Abstract
A panel of human serum samples spiked with various amounts of Aspergillus fumigatus genomic DNA was distributed to 23 centers within the European Aspergillus PCR Initiative to determine analytical performance of PCR. Information regarding specific methodological components and PCR performance was requested. The information provided was made anonymous, and meta-regression analysis was performed to determine any procedural factors that significantly altered PCR performance. Ninety-seven percent of protocols were able to detect a threshold of 10 genomes/ml on at least one occasion, with 83% of protocols reproducibly detecting this concentration. Sensitivity and specificity were 86.1% and 93.6%, respectively. Positive associations between sensitivity and the use of larger sample volumes, an internal control PCR, and PCR targeting the internal transcribed spacer (ITS) region were shown. Negative associations between sensitivity and the use of larger elution volumes (≥100 μl) and PCR targeting the mitochondrial genes were demonstrated. Most Aspergillus PCR protocols used to test serum generate satisfactory analytical performance. Testing serum requires less standardization, and the specific recommendations shown in this article will only improve performance.
INTRODUCTION
PCR testing can be used as a screening tool to exclude invasive aspergillosis (IA). High-frequency sampling is required, favoring easily obtainable specimens such as blood. In contrast, invasive specimens, such as those from bronchoalveolar lavage (BAL) or tissue, are used to confirm the presence of IA once disease is already suspected.
The European Aspergillus PCR Initiative (EAPCRI) reported on the process of standardizing molecular methods for the detection of Aspergillus in whole blood (WB) and highlighted the importance of the nucleic acid extraction protocol in achieving satisfactory analytical sensitivity (11). Compared to alternative specimens (serum and plasma), WB processing is technically demanding, requiring greater standardization. Many centers routinely test other specimens, and it is important to evaluate the processes in use and provide a standardized approach for Aspergillus PCR testing of these specimen types. Aspergillus PCR assays have been used to successfully detect DNA in serum. Although the methodology varies, the pooled sensitivity and specificity of serum PCR were 72% and 96.5%, respectively (2, 3, 10, 12, 14).
This article describes the EAPCRI process to evaluate the analytical performance of methods currently in use for the detection of Aspergillus DNA in serum through the blinded distribution of a simulated serum panel to 23 centers in Europe, the United States, and Australia for molecular testing. Statistical analysis of the results returned by each participant was performed anonymously to determine factors that affected analytical performance with an emphasis on analytical sensitivity to maintain the optimal negative predictive value required for a screening assay.
MATERIALS AND METHODS
The study followed the STARD guidelines where applicable (1).
Participants.
The participants comprised a core of eight laboratories and an extended group of a further 15 laboratories. To maintain impartiality throughout the analytical process, all centers were designated by a numerical code to allow blinded review of individual methodological procedures, determination of performance, and statistical analysis.
DNA source material.
Conidia were harvested from a sporulating culture of Aspergillus fumigatus (ATCC strain 1022) using a wet microbiological loop and resuspended in sterile water containing a drop of Tween 20 to prevent clumping. Conidia were serially diluted in sterile water and quantified using a Fuchs-Rosenthal counting chamber. A volume of suspension containing the equivalent to 1.2 × 106 conidia was harvested by centrifugation at 10,000 × g for 5 min. The supernatant was discarded, and conidia were exposed to mechanical disruption using the equivalent of 20 μl of ceramic beads (Roche, Burgess Hill, United Kingdom) and 30 s of bead-beating using a minibead beater (Biospec Products). After pulse centrifugation, the beads were washed with 200 μl molecular-grade water and DNA was extracted using the Roche High Pure Template DNA kit as per the manufacturer's instructions with an elution volume of 100 μl. The DNA was serially diluted using molecular-grade Tris-EDTA buffer (Sigma-Aldrich, United Kingdom) to attain a range of DNA concentrations (Table 1).
Table 1.
