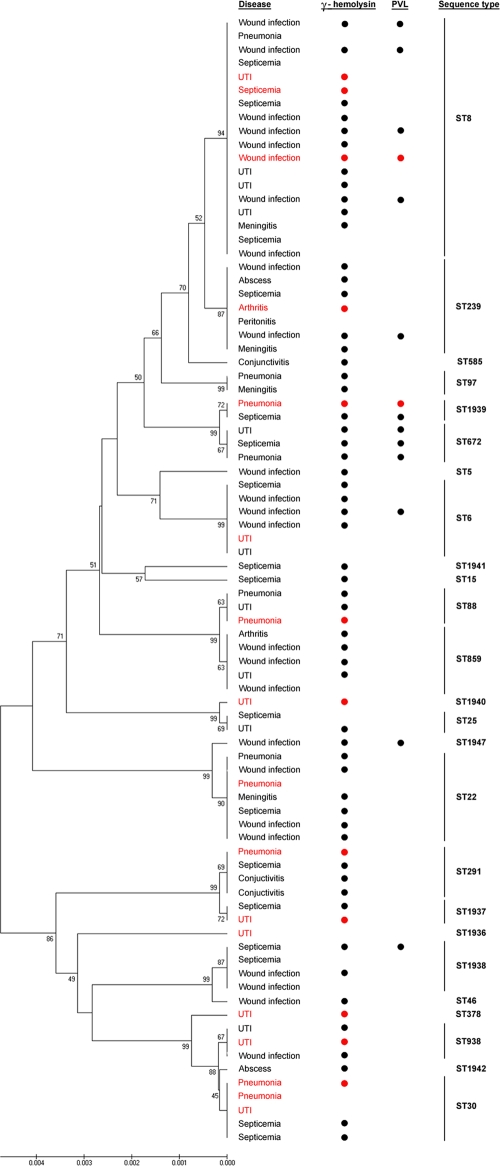
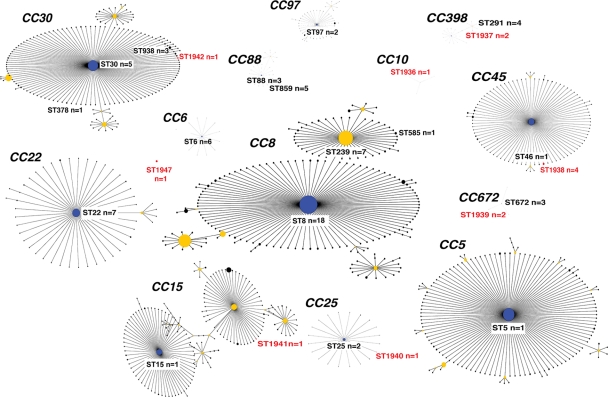
Susceptibility testing, together with PCR targeting the mec gene, revealed that 20% of the isolates were methicillin resistant. Resistance to methicillin was identified in 6 different STs (ST15, ST25, ST239, ST291, ST378, ST859) (Table 1), and three of these STs (ST15, ST239, ST378) comprised only MRSA isolates; the other three STs (ST25, ST291, ST859) had mixed populations of MRSA and MSSA isolates (Table 1). All isolates were susceptible to vancomycin. Sixty-eight (82%) isolates showed a 937-bp amplicon, indicating the presence of the hlg gene, which encodes γ-hemolysin. This gene was found in all STs except ST1936 (Fig. 1). Another 14 (17%) isolates were positive for PVL genes (Fig. 1). The majority of these isolates were found among HA-SA isolates, whereas only two PVL-positive isolates were detected among CA-SA isolates (Fig. 1). MLST resolved the 83 isolates into 25 STs (Fig. 1), including four major international epidemic S. aureus (E-SA) lineages, ST5, ST8, ST239, and ST22. Three E-SA lineages, ST8, ST239, and ST22, were the most commonly identified STs among the tested population, resulting in an overall prevalence of 22% (18 isolates), 8.4% (7 isolates), and 8.4% (7 isolates), respectively. After these three E-SA lineages, ST6, ST30, and ST859 were most commonly found; ST6 was identified in six different patients, while ST30 and ST859 were each detected in five patients. Eight STs were novel (ST1936, ST1937, ST1938, ST1939, ST1940, ST1941, ST1942, and ST1947) (Table 1). Figure 2 shows the ancestral source of the 83 S. aureus isolates from this study. The 82 isolates belonged to 13 different clonal complexes, while one isolate, represented by ST1947, was identified as a singleton (could not be assigned to any known CC). Most of the isolates belonged to CC8 (31%; n = 26), which included two E-SA lineages, ST8 and ST239. Ten isolates belonged to CC30 (12%), followed by CC88 (9.6%; n = 8), CC22 (8.4%; n = 7), CC6 (7.2%; n = 6), CC398 (7.2%; n = 6), CC672 (6%; n = 5), and CC45 (6%; n = 5). Another five minor CCs identified in this study were CC25 (3.6%; n = 3), CC15 (2.4%; n = 2), CC97 (2.4%; n = 2), CC5 (1.2%; n = 1), and CC10 (1.2%; n = 1).
