Abstract
DNA-based human papillomavirus (HPV) assays show high sensitivity but poor specificity in detecting high-grade cervical lesions. Assays detecting mRNA of the oncoproteins E6 and E7 show higher specificity but lack either detection of all high-risk HPV genotypes or the capacity to specify the detected genotypes. Therefore, a real-time PCR assay detecting type-specific E6/E7 mRNA was developed and the clinical performance evaluated. A total of 210 cervical LBC (liquid-based cytology) samples from 204 women were analyzed for HPV DNA and mRNA with the in-house real-time PCR as well as PreTect HPV-Proofer. The sensitivity of real-time PCR mRNA detection to identify histologically confirmed CIN2+ (cervical intraepithelial neoplasia, grade 2 or higher) was 0.91, compared to 0.95 for DNA analysis. The specificity was 0.68 compared to 0.38, and the positive predictive value (PPV) was higher for mRNA (0.67 versus 0.52) without any loss in negative predictive value (NPV). The sensitivity of the real-time PCR mRNA test was somewhat higher than that for PreTect HPV-Proofer (0.83 versus 0.75) in analyses for the same genotypes. The specificities were similar (0.76 versus 0.77). In analyses for mRNA of the eight most common genotypes in cervical cancer (HPV16, -18, -31, -33, -35, -45, -52, and -58), the sensitivity of detection of CIN2+ lesions was 0.87 and the specificity 0.74, with a PPV of 0.70. In conclusion, real-time PCR for detection of HPV E6/E7 mRNA transcripts can be a sensitive and specific tool in screening and investigation of cervical neoplasia. The composition of HPV types in mRNA testing needs to be further investigated to optimize sensitivity and specificity.
INTRODUCTION
Cervical cancer is closely associated with infection of human papillomaviruses (HPV), but only a small proportion of these infections cause cancer. There are at least 12 oncogenic genotypes (HPV16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, and -59) associated with a high risk of cervical cancer (HR-HPV genotypes) and a number of genotypes that probably also have oncogenic properties (18). The viral proteins E6 and E7 are considered to be responsible for transformation of the infected epithelial cell, as well as the maintenance of the malignant phenotype. The proteins can affect many cellular proteins, such as the tumor suppressor proteins pRB and p53 (reviewed by Ghittoni et al. [11] and McLaughlin-Drubin and Munger [16]) in a manner that leads to extension of the cellular life span (including resistance to apoptosis), DNA synthesis, genomic instability, and interference with antiviral and antitumor immune responses. The mechanisms that determine whether an HPV infection will be cleared by the immune system or become persistent and cause transformation are not well understood. However, integration of the viral genome into the cellular genome seems to be an important event. Usually, the viral gene coding for E2, a regulator of E6/E7 transcription, is lost during integration. Thus, integration typically leads to overexpression of E6/E7, which may facilitate tumor progression (19). Moreover, common fragile sites are frequently targeted for viral integration, possibly causing genomic instability (25).