Composition and corresponding positivity of the EAPCRI serum panela
| Initial A. fumigatus DNA concn (genomes/μl)b | Concn (μl) added to 30 ml serum | Serum burden (genomes/ml) | 95% variability in 1-ml samplec | Positivity (no. positive/tested)d | Mean Cq (95% CI) |
|---|---|---|---|---|---|
| 12,000 | 25 | 10,000 | 9,803–10,197 | 45/45 | 27.8 (27.0–28.6) |
| 1,200 | 25 | 1,000 | 938–1,062 | 45/45 | 31.1 (30.3–31.9) |
| 12.5 | 500 | 456–544 | 45/45 | 32.7 (31.9–33.5) | |
| 120 | 25 | 100 | 80–120 | 44/45 | 34.7 (33.9–35.7) |
| 12.5 | 50 | 36–64 | 44/47 | 36.0 (35.2–36.9) | |
| 12 | 25 | 10 | 4–16 | 43/47 | 37.5 (36.7–38.3) |
| 12.5 | 5 | ≤1–9 | 38/46 | 38.1 (37.2–39.0) | |
| 1.2 | 25 | 1 | 0–3 | 12/47 | 38.8 (37.1–40.5) |
| 0 | 0 | 0 | 0 | 6/94 | 37.1 (35.3–38.8) |
Each panel was composed of one aliquot of each fungal burden and two negative control serum samples.
DNA extracted from 1.2 × 106 A. fumigatus conidia and serially diluted to attain desired concentrations assuming one conidium contains a single genome and 100% DNA extraction efficiency when initially extracting DNA. Initial DNA eluted in 100 μl.
Shown is the 95% confidence interval calculated on the basis of taking a 1-ml specimen from an initial 30-ml volume where the mean concentration will be as stated in the third column and assuming a Gaussian distribution.
Positivity defined as number of positive results per number of PCR replicates for each burden. Excluded are results from contaminated methods 10 and 21 and from unsuitable method 11 (Table 2).
Specimens.
Blood was obtained by consent from healthy volunteers, screened for the presence of infectious agents as per the protocol of the Institute of Transfusional Medicine, Wuerzburg University (Wuerzburg, Germany), and pooled. The serum was tested for the presence of contaminating Aspergillus DNA before processing (13). After being divided into 30-ml aliquots, two aliquots were retained to provide negative-control material, while eight of the aliquots were spiked with various concentrations of A. fumigatus DNA (Table 1). The 30-ml serum batches were further divided into 1-ml aliquots. To avoid airborne contamination, all processing of material took place in a category 2 laminar flow cabinet.
Panels.
Each panel consisted of 10 1-ml serum samples (eight containing A. fumigatus DNA and two negative) and a PCR control consisting of A. fumigatus DNA equivalent to 10 genomes/μl to monitor PCR performance. The expediency of the panel was tested by an “in-house” Aspergillus-specific real-time PCR (13) and by the commercially available MycAssay Aspergillus (Myconostica, Manchester, United Kingdom). After developing the panels, the distributing center froze all panels at −80°C with no freeze-thaw intervals. To validate panels, initial blind testing was performed by the core group (n = 8) before distribution and blind testing by additional centers (n = 15). The panels were circulated on dry ice for next day delivery by courier. Participating centers were asked to confirm receipt, comment on the state of the panel (frozen or thawed), and keep specimens frozen at −80°C until testing. Centers were requested to return results within a designated time frame and provide detailed protocols for their DNA extraction and PCR amplification systems. The information required included the sample volume used, extraction method, DNA elution volume, PCR method, PCR target, PCR template input volume, PCR total reaction volume, PCR amplification platform, and internal control PCR results.
The acceptable threshold for the PCR performance panel was equivalent to 10 genomes/ml serum. It was essential that all methods evaluated were able to attain this level of detection. Methods not performing to this standard were considered suboptimal. Two concentrations below this threshold were included (5 and 1 genome/ml serum), but the lower 95% confidence interval (95% CI) for the possible concentration in each 1-ml aliquot was below 1 genome per 1-ml sample at these concentrations (Table 1).
DNA extraction.
All participants were encouraged to use their own methodology. No recommendations were provided. A total of 15 different extraction systems were used (Table 2).
Table 2.
