Abstract
We present here the first report of disseminated skin Mycobacterium infections in two liver transplant recipients, in which hsp65 gene sequencing was used for rapid species identification. Both patients had hepatitis B virus-related cirrhosis and diabetes mellitus and presented with progressive generalized, nodular skin lesions. In one patient, a 50-year-old woman who had frequent contact with marine fish, an acid-fast bacillus was isolated from skin biopsy tissue after 2 months of culture. While awaiting phenotypic identification results, hsp65 gene sequencing showed that it was most closely related to that of Mycobacterium marinum with 100% nucleotide identity. The patient was treated with oral rifampin, ethambutol, and moxifloxacin. In the other patient, a 59-year-old woman, direct PCR for Mycobacterium using hsp65 gene from skin biopsy tissue was positive, with the sequence most closely related to that of M. haemophilum with 100% nucleotide identity. Based on PCR results, the patient was treated with clarithromycin, ethambutol, moxifloxacin, and amikacin. A strain of M. haemophilum was only isolated after 3 months. Skin lesions of both patients resolved after 1 year of antimycobacterial therapy. Nontuberculous Mycobacterium infections should be considered in liver transplant recipients presenting with chronic, nodular skin lesions. This report highlights the crucial role of hsp65 gene PCR and sequencing on both cultured isolates and direct clinical specimens for rapid diagnosis of slow-growing Mycobacterium infection.
INTRODUCTION
Organ and tissue transplant recipients are predisposed to infections due to opportunistic pathogens, including various Mycobacterium species. While Mycobacterium tuberculosis is the most common Mycobacterium species associated with infections, nontuberculous Mycobacterium infections have been reported at an estimated incidence of 0.16 to 2.8% of transplant recipients in the United States (8). Nontuberculous Mycobacterium infections in these patients can be difficult to diagnose and often difficult to treat and are thus associated with significant morbidity and mortality.
Identification of nontuberculous mycobacteria traditionally relies on isolation of the Mycobacterium and subsequent identification by phenotypic conventional biochemical tests or whole-cell fatty acid analysis. However, these methods are associated with a number of drawbacks. First, these bacteria are often slow growing, so the turnaround time for identification is long when conventional biochemical tests are used. Second, noncultivable species or isolates with ambiguous biochemical profiles are sometimes encountered, making them unidentifiable. As for whole-cell fatty acid analysis using gas chromatography, the special equipment and expertise required are not available in most routine clinical microbiology laboratories. Newer technologies, such as liquid chromatography-nuclear magnetic resonance/mass spectrometry, are being developed, which may help diagnosis of mycobacterial infections in the future (2).
Molecular methods, especially PCR and sequencing, have revolutionized the rapid identification and classification of bacteria, including Mycobacterium species (20, 39). For example, 16S rRNA gene analysis has been used for this purpose, providing rapid diagnosis and guiding prompt antibiotic treatment (37, 38). However, some Mycobacterium species cannot be differentiated from one another by 16S rRNA gene sequencing, e.g., between M. avium-intracellulare and M. paratuberculosis, between M. chelonae and M. abscessus, between M. kansasii and M. gastri, between M. malmoense and M. szulgai, between M. marinum and M. ulcerans, between M. mucogenicum and M. phocaicum, and among M. tuberculosis complex isolates (39). As a result, other gene targets have been explored for differentiation of some Mycobacterium species. We present here the first report of disseminated skin Mycobacterium infections in two liver transplant recipients, in which rapid species identification was enabled by hsp65 gene sequencing. The crucial role of hsp65 gene sequencing in the rapid diagnosis of nontuberculous Mycobacterium infections and the choice of an appropriate antibiotic therapy is also discussed.
MATERIALS AND METHODS
Patients and microbiological methods.
Clinical specimens were collected and handled according to standard protocols (34). Bacterial strains were identified by standard conventional methods (34). Antibiotic susceptibility tests were performed by proportion method according to Clinical and Laboratory Standards Institute (5, 34). For M. haemophilum, a test inoculum supplemented with hemin was used.
DNA extraction, PCR, and sequencing of hsp65 genes.