Table 1.
Characteristics of 83 individual S. aureus isolates from Alzahra Hospital in Infahan, Iran
| ST | Allelic profile | Clonal complex | Antimicrobial susceptibility (no. of MRSA isolates/ no. of MSSA isolates) | Virulence profile |
Epidemiology (no. of HA isolates/no. of CA isolates) | |
|---|---|---|---|---|---|---|
| No. (%) of γ-hemolysin isolates | No. (%) of PVL isolates | |||||
| 5 | 1-4-1-4-12-1-10 | 5 | 0/1 | 1 (100) | 0 (0) | 1/0 |
| 6 | 12-4-1-4-12-1-3 | 6 | 0/6 | 4 (66) | 1 (17) | 5/1 |
| 8 | 3-3-1-1-4-4-3 | 8 | 0/18 | 14 (77) | 5 (28) | 15/3 |
| 15 | 13-13-1-1-12-11-13 | 15 | 1/0 | 1 (100) | 0 (0) | 1/0 |
| 22 | 7-6-1-5-8-8-6 | 22 | 0/7 | 6 (85) | 0 (0) | 6/1 |
| 25 | 4-1-4-1-5-5-4 | 25 | 1/1 | 1 (50) | 0 (0) | 2/0 |
| 30 | 2-2-2-2-6-3-2 | 30 | 0/5 | 3 (60) | 0 (0) | 2/3 |
| 46 | 10-14-8-6-14-3-2 | 45 | 0/1 | 1 (100) | 0 (0) | 1/0 |
| 88 | 22-1-14-23-12-4-31 | 88 | 0/3 | 3 (100) | 0 (0) | 2/1 |
| 97 | 3-1-1-1-1-5-3 | 97 | 0/2 | 2 (100) | 0 (0) | 2/0 |
| 239 | 2-3-1-1-4-4-3 | 8 | 7/0 | 6 (85) | 1 (14) | 6/1 |
| 291 | 3-37-19-2-20-26-32 | 398 | 3/1 | 4 (100) | 0 (0) | 3/1 |
| 378 | 2-2-2-1-6-3-2 | 30 | 1/0 | 1 (100) | 0 (0) | 0/1 |
| 585 | 2-1-1-1-4-4-3 | 8 | 0/1 | 1 (100) | 0 (0) | 1/0 |
| 672 | 4-3-1-1-11-72-11 | 672 | 0/3 | 3 (100) | 3 (100) | 3/0 |
| 859 | 79-1-14-23-12-4-31 | 88 | 4/1 | 4 (80) | 0 (0) | 5/0 |
| 938 | 2-2-2-6-6-3-2 | 30 | 0/3 | 2 (66) | 0 (0) | 2/1 |
| 1936a | 8-7-6-6-9-9-7 | 10 | 0/1 | 0 (0) | 0 (0) | 0/1 |
| 1937a | 3-37-19-6-20-26-32 | 398 | 0/2 | 2 (100) | 0 (0) | 1/1 |
| 1938a | 10-14-8-6-10-3-220 | 45 | 0/4 | 2 (50) | 1 (25) | 4/0 |
| 1939a | 4-3-1-1-11-4-11 | 672 | 0/2 | 2 (100) | 2 (100) | 1/1 |
| 1940a | 213-1-4-1-5-5-4 | 25 | 0/1 | 1 (100) | 0 (0) | 0/1 |
| 1941a | 7-1-1-1-1-1-1 | 15 | 0/1 | 1 (100) | 0 (0) | 1/0 |
| 1942a | 2-2-259-2-6-3-2 | 30 | 0/1 | 1 (100) | 0 (0) | 1/0 |
| 1947a | 1-6-1-5-8-8-64 | None | 0/1 | 1 (100) | 1 (100) | 1/0 |
Novel ST.