Methods used to test the EAPCRI serum panela
| Method | Sample vol (ml) | Available target at threshold (95% CI)b | DNA extraction method | Elution vol (μl) | Internal control PCR used (yes/no) | PCR platform (target) | PCR vol (μl) |
Positivity to thresholdc/overalld (%) | False positivity (%)e | |
|---|---|---|---|---|---|---|---|---|---|---|
| Input | Final | |||||||||
| 1 | 1 | 10 (4–16) | Roche High Pure LV | 60 | Yes | LC 2.0 (28S) | 10 | 20 | 100/93.75 | 0 |
| 2 | 0.4 | 4 (1–7) | Qiagen EZ1 Virus 2.0 | 60 | Yes | LC 2.0 (28S) | 10 | 20 | 100/87.5 | 0 |
| 3 | 0.5 | 5 (2–8) | bioMérieux EasyMag | 60 | Yes | LC 2.0 (28S) | 10 | 20 | 100/93.75 | 0 |
| 4 | 1 | 10 (4–16) | Roche MagNA Pure LV | 50 | Yes | LC 480 (28S) | 25 | 50 | 100/93.75 | 0 |
| 5 | 1 | 10 (4–16) | Roche MagNA Pure LV | 50 | Yes | LC 480 (Mito) | 5 | 20 | 100/87.5 | 0 |
| 6 | 1 | 10 (4–16) | Roche MagNA Pure LV | 50 | Yes | LC 480 (28S) | 5 | 20 | 100/93.75 | 0 |
| 7 | 0.2 | 2 (0–5) | Qiagen Qiamp DNA | 50 | Yes | LC 480 (28S) | 2–4 | 20 | 100/100 | 0 |
| 8 | 0.2 | 2 (0–5) | Roche MagNA Pure NA | 50 | No | LC 2.0 (18S) | 10 | 20 | 92.3/70.6 | 0 |
| 9 | 0.2 | 2 (0–5) | Roche MagNA Pure LV | 50 | No | LC 2.0 (18S) | 10 | 20 | 92.3/82.4 | 25 |
| 10 | 0.6 | 6 (3–9) | bioMérieux EasyMag | 50 | No | LC 2.0 (18S) | 10 | 20 | 100/100 | 100 |
| 11 | 1 | 10 (4–16) | Cepheid GeneExpert | 100 | Yes | GeneExpert (18S) | 25 | 50 | 50/37.5 | 0 |
| 12 | 1 | 10 (4–16) | Qiagen Ultrasens Virus | 35 | Yes | StepOnePlus (ITS2) | 10 | 20 | 100/93.75 | 0 |
| 13 | 1 | 10 (4–16) | Qiagen Circulating NA kit | 50 | Yes | 7500 FAST (ITS) | 7 | 20 | 100/100 | 0 |
| 14 | 0.2 | 2 (0–5) | Qiagen EZ1 tissue | 50 | Yes | 7000 (A. fumigatus28S) | 7.5 | 20 | 100/81.25 | 0 |
| 15 | 0.2 | 2 (0–5) | Qiagen EZ1 tissue | 50 | Yes | 7000 (28S) | 7.5 | 20 | 91.6/75 | 25 |
| 16 | 0.8 | 8 (5–11) | Qiagen Qiamp DNA kit | 100 | Yes | LC 1.2 (28S) | 5 | 20 | 91.6/75 | 0 |
| 17 | 0.22 | 2 (0–5) | bioMérieux EasyMag | 110 | Yes | 7900 FAST (28S) | 10 | 30 | 100/81.25 | 0 |
| 18 | 0.5 | 5 (2–8) | bioMérieux EasyMag | 110 | Yes | 7900 FAST (28S) | 10 | 30 | 100/87.5 | 0 |
| 19 | 1 | 10 (4–16) | Promega Wizard Genomic DNA | 50 | Yes | Rotorgene 6000 (28S) | 5 | 30 | 92.3/73.7 | 33.3 |
| 20 | 1 | 10 (4–16) | Qiagen QIAsymphony | 60 | No | 7500 FAST (ITS1) | 5 | 25 | 100/87.5 | 0 |
| 21 | 0.4 | 4 (1–7) | Qiagen QIAmp DNA blood | 60 | Yes | Mx3000p (28S) | 4 | 20 | 100/100 | 100 |
| 22 | 1 | 10 (4–16) | Roche MagNa Pure LV | 50 | Yes | LC 1.2 (18S) | 10 | 20 | 100/100 | 0 |
| 23 | 0.4 | 4 (1–7) | Qiagen EZ1 Virus 2.0 | 60 | Yes | SmartCycler (18S) | 10 | 25 | 100/87.5 | 50 |
| 24 | 1 | 10 (4–16) | Promega Maxwell | 30 | Yes | 7900 FAST (18S) | 5 | 20 | 83.3/75 | 0 |
| 25 | 0.5 | 5 (2–8) | Roche High Pure Template DNA | 100 | Yes | LC 480 (28S) | 20 | 50 | 100/100 | 0 |