Bacterial DNA from tissues was extracted by using a DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Bacterial DNA extraction from bacterial colonies was performed as described previously with modification (15, 36). Briefly, 200 μl of bacterial colonies suspended in sterile water was incubated at 95°C for 30 min, followed by the addition of 200 μl of 24:1 (vol/vol) chloroform-isoamyl alcohol. The mixture was centrifuged at 16,100 × g for 5 min, and the supernatant was collected. Genomic DNA was extracted with the addition of a 1/10 volume of 3 M ammonium acetate and a 2.5 volume of 95% ethanol. After centrifugation for 5 min, the supernatant was removed, and the pellet was air dried. The DNA pellet was resuspended in 100 μl of sterile MilliQ water. PCR amplification and DNA sequencing of the hsp65 genes were performed according to previously published protocols (15, 28, 36). LPW13636 (5′-ACCAACGATGGTGTGTCCAT-3′) and LPW13637 (5′-CTTGTCGAACCGCATACCCT-3′) (Sigma-Aldrich, Steinheim, Germany) were used as the PCR primers. Both strands of the PCR products were sequenced using the PCR primers.
Phylogenetic characterization.
The sequences of the PCR products were compared to known hsp65 gene sequences in the GenBank by multiple sequence alignment using CLUSTAL X 1.83 (29). The phylogenetic relationships to other closely related Mycobacterium species were determined by using the neighbor-joining method. A total of 401 nucleotide positions were included in the analysis.
Nucleotide sequence accession numbers.
The hsp65 gene sequences of patient isolates 1 and 2 have been registered within the GenBank sequence database under accession numbers JF775378 and JF775377, respectively.
RESULTS
Patients.
Case 1 was a 50-year-old woman was admitted to hospital in April 2010 because of progressive multiple painless, erythematous, ulcerated, or crusted, nodular skin lesions for 3 months (Fig. 1a). The skin lesions first appeared on the tip of nose and then spread to the face, upper limbs, and right leg after receiving pulsed intravenous methylprednisolone for acute rejection. She had history of hepatitis B virus (HBV)-related cirrhosis with cadaveric liver transplant done in October 2007. She also had diabetes mellitus on insulin therapy and was put on triple immunosuppressants, including mycophenolate mofetil (MMF), tacrolimus, and prednisolone for graft rejection. She was also receiving adefovir and lamivudine for HBV reactivation. She was afebrile all along. Pancytopenia was noted on admission, with a total white cell count of 1.22 × 109/liter, a neutrophil count of 0.93 × 109/liter, a lymphocyte count of 0.19 × 109/liter, a monocyte count of 0.08 × 109/liter, a hemoglobin level of 7.4 g/dl, and a platelet count of 55 × 109/liter. Her renal function was impaired with serum urea at 13.8 mmol/liter and creatinine at 125 μmol/liter. Liver enzymes were within normal ranges. Bone marrow biopsy specimens showed hypoplastic marrow with reduced granulopoiesis. Skin biopsy specimens of the face and left arm lesions for Gram smear, bacterial, and fungal cultures were negative. Histology showed florid histiocytic infiltrates throughout the dermis associated with scattered multinucleated giant cells and a rim of lymphocytes suggestive of granulomatous dermatitis. However, special histochemical stains, including periodic acid-Schiff, Gram, Giemsa, Ziehl-Neelsen, Wade-Fite, Grocott, mucicarmine, and Wathin-Starry stains, were negative. Since she had been a fisherman many years ago and had often handled and consumed seafood and marine fish, treatment with empirical antimycobacterial agents, including oral rifampin at 300 mg daily, ethambutol at 800 mg every 36 h, and moxifloxacin at 400 mg daily, was commenced 2 days after admission, along with titration of tacrolimus dosage. A dose of granulocyte colony-stimulating factor was also given for neutropenia. Blood and bone marrow cultures for mycobacteria were negative.
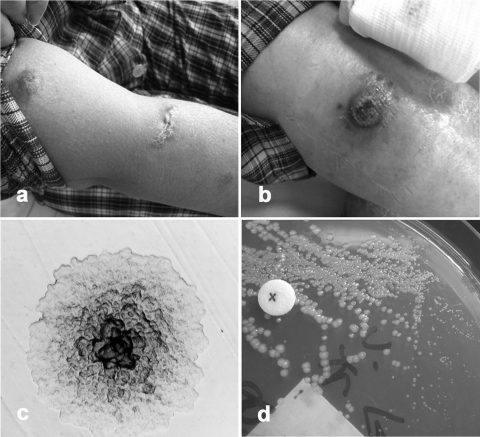
Fig. 1.