There are numerous commercial tests available for HR-HPV DNA detection but only a few based on detection of oncogenic mRNA. DNA detection tests are highly sensitive for detection of high-grade cervical intraepithelial neoplasia (CIN) and have been shown to be a valuable tool in triage of atypical squamous cells of uncertain significance (ASCUS) and follow-up after treatment (5). Furthermore, the use of HPV DNA tests in primary screening have been shown in several studies to be more sensitive than conventional cytology in detecting cervical cancer and severe precancerous lesions and may serve to prolong the screening interval (1, 7, 17). However, the specificities of HPV DNA tests for identification of cervical neoplasia are lower than those for cytology, especially among younger women (24). Therefore, HPV-positive women need to be triaged before referral for further investigations, such as colposcopy, but the preferable triage is yet to be established. Cytology could be an alternative, as well as detection of HPV E6/E7 mRNA or cellular tumor markers such as p16 (4). One commercially available mRNA test is the PreTect HPV-Proofer (NorChip, Klokkarstua, Norway), also called NucliSENS EasyQ (bioMérieux, Marcy l'Etoile, France), which detects mRNA of the five most common HPV types, HPV16, -18, -31, -33, and -45, based on the nucleic acid sequence-based amplification (NASBA) technique. The specificity of the test is higher than that of DNA tests (21, 24), but the sensitivity is lower, and mainly due to the fact that it does not detect all HR-HPV types, it can never be as sensitive as a DNA test. The other commercially available mRNA test for HPV is Aptima (Gen-Probe, San Diego, CA), which detects mRNA of 12 HR-HPV types as well as mRNA of HPV66 and HPV68, based on transcription-mediated amplification (TMA). However, Aptima does not specify the individual HPV types detected. The test has a sensitivity similar to that of a DNA test, but with higher specificity for detection of dysplasia (8, 20, 24). PreTect HPV-Proofer is more specific than Aptima. The reason for the higher sensitivity and lower specificity of Aptima than PreTect HPV-Proofer could be that Aptima detects mRNA of more genotypes, some more common in low-grade lesions. However, Aptima also detects HPV DNA, even though it is more sensitive for mRNA (10). Similarly, there have been reports that the NASBA technique can detect DNA (3, 22), causing false-positive results.
We previously developed a real-time PCR test based on amplification of E6/E7 DNA of 12 HR-HPV and 2 LR-HPV genotypes (15). The performance of the method has been validated by showing agreement with a linear array assay (Roche) (15) and by exhibiting 100% proficiency in the WHO LabNet proficiency panel study in 2009 (9). We have adapted this assay to detect only mRNA by adding a DNase-digesting step and a reverse transcription step. In this study, we evaluated the clinical performance of this type-specific HPV mRNA test and correlated the results with the HPV DNA analysis (using the same primers and probes) and the mRNA test PreTect HPV-Proofer.
MATERIALS AND METHODS
Samples.
Liquid-based cytology (LBC) samples were collected in PreservCyt medium (Cytyc, Marlborough, MA) from 204 women who were undergoing gynecological screening (n = 51, including 26 who were pregnant) or had been admitted to a referral center for investigation because of abnormalities in cervical cytology (n = 153, including 25 who were pregnant). The ages of the women ranged between 21 and 79 years, with median and mean ages of 32 and 34 years, respectively. Five of the women provided two or three samples, resulting in 210 samples. All women received information of the study design and provided written consent. Approval was obtained from the local ethics committee. Neoplasias were evaluated by colposcopy-directed biopsies and/or total excisional biopsies (conization) and subsequent histological examination. An expert pathologist re-evaluated all histological samples. If the second diagnosis differed from the original diagnosis by more than one level of severity, the pathologist confirmed the diagnosis with another pathologist.
DNA and RNA extraction.
DNA or total nucleic acid (NA) was extracted using a MagNA Pure LC instrument (Roche). For DNA analysis, 250 to 500 μl of the LBC sample was used for extraction with the DNA I protocol. For mRNA analysis, 3 to 5 ml of the sample was briefly centrifuged and pelleted cells were resuspended in 1 ml of RLT lysis buffer (Qiagen, Hilden, Germany) for extraction with the total-NA large-volume protocol. To ensure the quality of mRNA, the LBC samples were not allowed a storage period longer than 30 days before resuspension in lysis buffer and total-NA extraction. After lysis treatment, some samples were stored at −70°C before extraction. Prior to analysis, extracted NAs were stored at −70°C.
Real-time PCR.