Fig. 1.
Phylogenies of concatenated sequences from the 83 isolates of S. aureus. Each isolate was presented by the type of infection that it caused. Bullets identify isolates that are γ-hemolysin and PVL positive. Vertical bars on the far right identify groups of isolates with the same sequence types. Community-acquired S. aureus isolates are indicated by a red color, while hospital-acquired S. aureus isolates are black. The phylogenetic tree was inferred according to the unweighted pair group method with arithmetic mean using the matrix of pairwise differences.
Fig. 2.
Population snapshot of clinical S. aureus lineages associated with their clonal complexes. All STs identified in this study are marked, together with their clonal complexes (CC). The number of isolates (n) belonging to each STs is included. Novel STs are indicated by a red color. Primary and subgroup ancestors are coded by blue and yellow, respectively. Each dot represents a single ST, and the size of each dot is proportional to the number of isolates globally deposited. Sequence types not identified in this study have been removed from their clonal complexes for clarity.
Our analysis of 83 clinical isolates revealed that an epidemic S. aureus strain, ST8-MSSA, was the most common cause of infection among the patients from the Alzahra Hospital. The ST8 strain was found in different hospital wards—pediatric, surgery, intensive care, and internal medicine—as well as in the community environment. Further, the ST8 strain was associated with a variety of infections, including pneumonia, septicemia, urinary tract infection (UTI), meningitis, and wound infections, indicating the presence of an unusually adaptable and virulent strain. After ST8-MSSA, two other major epidemic S. aureus lineages, ST239 and ST22, were most frequently identified as the cause of infections in the Alzahra Hospital. All ST239 isolates were MRSA. Ko et al. (9), studying the evolutionary pattern of MRSA clones in Asia, identified two major genotypes of MRSA isolates, CC5 and CC239. These two MRSA genotypes were clearly demarcated by geographic region. Most Korean and Japanese MRSA isolates were identified as ST5, while ST239 was the predominant MRSA strain in China, Indonesia, Singapore, Sri Lanka, and Vietnam. Our results show that ST239 is the major MRSA strain at the Alzahra Hospital in Isfahan, Iran, further indicating that this major international EMRSA clone has most likely disseminated throughout mainland Asia. In the present study, all isolates of another major epidemic S. aureus lineage, ST22, belonged to an MSSA clone. The majority of ST22 isolates were hospital acquired and were associated with wound infections, pneumonia, meningitis, and septicemia. Our findings also point to the importance of the epidemic MSSA clones from the clinical and epidemiological point of view, further suggesting that the overall genetic background of these S. aureus clones plays a crucial role in the dissemination of this human pathogen and its clinical severity, while the SCCmec has probably a more limited role in this. Surprisingly, the population of S. aureus from the Alzahra Hospital showed profound clonality where, out of 83 isolates, 82 were placed into 13 different clonal complexes. The majority of isolates (76%) belonged to six clonal complexes, CC8, CC22, CC30, CC88, CC6, and CC398, which have been identified worldwide, indicating that these international epidemic S. aureus lineages are a serious health threat to patients in Isfahan. We observed an unusually high prevalence (19. 7%) of pvl genes among the clinical MSSA isolates. Typically, Panton-Valentine leukocidin, synergohymenotropic toxin (15), is associated with CA-MRSA strains worldwide (19). In the present study, the PVL trait was found within seven different S. aureus genotypes, which correlate more to European isolates than to American isolates. The existence of distantly related S. aureus PVL-positive lineages from Europe has been previously reported (18), whereas in the United States a single major PVL clone is predominant (12).