(a and b) Clinical photos showing multiple, erythematous, nodulo-ulcerative skin lesions on limbs from patients 1 (a) and 2 (b). (c) A distinct microcolony of M. haemophilum (isolate 2) subcultured on Middlebrook 7H10 agar observed under a low-power light microscope after 2 weeks of incubation at 31°C. (d) Growth enhancement of M. haemophilum (isolate 2) around factor X disk on Middlebrook 7H10 agar after 3 weeks of incubation at 31°C, indicating hemin dependence.
Mycobacterial culture of the skin biopsy tissue was positive for an acid-fast bacillus (isolate 1) on Lowenstein-Jensen solid medium 4 weeks later. PCR of the hsp65 gene was performed for rapid identification to the species level. The isolate grew better at 31°C than at 37°C, appearing as yellow-pigmented colonies following exposure to light, suggesting that it was a photochromogen. It was positive for Tween hydrolysis, urease, and pyrazinamidase but was negative for nitrate reduction. The isolate was phenotypically identified as M. marinum 2 months from the date of the skin biopsy. Antimicrobial susceptibility tests showed that the isolate was susceptible to clarithromycin, amikacin, rifampin, ethambutol, and moxifloxacin. Her skin lesions resolved with antibiotics, which were given for a total of 1 year.
Case 2 was a 59-year-old woman was admitted to hospital in April 2010 because of progressive multiple painless, erythematous, nodular skin lesions for several months (Fig. 1b). The skin lesions were distributed on face and trunk. She also had a history of HBV-related cirrhosis with cadaveric liver transplant done in April 2002, for which she was receiving MMF and lamivudine. She had moderate obesity and diabetes mellitus on oral hypoglycemic agents and history of esophageal variceal bleeding and duodenal ulcer. She was afebrile all along. She also had mild pancytopenia with a total white cell count of 3.43 × 109/liter, a neutrophil count of 2.48 × 109/liter, a lymphocyte count of 0.52 × 109/liter, and a monocyte count of 0.18 × 109/liter. The hemoglobin level was 10.4 g/dl, and the platelet count was 131 × 109/liter. Her renal function was impaired with serum urea at 10.6 mg/dl and creatinine at 1.3 mg/dl. Liver enzymes were within normal ranges. Skin biopsy specimens of the right arm and back lesions for Gram smear, bacterial, and fungal cultures were negative but positive for acid-fast bacilli on Ziehl-Neelsen stain. Histology showed suppurative granulomatous inflammation with numerous acid-fast bacilli identified with the Ziehl-Neelsen stain. Direct PCR of the skin tissue for hsp65 gene was performed. Based on the sequencing result, antimycobacterial agents, including oral clarithromycin at 600 mg twice daily, ethambutol at 800 mg daily, and moxifloxacin at 400 mg daily was commenced 1 week after admission, together with intravenous amikacin for the initial 3 weeks. Blood cultures for mycobacteria were negative.
In view of the positive results from the PCR and Ziehl-Neelsen staining of the direct tissue specimen, mycobacterial culture of the skin biopsy tissue was performed on Lowenstein-Jensen solid medium, chocolate agar, Middlebrook 7H10 agar with X factor (hemin) disks, and MGIT broth medium. However, all solid medium cultures were overgrown by bacteria. Pure colonies of acid-fast bacilli (isolate 2) were only finally isolated after subcultures of NaOH-decontaminated sediments from MGIT broth on Middlebrook 7H10 agar 3 months later. The isolate was initially detected as microcolonies observed around the factor X (hemin) disk, from which subculture was performed for isolation (Fig. 1c). The isolate grew on chocolate agar as rough, nonpigmented colonies after 2 weeks of incubation at 31°C. Growth was enhanced by addition of factor X (hemin) disk (Fig. 1d). It does not grow at 37°C or on Lowenstein-Jensen medium. It was negative for nitrate reduction. The isolate was phenotypically identified as M. haemophilum more than 4 months from the date of the tissue biopsy. Antimicrobial susceptibility test by proportion method showed that the isolate had a proportion of resistant mutants of <1% to rifampin, ethionamide, kanamycin, ofloxacin, and cycloserine but >1% to ethambutol, isoniazid, streptomycin, and para-aminosalicylic acid. The MIC of the isolate to clarithromycin was determined to be <0.015 μg/ml by the Etest method. Since the patient was intolerant to moxifloxacin, she was continued with oral clarithromycin and ethambutol and her skin lesions resolved after 1 year of antimycobacterial therapy.