| 26 | 1 | 10 (4–16) | Roche MagNA Pure Compact | 50 | Yes | 7500 FAST (18S) | 5 | 50 | 100/87.5 | 0 |
| 27 | 0.2 | 2 (0–5) | Qiagen QIAmp DNA kit | 65 | Yes | SmartCycler (18S) | 10 | 25 | 100/87.5 | 50 |
| 28 | 0.4 | 4 (1–7) | Qiagen QIAmp DNA kit | 65 | Yes | SmartCycler (18S) | 10 | 25 | 100/87.5 | 0 |
| 29 | 0.2 | 2 (0–5) | Roche MagNA Pure NA | 100 | No | 9700 (MitoC) | 5 | 20 | 100/87.5 | 0 |
| Mean | 0.62 | 6 (3–9) | 62.9 | 9.3f | 26.6 | 92.7/86.5g | 13.5g | |||
| Total | 24/29 | |||||||||
Results for methods in boldface represent (i) procedures with less than 100% reproducibility at the designated threshold of 10 genomes/ml (n = 5 protocols: 8, 9, 11, 15, and 16), (ii) positivity rates below the overall positivity of 86.1% (95% CI, 82.1 to 89.5%) (n = 8 protocols: 8, 11, 14, 15, 16, 17, 19, and 24), or (iii) false-positivity rates (1 − specificity) that result in specificity being below the overall specificity of 93.6% (95% CI, 86.6 to 97.6%) (n = 7 protocols: 9, 10, 15, 19, 21, 23, and 27). Please note that despite reproducibly detecting the designating threshold, protocols 19 and 24 achieved 66.7% and 0% positivity, respectively, when testing the 50-genome/ml sample, resulting in <100% positivity to threshold. Please note that the overall sensitivity and specificity (stated above) were calculated excluding the data from methods 10, 11, and 21. LV, large volume; NA, nucleic acid; LC 2.0, Roche LightCycler 2.0; LC 480, Roche LightCycler 480; GeneExpert, Cepheid GeneExpert; StepOnePlus, ABI StepOnePlus; 7500 FAST, ABI 7500 FAST; 7000, ABI 7000; 7900 FAST, ABI 7900 FAST; Rotorgene 6000, Corbett Rotorgene 6000; LC 1.2, Roche LightCycler 1.2; Mx3000p, Stratagene Mx3000p; SmartCycler, Cepheid SmartCycler; 9700, ABI 9700; 28S, 28S rRNA gene; Mito, mitochondrial gene; 18S, 18S rRNA gene; ITS2, internal transcribed spacer 2; ITS, internal transcribed spacer; ITS1, Internal Transcribed Spacer 1; MitoC, mitochondrial cytochrome B.
The available burden and 95% CI when testing sample volumes less than 1 ml were determined assuming that the 1-ml aliquot provided contained the mid-point value of 10 genomes/ml.
Positivity to threshold represents the number of positive PCR results per test performed for each sample at, and above, the designated threshold of 10 genomes/ml (10,000, 1,000, 500, 100, 50, and 10 genomes/ml).
Overall positivity represents the number of positive PCR results per tests performed for the entire panel excluding the negative samples (10,000, 1,000, 500, 100, 50, 10, 5, and 1 genome/ml).
False positivity represents the number of positive PCR results per test performed for both negative samples.
Calculated using a mid-point value of 3 μl for method 7.
Includes results for all 29 protocols listed in the table.
PCR amplification.