The TaqMan real-time PCR assay targets 12 high-risk (HPV16, -18, -31, -33, -35, -39, -45, -51, -52, -56, -58, -59) and two low-risk (HPV6 and -11) types using E6/E7 region primers and probes in a duplex format (15). Detection of the human β-globin gene serves as a control for sample sufficiency. Briefly, 10 μl of extracted DNA was added to 25 μl universal PCR master mix (Roche Diagnostics, Branchburg, NJ) with 0.3 μM primers and 0.2 μM probes, supplemented with nuclease-free water to a final volume of 40 μl. After uracil DNA glycosylase activation at 50°C for 2 min and initial denaturation at 95°C for 10 min, the PCR for DNA detection was run for 45 cycles (15 s at 95°C, 60 s at 58°C) on an ABI 7300 instrument (Applied Biosystems, Carlsbad, CA). The threshold cycle (CT) value for each reaction was recorded (a low CT value indicates a large amount of target). Only samples yielding a CT value for β-globin below 36 were included in analysis. The modified method used for HPV mRNA detection included a DNase digestion step, using an Ambion Turbo DNA-free kit (Applied Biosystems). Ten microliters of the DNase-treated sample was added to a one-step RT-PCR master mix containing 1 μl ribonuclease inhibitor (RNase Out) and 1 μl SuperScript (all from Invitrogen, Carlsbad, CA), supplemented with nuclease-free water to a final reaction volume of 50 μl, including 0.3 μM primers and 0.2 μM probes. The PCR program was identical to that for DNA except for an initiating step of reverse transcription at 48°C for 30 min. To ascertain that no remaining HPV DNA was present, the DNase-treated samples were also run with the DNA detection protocol, i.e., without the RT step. mRNA detection was accepted only if the corresponding DNA was not detected or was detected with a CT value more than 10 cycles above the CT value for mRNA. The E6/E7 gene is transcribed into one full-length mRNA transcript coding for both proteins but is also spliced to an E7-encoding transcript (HPV16 is the only genotype transcribed into two spliced transcripts as well as a full-length one) (23). Our real-time PCR detects all transcripts, both full-length and spliced, except for genotypes 33, 52, 58, and 59, for which only the full-length transcripts are detected. For HPV16, the shorter of the two spliced transcripts is not detected.
PreTect HPV-Proofer.
Detection of E6/E7 mRNA of genotypes 16, 18, 31, 33, and 45 was performed using the PreTect HPV-Proofer kit according to the manufacturer's guidelines. Briefly, the analysis is based on the NASBA technique with isothermal amplification of mRNA in a duplex format, measured in real time. Five microliters of total-NA extract was added to 10 μl of a master mix with primers, molecular beacon probes, and KCl. After incubation for 2 min at 65°C and 2 min at 41°C, 5 μl of enzyme was added and spun down before amplification at 41°C. Analysis of the cellular U1A transcript was included in the test to determine the validity of the results.
Statistics.
The sensitivity, specificity, positive predictive value, and negative predictive value of each test algorithm were calculated with histologically confirmed CIN2+ as the gold standard, but calculations were also made for CIN3+. Calculations of 95% confidence intervals (95% CI) were based on the normal approximation to the binomial distribution as suggested by Harper and Reeves (12).
RESULTS
Cytological and histological diagnoses.
For 155 (74%) of the 210 LBC samples, histological evaluations from biopsies and/or total excised specimens taken at the same time were available (Table 1). For 53 samples with benign cytology (51 of them from women in screening) and two samples with ASCUS in cytology, no histological data were available. The histological diagnosis “benign” (n = 32) includes inflammation (n = 11), metaplasia (n = 2), ulcus (n = 1), and HPV infection without signs of CIN (n = 5). The diagnosis “CIN3” (n = 31) includes adenocarcinoma in situ (n = 3). Histological data were available for 50 cytologically benign samples, showing CIN1 or worse in 24 cases (48%), including six cases of CIN3 and four cases of cancer. These 24 represent 16% of the women in this study undergoing investigation for dysplasia, mostly due to earlier atypical cytology. Overall, histological examination tended to upgrade the cytological diagnoses to a more severe grade of dysplasia.
Table 1.
Cytological and histological diagnoses of 210 LBC samples
| Diagnosis | No. of samples (%) [median patient age, yrs] |
|
|---|---|---|
| Cytology (n = 210) | Histology (n = 155)a | |
| Benign | 106 (50) [31] | 34 (22) [32] |
| ASCUS | 37 (18) [30] | |
| ASC-H | 2 (1.0) [31] | |
| Glandular dysplasia | 3 (1.4) [30] | 1 (0.6) [46] |
| CIN1 | 15 (7.1) [31] | 33 (21) [31] |
| CIN2 | 11 (5.2) [29] | 28 (18) [29] |
| CIN3 | 19 (9.0) [35] | 31 (20) [31] |
| Adenocarcinoma | 3 (1.4) [47] | 7 (4.5) [39] |
| Squamous cell carcinoma | 14 (6.7) [46] | 21 (14) [47] |
Histology was not done on 55 samples from patients with a median age of 31 years.