PCR and sequencing of hsp65 gene and phylogenetic characterization.
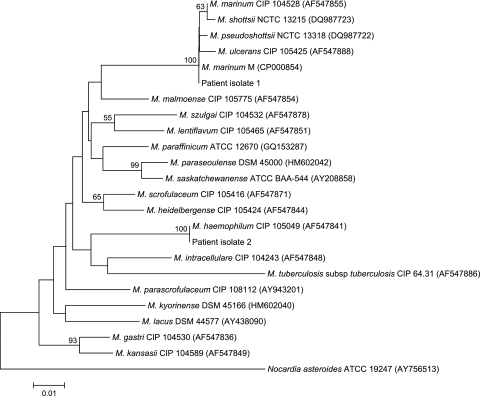
PCR of the hsp65 genes of the two isolates showed bands at about 441 bp. The hsp65 gene sequence of isolate 1 possessed no base difference compared to that of M. marinum strain M (GenBank accession no. CP000854), a one (0.2%)-base difference compared to that of M. pseudoshottsii strain JCM 15466 (GenBank accession no. AB548711), and two (0.5%)-base difference compared to that of the M. ulcerans strain ATCC 19423 (GenBank accession no. AB548728) and the M. shottsii strain NCTC 13215 (GenBank accession no. AM902955) (Fig. 2). The hsp65 gene sequence of isolate 2 possessed no base difference compared to that of M. haemophilum CIP 105049 (GenBank accession no. AF547841), a 21 (5.2%)-base difference compared to that of M. scrofulaceum strain CIP 105416 (GenBank accession no.AF434733), and a 23 (5.7%)-base difference compared to that of M. parascrofulaceum strain ATCC BAA-614 (GenBank accession no. GQ153295), M. kansasii strain CIP 104589 (GenBank accession no. AF292375), M. intracellulare strain ATCC 13950 (GenBank accession no. GQ153290), M. paraffinicum strain ATCC 12670 (GenBank accession no. GQ153287), M. lacus strain DSM 44577 (GenBank accession no. AY438090), and M. lentiflavum strain CIP 105465 (GenBank accession no. AF547851), indicating that the isolate was a strain of M. haemophilum. (Fig. 2).
Fig. 2.
Phylogenetic tree showing the relationships of the hsp65 genes of patient isolates to related mycobacteria. The tree was constructed by using the neighbor-joining method, and the Nocardia asteroides hsp65 gene sequence was used as outgroup. Bootstrap values were calculated from 1,000 trees. The scale bar indicates the estimated number of substitutions per 100 bases using the Jukes-Cantor correction. Names and accession numbers are given as cited in the GenBank database.
DISCUSSION
We present here the first report of disseminated Mycobacterium skin infections in two liver transplant recipients. M. marinum and M. haemophilum have rarely been reported as pathogens after transplantation. M. marinum has been reported to cause infections after renal and lung transplant, with most cases presenting as erythematous cutaneous, sometimes disseminated, nodules, with sporotrichoid spread (10, 19, 31). M. haemophilum has also rarely been associated with infections in immunocompromised patients, including renal, heart, lung, and bone marrow transplant recipients (12, 16, 17, 21, 26, 35). Although infections due to other nontuberculous Mycobacterium species have been occasionally reported in liver transplant recipients (4, 18), disseminated infections due to M. marinum and M. haemophilum in these patients have not been reported. In this report, both patients had received liver transplant more than 2 years ago and were receiving heavy immunosuppressants with lymphopenia, who presented with disseminated skin lesions. Although the two isolates were not recovered from blood or other body tissues which may be have been limited by their optimal growth temperatures, the two patients likely had systemic dissemination of the infections, as manifested by the generalized distribution of the skin lesions. The presentation of chronic, nodular skin lesions in these patients should raise the suspicion of disseminated infection due to M. marinum or M. haemophilum.