In summary, we determined that the major international EMSSA clones, ST8, ST22, ST30, and ST6, were the causative agents of most of the infections which occurred in the Alzahra Hospital in Isfahan, Iran. In addition, we report a surprisingly high percentage of the PVL-positive MSSA strains that may pose a new threat in terms of the pathogenicity and epidemiology of S. aureus in the region.
Nucleotide sequence accession numbers.
Nucleotide sequences representing each allele were deposited in the GenBank database. The accession numbers of submitted sequences ranged from JF739183 to JF739250.
Acknowledgments
This work was supported by the University of Saskatchewan, Saskatoon, Canada (to J.-A.R.D.), and the Isfahan University of Medical Sciences, Isfahan, Iran (support for sabbatical leave to S.A.H.). S.V. and S.S. were supported by the Saskatchewan Health Research Foundation (grant number 9127, the 2007-2008 Health Research Team Grant Competition, Research Alliance for the Prevention of Infectious Disease [RAPID]). N.T., K.M., and K.M. were supported by the Isfahan University of Medical Sciences.
We thank Daniela Vidovic (University of Saskatchewan) for her excellent artwork.
Footnotes
Published ahead of print on 21 September 2011.
REFERENCES
- 1. Enright M. C., Day N. P. J., Davies C. E., Peacock S. J., Spratt B. G. 2000. Multilocus sequence typing for the characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Enright C. M., et al. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. U. S. A. 99:7687–7692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Feil E. J., Li B. C., Aanensen D. M., Hanage W. P., Spratt B. G. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garner J. S., Jarvis W. R., Emori T. G., Horan T. C., Hughes J. M. 1988. CDC definitions for nosocomial infections, 1988. Am. J. Infect. Control 16:128–140 [DOI] [PubMed] [Google Scholar]
- 5. Hiramatsu K., Cui L., Kuroda M., Ito T. 2001. The emergence and evolution of methicillin-resistant Staphylococcus aureus. Trends Microbiol. 9:486–493 [DOI] [PubMed] [Google Scholar]
- 6. Jevons M. P. 1961. “Celbenin”-resistant staphylococci. Brit. Med. J. 1:124–125 [Google Scholar]
- 7. Kennedy A. D., et al. 2008. Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc. Natl. Acad. Sci. U. S. A. 105:1327–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111–120 [DOI] [PubMed] [Google Scholar]
- 9. Ko S. K., et al. 2005. Distribution of major genotypes among methicillin-resistant Staphylococcus aureus clones in Asian countries. J. Clin. Microbiol. 43:421–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lina G., et al. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128–1132 [DOI] [PubMed] [Google Scholar]
- 11. Mongkolrattanothai K., Boyle S., Kahana M. D., Daum R. S. 2003. Severe Staphylococcus aureus infections caused by clonally related community-acquired methicillin-susceptible and methicillin-resistant isolates. Clin. Infect. Dis. 37:1050–1058 [DOI] [PubMed] [Google Scholar]
- 12. Moran G. J., et al. 2006. Methicillin-resistant Staphylococcus aureus infections among patients in the emergency department. N. Engl. J. Med. 355:666–674 [DOI] [PubMed] [Google Scholar]
- 13. National Committee for Clinical Laboratory Standards 2003. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A6, 6th ed. NCCLS, Wayne, PA [Google Scholar]
- 14. National Committee for Clinical Laboratory Standards 2003. Performance standards for antimicrobial susceptibility testing; 13th informational supplement, vol. 23, no. 1. M100-S13. NCCLS, Wayne, PA [Google Scholar]
- 15. Supersac G., Prevost G., Piemont Y. 1993. Sequencing of leucocidin R from Staphylococcus aureus P83 suggests that staphylococcal leucocidins and gamma-hemolysin are members of a single, two-component family of toxins. Infect. Immun. 61:580–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 17. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tristan A., et al. 2007. Global distribution of Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 13:594–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vandenesch F., et al. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Pantone-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]