HPV DNA and mRNA type distribution.
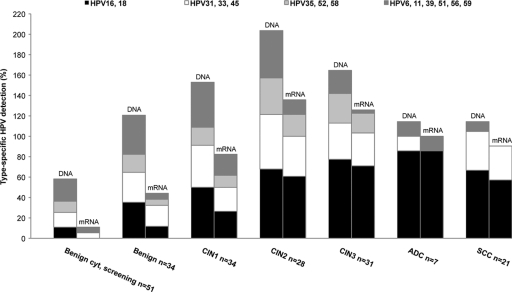
Type-specific rates of detection of HPV DNA and mRNA according to histology are shown in Fig. 1 (one sample with glandular dysplasia was included in the CIN1 group). HPV16 was the most prevalent type in the 87 samples histologically classified as CIN2+ (45 [52%]), followed by HPV18 (18 [21%]), HPV31 (17 [20%]), HPV33 (13 [15%]), HPV52 (12 [14%]), HPV39 (6 [7%]), and HPV45, HPV51, and HPV56 (5 [6%]). The five most common HPV types in the 28 samples with cancer were HPV16, HPV18, HPV33, HPV45, and HPV31, in that order.
Fig. 1.
Type-specific detection of HPV DNA and mRNA, distributed by histology. Each HPV type in a multiple infection is counted, which may result in a cumulative percentage of more than 100. ADC, adenocarcinoma; SCC, squamous cervical carcinoma.
For analysis of HPV mRNA, the picture was similar. The most common genotype in CIN2+ samples expressing E6/E7 mRNA was HPV16 (41 [47%]), followed by HPV18 (16 [18%]), HPV31 (15 [17%]), HPV33 (8 [9%]), HPV52 (7 [8%]) and HPV45 (5 [6%]). Consequently, 40% of HPV56 infections, 33% of HPV39 infections, and 0% of HPV59 infections in CIN2+ lesions showed expression of E6/E7 mRNA, in comparison to HPV45 (100%), HPV35 (100%), HPV16 (91%), HPV18 (89%), and HPV31 (88%) infections. In 24% (28/118) of mRNA-positive samples (68% of them CIN2+), mRNAs of multiple genotypes were present. The samples expressing mRNAs of two or more genotypes represent 22% of all CIN2+ samples.
In four CIN3+ samples (three cancers), no HR-HPV mRNA could be found. In two of these samples (both cancers), HR-HPV DNA was undetectable. (The samples tested positive for HPV68 or HPV70, respectively, with other methods). The two DNA-positive CIN3+ samples with undetectable mRNA were from two single infections with HPV16 or HPV33, respectively.
Of 34 samples with benign histology, 24 (71%) were HPV positive and 12 (35%) expressed E6/E7 mRNA. However, all these women had a history of dysplasia. In a screening cohort of 51 women (median age, 31) with benign cytology (no histology available), the prevalence of HPV infection was 43% (22 positive), and 9.8% (5) showed expression of E6/E7 mRNA.
Overall, there was good agreement between DNA and mRNA testing, and of all 258 HPV types detected, 118 were identified by both DNA and mRNA testing (46%). As expected, the DNA analysis had a higher detection rate and identified 140 HPV types that were not detected by mRNA testing. Conversely, mRNA was detected in 5 samples in which the same genotype was not detected by the DNA assay.
Sensitivity, specificity, PPV, and NPV.