The present report highlights the role of PCR and sequencing in the rapid diagnosis of infections due to these slow-growing mycobacteria. 16S rRNA sequencing is known to be limited by the lack of sequence divergence among some closely related mycobacterial species. For example, the 16S rRNA gene of M. marinum shared ≥99.6% identities to that of M. ulcerans, M. pseudoshottsii, and M. shottsii; whereas the 16S rRNA gene of M. haemophilum shared 99.4% identity to that of M. malmoense (data not shown), in line with previous studies suggesting that 16S rRNA gene alone may not be sufficiently accurate for identification of these species (30, 33). Alternative gene targets, such as hsp65, rpoB, sodA, gyrB, dnaK, and 16S-23S rRNA internal transcribed spacer, have therefore been studied for the identification of Mycobacterium species (1, 7, 13, 24, 25, 27). hsp65 gene analysis has been used for the identification of nontuberculous Mycobacterium species such as M. marinum, M. haemophilum, M. chelonae, M. fortuitum, and M. avium (3, 6, 9, 11, 14). In the present study, PCR and sequencing of hsp65 was useful in rapid diagnosis and guiding empirical antimicrobial treatment well before the culture results were available. In particular, direct PCR of the hsp65 gene from the skin biopsy tissue was positive in the second patient, achieving rapid diagnosis without awaiting culture and phenotypic identification which had taken more than 4 months. Although in the first patient, the hsp65 gene of the M. marinum isolate only possessed a one-base difference compared to that of M. pseudoshottsii, and a two-base difference compared to that of M. ulcerans and M. shottsii, reflecting the close relatedness of these species (22, 23, 32), the present PCR was still helpful in rapid diagnosis. hsp65 gene analysis suggested that the isolate belonged to M. marinum, a much more commonly encountered species than M. pseudoshottsii, M. ulcerans, and M. shottsii in our population, which is also compatible with the patient's frequent exposure to aquatic environments. Although limitations in applying hsp65 gene analysis for species identification should be noted, the present report illustrates its role as a fast and effective diagnostic method, and this approach should be considered in severe infections due to slow-growing mycobacteria.
The ideal choice and optimal duration of treatment of nontuberculous Mycobacterium infections in transplant recipients is often difficult to determine. M. marinum is susceptible in vitro to antibiotics such as ethambutol, rifamycins, macrolides, and moxifloxacin, whereas M. haemophilum is known to be most susceptible in vitro to clarithromycin, ciprofloxacin, amikacin, and rifabutin/rifampin (19). As in most reported cases of M. marinum and M. haemophilum infections in other transplant recipients (19, 26), both of our patients responded to antimycobacterial treatment without surgical excision.
While the source of M. marinum infection in the first patient is likely related to aquatic waters or fish, the origin of the M. haemophilum isolate in the second patient remains obscure. Although believed to be ubiquitous, the natural habitat and mode of transmission of M. haemophilum is much less understood. Earlier reports speculating water and aerosolized pentamidine as sources were not supported by more concrete evidence (26). A recent report also described its association with dental manipulation (17). Further studies are required to investigate the source of M. haemophilum infections in humans.
Because M. haemophilum is difficult to isolate, the incidence of M. haemophilum infections may have been underestimated. M. haemophilum (which means “blood-loving” Mycobacterium) does not grow in routine mycobacterial cultures such as Lowenstein-Jensen or Middlebrook 7H11 medium unless the medium is supplemented with ferric iron-containing compounds (26). Similar to M. marinum, it grows best at 28 to 32°C, which also reflects its predilection for skin involvement on the face and extremities in a high proportion of cases (26). In the present study, the isolation of pure colonies of M. haemophilum for phenotypic identification was time-consuming. Clinical suspicion of M. haemophilum infection should be raised in patients presenting with features suggestive of nontuberculous mycobacteriosis, where routine mycobacterial cultures are negative.
ACKNOWLEDGMENTS
This study was partly supported by the Committee for Research and Conference Grant, The University of Hong Kong, and by the HKSAR Research Fund for the Control of Infectious Diseases (Commissioned Study) of the Health, Welfare, and Food Bureau.
We thank Sze-Tat Fun for excellent technical assistance.