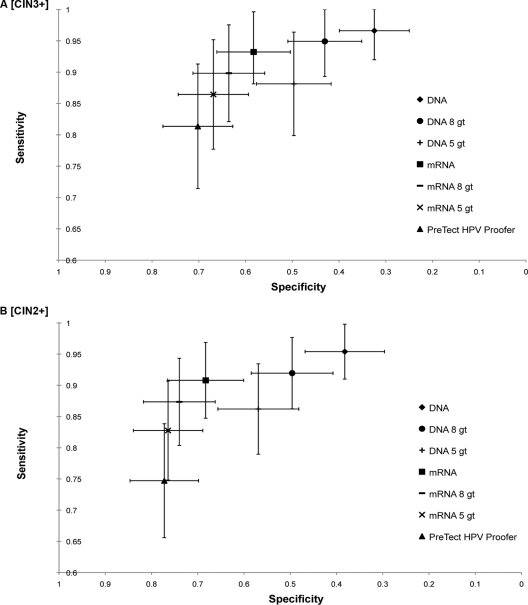
The sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of the HPV DNA assay, the HPV mRNA assay, and the PreTect HPV-Proofer assay for detecting CIN3+ and CIN2+ lesions were calculated. Furthermore, calculations were made for the two in-house tests (DNA and mRNA) when only the five HPV types most common in cervical cancer (HPV16, -18, -31, -33, and -45 [2]), which are the five genotypes in the PreTect HPV-Proofer assay, were included. However, recent data suggest that in different parts of the world, the most common HPV types in cervical cancer may vary (6, 14). HPV16 and -18 are the most common worldwide, and with a few exceptions, the most common genotypes after HPV16 and -18 are HPV31, -33, -35, -45, -52, and -58, in varying order. We therefore calculated sensitivity, specificity, PPV, and NPV for these eight genotypes as well. Since it was not possible to differentiate between low and moderate grades of neoplasia in glandular cells, one sample with glandular neoplasia that did not reach the level of adenocarcinoma in situ was included as a CIN1 sample in the calculations tabulated in Table 2. The sensitivity and specificity results are illustrated in Fig. 2.
Table 2.
Sensitivity, specificity, PPV, and NPV of different test algorithms for the detection of histologically confirmed CIN3+ or CIN2+ lesions
| Test and lesion typea | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) |
|---|---|---|---|---|
| HPV DNA | ||||
| CIN3+ | 0.97 (0.92–1.00) | 0.32 (0.25–0.40) | 0.36 (0.28–0.43) | 0.96 (0.91–1.00) |
| CIN2+ | 0.95 (0.91–1.00) | 0.38 (0.30–0.47) | 0.52 (0.44–0.60) | 0.92 (0.85–1.00) |
| HPV DNA 8 gt | ||||
| CIN3+ | 0.95 (0.89–1.00) | 0.43 (0.35–0.51) | 0.39 (0.31–0.47) | 0.96 (0.91–1.00) |
| CIN2+ | 0.92 (0.86–0.98) | 0.50 (0.41–0.58) | 0.56 (0.48–0.64) | 0.90 (0.82–0.97) |
| HPV DNA 5 gt | ||||
| CIN3+ | 0.88 (0.80–0.96) | 0.50 (0.42–0.58) | 0.41 (0.32–0.49) | 0.91 (0.85–0.98) |
| CIN2+ | 0.86 (0.79–0.93) | 0.57 (0.48–0.66) | 0.59 (0.50–0.67) | 0.85 (0.78–0.93) |
| HPV mRNA | ||||
| CIN3+ | 0.93 (0.87–1.00) | 0.58 (0.50–0.66) | 0.47 (0.38–0.56) | 0.96 (0.91–1.00) |
| CIN2+ | 0.91 (0.85–0.97) | 0.68 (0.60–0.77) | 0.67 (0.58–0.75) | 0.91 (0.86–0.97) |
| HPV mRNA 8 gt | ||||
| CIN3+ | 0.90 (0.82–0.98) | 0.64 (0.56–0.71) | 0.49 (0.40–0.59) | 0.94 (0.90–0.99) |
| CIN2+ | 0.87 (0.80–0.94) | 0.74 (0.66–0.82) | 0.70 (0.62–0.79) | 0.89 (0.83–0.95) |
| HPV mRNA 5 gt | ||||
| CIN3+ | 0.86 (0.78–0.95) | 0.67 (0.59–0.74) | 0.50 (0.41–0.60) | 0.93 (0.88–0.98) |
| CIN2+ | 0.83 (0.75–0.91) | 0.76 (0.69–0.84) | 0.71 (0.62–0.80) | 0.86 (0.80–0.93) |
| PreTect HPV Proofer | ||||
| CIN3+ | 0.81 (0.71–0.91) | 0.70 (0.63–0.77) | 0.51 (0.41–0.62) | 0.91 (0.85–0.96) |
| CIN2+ | 0.75 (0.66–0.84) | 0.77 (0.70–0.85) | 0.70 (0.61–0.79) | 0.81 (0.74–0.88) |
8 gt, eight HPV genotypes (HPV16, -18, -31, -33, -35, -45, -52, and -58); 5 gt, five HPV genotypes (HPV16, -18, -31, -33, and -45).
Fig. 2.
Clinical sensitivity and specificity (with 95% confidence intervals) of the different test algorithms for detection of CIN3+ lesions (A) and CIN2+ lesions (B).
DISCUSSION
This study aimed to evaluate the clinical performance of a real-time PCR assay that detects mRNA transcripts coding for the oncogenic proteins E6 and E7 of 12 high-risk and 2 low-risk HPV types, using the primers and probes described previously for HPV DNA. For 210 LBC samples with various grades of cervical neoplasia, there was good agreement between HPV mRNA and HPV DNA results, although the detection rate was higher with the DNA assay, as expected.
Our assay for mRNA detection, which includes a step that verifies that the mRNA signal is not due to detection of DNA, had a sensitivity of detection of CIN2+ and CIN3+ that was only slightly lower than for DNA detection (0.91 versus 0.95 and 0.93 versus 0.97, respectively), but the NPV for the mRNA was not lower than that of the DNA test. Importantly, the specificity was higher for mRNA than for DNA detection (0.68 versus 0.38 for CIN2+ lesions and 0.58 versus 0.32 for CIN3+ lesions). These results are in agreement with the study by Szarewski et al., in which several HPV DNA and mRNA tests were compared (24). The sensitivity and specificity of the PreTect HPV-Proofer assay (detecting five HR-HPV) for CIN2+ were 0.74 and 0.73 in their study, compared with 0.75 and 0.77 in our evaluation.
When our assay for mRNA typing was compared with the PreTect HPV-Proofer assay, the sensitivity of real-time PCR was somewhat higher (0.83 versus 0.75 for CIN2+ lesions and 0.86 versus 0.81 for CIN3+ lesions, when the same five genotypes were tested for). This may reflect a higher analytical sensitivity of real-time PCR than NASBA (because samples negative by PreTect HPV-Proofer but positive by real-time PCR in general contained small amounts of virus, as indicated by high CT values [data not shown]). The genotype most commonly detected by real-time PCR but not PreTect HPV-Proofer was HPV31, and this is in agreement with calculations of analytical sensitivity of the NucliSENS EasyQ assay (based on the same platform as PreTect HPV-Proofer) showing that the sensitivity of detection of HPV31 mRNA is 10 to 100 times lower than that of the other types (13). Moreover, our in-house real-time PCR detects not only full-length mRNA but also spliced mRNA transcripts of most genotypes, in contrast to PreTect HPV-Proofer (13), which may increase the sensitivity of the real-time PCR assay. A high analytical sensitivity might confer a risk of detecting small amounts of HPV mRNA not significant for disease, but the CIN2+ specificity of the real-time PCR was equal to that for PreTect HPV-Proofer in analyses for the same genotypes.