Footnotes
Published ahead of print on 31 August 2011.
REFERENCES
- 1. Adekambi T., Berger P., Raoult D., Drancourt M. 2006. rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56:133–143 [DOI] [PubMed] [Google Scholar]
- 2. Banday K. M., et al. 2011. Use of urine volatile organic compounds to discriminate tuberculosis patients from healthy subjects. Anal. Chem. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 3. Castro-Silva A. N., et al. 2011. Cutaneous Mycobacterium haemophilum infection in a kidney transplant recipient after acupuncture treatment. Transpl. Infect. Dis. 13:33–37 [DOI] [PubMed] [Google Scholar]
- 4. Clark D., Lambert C. M., Palmer K., Strachan R., Nuki G. 1993. Monoarthritis caused by Mycobacterium avium complex in a liver transplant recipient. Br. J. Rheumatol. 32:1099–1100 [DOI] [PubMed] [Google Scholar]
- 5. Clinical and Laboratory Standards Institute. 2003. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard M24-A. Clinical and Laboratory Standards Institute, Wayne, PA: [PubMed] [Google Scholar]
- 6. Dai J., Chen Y., Lauzardo M. 2011. Web-accessible database of hsp65 sequences from Mycobacterium reference strains. J. Clin. Microbiol. 49:2296–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dai J., et al. 2011. Multiple-genome comparison reveals new loci for Mycobacterium species identification. J. Clin. Microbiol. 49:144–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Doucette K., Fishman J. A. 2004. Nontuberculous mycobacterial infection in hematopoietic stem cell and solid organ transplant recipients. Clin. Infect. Dis. 38:1428–1439 [DOI] [PubMed] [Google Scholar]
- 9. Ena P., et al. 2001. Rapid identification of cutaneous infections by nontubercular mycobacteria by polymerase chain reaction-restriction analysis length polymorphism of the hsp65 gene. Int. J. Dermatol. 40:495–499 [DOI] [PubMed] [Google Scholar]
- 10. Farooqui M. A., Berenson C., Lohr J. W. 1999. Mycobacterium marinum infection in a renal transplant recipient. Transplantation 67:1495–1496 [DOI] [PubMed] [Google Scholar]
- 11. Hsiao C. H., Lin Y. T., Lai C. C., Chou C. H., Hsueh P. R. 2010. Identification of nontuberculous mycobacterial infection by IS6110 and hsp65 gene analysis on lung tissues. Diagn. Microbiol. Infect. Dis. 68:241–246 [DOI] [PubMed] [Google Scholar]
- 12. Jang E. Y., et al. 2007. Case of pyomyositis due to Mycobacterium haemophilum in a renal transplant recipient. J. Clin. Microbiol. 45:3847–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kasai H., Ezaki T., Harayama S. 2000. Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38:301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim K., et al. 2010. Development and application of multiprobe real-time PCR method targeting the hsp65 gene for differentiation of Mycobacterium species from isolates and sputum specimens. J. Clin. Microbiol. 48:3073–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau S. K. P., et al. 2004. Eggerthella hongkongensis sp. nov. and Eggerthella sinensis sp. nov., two novel Eggerthella species, account for half of the cases of Eggerthella bacteremia. Diagn. Microbiol. Infect. Dis. 49:255–263 [DOI] [PubMed] [Google Scholar]
- 16. Malouf M. A., Glanville A. R. 1999. The spectrum of mycobacterial infection after lung transplantation. Am. J. Respir. Crit. Care Med. 160:1611–1616 [DOI] [PubMed] [Google Scholar]
- 17. Minani T. J., Saubolle M. A., Yu E., Sussland Z. 2010. Mycobacterium haemophilum as a novel etiology of cervical lymphadenitis in an otherwise healthy adult patient. J. Clin. Microbiol. 48:2636–2639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neau-Cransac M., Dupon M., Carles J., Le Bail B., Saric J. 1998. Disseminated Mycobacterium avium infection after liver transplantation. Eur. J. Clin. Microbiol. Infect. Dis. 17:744–746 [DOI] [PubMed] [Google Scholar]
- 19. Pandian T. K., Deziel P. J., Otley C. C., Eid A. J., Razonable R. R. 2008. Mycobacterium marinum infections in transplant recipients: case report and review of the literature. Transpl. Infect. Dis. 10:358–363 [DOI] [PubMed] [Google Scholar]