There have been suggestions that the high specificity of PreTect HPV-Proofer is mainly due to the fact that it analyzes the five most common genotypes and that a DNA test analyzing these five genotypes might be just as specific (3). However, this speculation is contradicted by our data, since specificity calculated for these five genotypes was higher for both CIN2+ and CIN3+ lesions in the mRNA than the DNA version of our real-time PCR (0.74 versus 0.57 and 0.64 versus 0.50, respectively), suggesting that presence of E6/E7 transcripts is important for disease. The high specificity by mRNA testing was illustrated by the finding that in a screening cohort of 51 women (median age, 31) with normal cytology (but with no histology available), HPV mRNA was detected in 9.8% and HPV DNA in 43%. The PPV of detection of both CIN2+ and CIN3+ was therefore higher for mRNA detection than for DNA detection (0.67 versus 0.52 and 0.47 versus 0.36, respectively), suggesting that mRNA testing may be a useful tool not only in triage but also in primary screening of cervical neoplasias. One should bear in mind that not all CIN2+ lesions will progress to cancer, and a hypothetical perfect test identifying only truly precancerous lesions would rate poorly with regard to sensitivity when CIN2+ in histology is used as the gold standard (as in this and most other studies).
The five most common genotypes present in CIN2+ and CIN3+ lesions were HPV16, -18, -31, -33, and -52 (in that order). In LBC samples from our patients with cancer, however, the five most common genotypes were HPV16, -18, -33, -45, and -31. This may reflect that the oncogenic properties of the genotypes vary. This idea was supported by the observation that some genotypes expressed E6/E7 mRNA more often than others. The eight genotypes most prone to express mRNA in CIN2+ lesions were, in descending order, HPV45, -35, -16, -18, -31, -33, -51, -52, and -58, the same genotypes (except for HPV51) most commonly found in cervical cancer worldwide (6, 14). The association of these eight genotypes with cancers may be a consequence of their potential to express oncogenic mRNA. Our finding encourages further and larger studies comparing mRNA and DNA detection rates for different HPV types.
We specifically evaluated the performance of the real-time PCR detection of mRNA for only the above-mentioned eight genotypes, which are most commonly observed in cancer. With this limitation, the sensitivity of the assay was somewhat higher than that of the assay for five genotypes, but the specificity did not substantially decrease and the PPV remained constant, suggesting that these eight types might constitute a good balance between sensitivity and specificity. This was also relevant for HPV DNA testing, since analyzing eight rather than all genotypes resulted in a significant increase in specificity at the expense of only a small amount of sensitivity, but without decreasing the high NPV.
Our data suggest that mRNA testing with real-time PCR may be a useful tool in investigation of as well as in primary screening for cervical neoplasias, and it might be worthwhile to consider which genotypes to include in further investigations to optimize sensitivity and specificity, especially in a postvaccine era, when it may be necessary to reconsider HPV testing strategies.
ACKNOWLEDGMENTS
We thank Monika Dohsé for technical assistance.
This study was supported by grants from the Western region R&D Fund, ALF-funds, the Capio Research Foundation, and the Assar Gabrielsson Foundation.
Footnotes
Published ahead of print on 21 September 2011.
REFERENCES
- 1. Anttila A., et al. 2010. Rate of cervical cancer, severe intraepithelial neoplasia, and adenocarcinoma in situ in primary HPV DNA screening with cytology triage: randomised study within organised screening programme. BMJ 340:c1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bosch F. X., et al. 2008. Epidemiology and natural history of human papillomavirus infections and type-specific implications in cervical neoplasia. Vaccine 26 (Suppl. 10):K1–K16 [DOI] [PubMed] [Google Scholar]
- 3. Boulet G. A., et al. Nucleic acid-sequence based amplification assay for human papillomavirus mRNA detection and typing: evidence for DNA amplification. J. Clin. Microbiol. 48:2524–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cuschieri K., Wentzensen N. 2008. Human papillomavirus mRNA and p16 detection as biomarkers for the improved diagnosis of cervical neoplasia. Cancer Epidemiol. Biomarkers Prev. 17:2536–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cuzick J., et al. 2008. Overview of human papillomavirus-based and other novel options for cervical cancer screening in developed and developing countries. Vaccine 26 (Suppl. 10):K29–K41 [DOI] [PubMed] [Google Scholar]
- 6. de Sanjose S., et al. 2010. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 11:1048–1056 [DOI] [PubMed] [Google Scholar]
- 7. Dillner J., et al. 2008. Long term predictive values of cytology and human papillomavirus testing in cervical cancer screening: joint European cohort study. BMJ 337:a1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dockter J., et al. 2009. Clinical performance of the APTIMA HPV assay for the detection of high-risk HPV and high-grade cervical lesions. J. Clin. Virol. 45 (Suppl. 1):S55–S61 [DOI] [PubMed] [Google Scholar]
- 9. Eklund C., Zhou T., Dillner J. 2010. Global proficiency study of human papillomavirus genotyping. J. Clin. Microbiol. 48:4147–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Getman D., et al. 2009. Efficiency of the APTIMA HPV assay for detection of HPV RNA and DNA targets. J. Clin. Virol. 45 (Suppl. 1):S49–S54 [DOI] [PubMed] [Google Scholar]
- 11. Ghittoni R., et al. 2010. The biological properties of E6 and E7 oncoproteins from human papillomaviruses. Virus Genes 40:1–13 [DOI] [PubMed] [Google Scholar]
- 12. Harper R., Reeves B. 1999. Reporting of precision of estimates for diagnostic accuracy: a review. BMJ 318:1322–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jeantet D., et al. 2009. NucliSENS EasyQ HPV v1 test—testing for oncogenic activity of human papillomaviruses. J. Clin. Virol. 45 (Suppl. 1):S29–S37 [DOI] [PubMed] [Google Scholar]
- 14. Li N., Franceschi S., Howell-Jones R., Snijders P. J., Clifford G. M. 2011. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide: variation by geographical region, histological type and year of publication. Int. J. Cancer 128:927–935 [DOI] [PubMed] [Google Scholar]
- 15. Lindh M., et al. 2007. Real-time Taqman PCR targeting 14 human papilloma virus types. J. Clin. Virol. 40:321–324 [DOI] [PubMed] [Google Scholar]
- 16. McLaughlin-Drubin M. E., Munger K. 2009. Oncogenic activities of human papillomaviruses. Virus Res. 143:195–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mesher D., et al. 2010. Long-term follow-up of cervical disease in women screened by cytology and HPV testing: results from the HART study. Br. J. Cancer 102:1405–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Munoz N., Castellsague X., de Gonzalez A. B., Gissmann L. 2006. Chapter 1: HPV in the etiology of human cancer. Vaccine 24 (Suppl. 3):S1–S10 [DOI] [PubMed] [Google Scholar]
- 19. Pett M., Coleman N. 2007. Integration of high-risk human papillomavirus: a key event in cervical carcinogenesis? J. Pathol. 212:356–367 [DOI] [PubMed] [Google Scholar]
- 20. Ratnam S., et al. 2011. Aptima HPV E6/E7 mRNA test is as sensitive as Hybrid Capture 2 assay but more specific at detecting cervical precancer and cancer. J. Clin. Microbiol. 49:557–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ratnam S., et al. 2010. Clinical performance of the PreTect HPV-Proofer E6/E7 mRNA assay in comparison with that of the Hybrid Capture 2 test for identification of women at risk of cervical cancer. J. Clin. Microbiol. 48:2779–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rodriguez-Lazaro D., Lloyd J., Ikonomopoulos J., Pla M., Cook N. 2004. Unexpected detection of DNA by nucleic acid sequence-based amplification technique. Mol. Cell. Probes 18:251–253 [DOI] [PubMed] [Google Scholar]
- 23. Sotlar K., et al. 2004. Detection of high-risk human papillomavirus E6 and E7 oncogene transcripts in cervical scrapes by nested RT-polymerase chain reaction. J. Med. Virol. 74:107–116 [DOI] [PubMed] [Google Scholar]
- 24. Szarewski A., et al. 2008. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol. Biomarkers Prev. 17:3033–3042 [DOI] [PubMed] [Google Scholar]
- 25. Thorland E. C., Myers S. L., Gostout B. S., Smith D. I. 2003. Common fragile sites are preferential targets for HPV16 integrations in cervical tumors. Oncogene 22:1225–1237 [DOI] [PubMed] [Google Scholar]