- 20. Peterson E. M., et al. 1989. Direct identification of Mycobacterium tuberculosis, Mycobacterium avium, and Mycobacterium intracellulare from amplified primary cultures in BACTEC media using DNA probes. J. Clin. Microbiol. 27:1543–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Plemmons R. M., McAllister C. K., Garces M. C., Ward R. L. 1997. Osteomyelitis due to Mycobacterium haemophilum in a cardiac transplant patient: case report and analysis of interactions among clarithromycin, rifampin, and cyclosporine. Clin. Infect. Dis. 24:995–997 [DOI] [PubMed] [Google Scholar]
- 22. Rhodes M. W., et al. 2005. Mycobacterium pseudoshottsii sp. nov., a slowly growing chromogenic species isolated from Chesapeake Bay striped bass (Morone saxatilis). Int. J. Syst. Evol. Microbiol. 55:1139–1147 [DOI] [PubMed] [Google Scholar]
- 23. Rhodes M. W., et al. 2003. Mycobacterium shottsii sp. nov., a slowly growing species isolated from Chesapeake Bay striped bass. Int. J. Syst. Evol. Microbiol. 53:421–424 [DOI] [PubMed] [Google Scholar]
- 24. Ringuet H., et al. 1999. hsp65 sequencing for identification of rapidly growing mycobacteria. J. Clin. Microbiol. 37:852–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roth A., et al. 1998. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36:139–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saubolle M. A., Kiehn T. E., White M. H., Rudinsky M. F., Armstrong D. 1996. Mycobacterium haemophilum: microbiology and expanding clinical and geographic spectra of disease in humans. Clin. Microbiol. Rev. 9:435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Simmon K. E., et al. 2007. Identification of an emerging pathogen, Mycobacterium massiliense, by rpoB sequencing of clinical isolates collected in the United States. J. Clin. Microbiol. 45:1978–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Telenti A., et al. 1993. Rapid identification of Mycobacteria to the species level by polymerase chain-reaction and restriction enzyme analysis. J. Clin. Microbiol. 31:175–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tønjum T., Welty D. B., Jantzen E., Small P. L. 1998. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J. Clin. Microbiol. 36:918–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Torres F., Hodges T., Zamora M. R. 2001. Mycobacterium marinum infection in a lung transplant recipient. J. Heart Lung Transplant. 20:486–489 [DOI] [PubMed] [Google Scholar]
- 32. Trott K. A., et al. 2004. Characterization of a Mycobacterium ulcerans-like infection in a colony of African tropical clawed frogs (Xenopus tropicalis). Comp. Med. 54:309–317 [PubMed] [Google Scholar]
- 33. Ucko M., et al. 2002. Strain variation in Mycobacterium marinum fish isolates. Appl. Environ. Microbiol. 68:5281–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Versalovic J., et al. 2011. Manual of clinical microbiology, 10th ed. ASM Press, Washington, DC [Google Scholar]
- 35. White M. H., Papadopoulos E. B., Small T. N., Kiehn T. E., Armstrong D. 1995. Mycobacterium haemophilum infections in bone marrow transplant recipients. Transplantation 60:957–960 [PubMed] [Google Scholar]
- 36. Woo P. C. Y., Fung A. M. Y., Lau S. K. P., Wong G. Y., Yuen K. Y. 2002. Diagnosis of pelvic actinomycosis by 16S rRNA gene sequencing and its clinical significance. Diagn. Microbiol. Infect. Dis. 43:113–118 [DOI] [PubMed] [Google Scholar]
- 37. Woo P. C., et al. 2000. Identification of Mycobacterium neoaurum isolated from a neutropenic patient with catheter-related bacteremia by 16S rRNA sequencing. J. Clin. Microbiol. 38:3515–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Woo P. C., et al. 2002. Relatively alcohol-resistant mycobacteria are emerging pathogens in patients receiving acupuncture treatment. J. Clin. Microbiol. 40:1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Woo P. C., Lau S. K., Teng J. L., Tse H., Yuen K. Y. 2008. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin. Microbiol. Infect. 14:908–934 [DOI] [PubMed] [Google Scholar]