Participating centers were asked to use their current amplification system. It was recommended that DNA extracts be tested in duplicate—preferably triplicate. The use of an internal control PCR was recommended. The use of nested-PCR assays was discouraged, as these were considered too prone to false-positive results and unlikely to attain widespread acceptance outside specialist molecular centers. Twenty-two combinations of PCR target, input volume, final reaction volume, and amplification platform were provided (Table 2).
Data analysis.
A data analysis subgroup collated the results and technical information and determined covariates for blinded statistical analysis by an independent party (Table 3).
Table 3.
Covariates investigated by linear regression methods in this study
| Covariate | Explanation |
|---|---|
| Sample vol | Vol of sample used for DNA extraction |
| DNA extraction system | Extraction platform used to extract DNA from sample—Roche, Qiagen, or combined other systemsa |
| Elution vol | Vol used to elute DNA at the end of extraction |
| PCR amplification system | Amplification platform used to perform real-time PCR—Roche, Applied Biosystems, or combined other systemsb |
| Template vol | Vol of DNA eluate used in each PCR |
| PCR vol | Final vol of each PCR |
| PCR target | Internal transcribed spacer target, 18S rRNA target, 28S rRNA target, or mitochondrial DNA target for Aspergillus PCR |
| Internal control | Internal control PCR used (yes/no) |
bioMérieux EasyMag and Promega Wizard and Maxwell systems. Note one Qiagen system and one method using the bioMérieux EasyMag were excluded due to systemic contamination. The Cepheid GeneExpert was excluded on the basis of not being designed for use with serum.
Cepheid SmartCycler and Corbett Rotorgene 6000. Note one method using the Stratagene Mx3000p and one using the Roche Lightcycler 2.0 were excluded due to systemic contamination. The Cepheid GeneExpert was excluded on the basis of not being designed for use with serum.
Statistical analysis.
(i) PCR positivity and genomic load.
PCR positivity was determined according to the reported real-time PCR quantification cycle (Cq value), using a universal positivity threshold of 45 cycles, and PCR results with a Cq value of >45 cycles were considered negative. In doing so, continuous Cq data were changed into qualitative positive and negative results, and positivity at each fungal burden was determined, along with the binomial exact 95% confidence interval. The specificity was calculated using the equation 1 − positivity rate for the serum samples with no fungal burden. In addition, a receiver operator curve (ROC) was constructed with PCR performance determined by a varying Cq threshold for positivity.
Mixed-model logistic regression analysis evaluated the probability of a positive result as a function of the genomic load using the PCR result as a binomial, positive or negative, outcome variable.
(ii) Linear analysis.
The correlation between threshold cycle number (Cq) of real-time PCR and genomic load (log10 transformed) was investigated by linear regression analysis. Samples with no fungal burden were excluded from the model, as data associated with a zero fungal burden would establish a discontinuity (genomes present/not present). To correctly evaluate the clustered nature of the data, a mixed (multilevel) random-coefficient model was used, grouping the various methods using the Stata command “xtmixed.” The basic model was Cq versus log10 genomes/ml.
From the slope parameter, the mean reaction efficiency was calculated. The formula used was relative efficiency = 10−1/slope − 1, where the 100% efficiency corresponds to the doubling (×2) of the target sequence per PCR cycle. In this context, a slope of −3.32193 would correspond to a (full) relative efficiency of 100%.
The same basic model was adapted to the analysis of covariates (sample volume, elution volume, template volume, PCR volume, extraction procedure, PCR platform, DNA sequence target, use of internal control). Each covariate was added singly to the above-described model (Cq versus log10 genomes/ml. A 4-level categorical variable was used for extraction evaluation, with the following levels: Roche, Qiagen, bioMérieux, and Promega. A similar coding criterion was used for the PCR platform (four levels: ABI, LightCycler, Rotorgene, and SmartCycler) and DNA sequence target (four levels: 28S, 18S, internal transcribed spacer [ITS], and mitochondria). A multivariate approach was performed using the covariates indicated above. After adjustment, the final model retained the independently significant covariates.
RESULTS
Twenty-two of the 23 centers requesting participation were able to return results, but one of these centers used a nested-PCR system, and the results were excluded from further analysis. A total of 29 different protocols (combined DNA extraction and PCR amplification) were provided by the remaining 21 centers (Table 2). Two centers contributed three protocols, four centers contributed two protocols, and 15 centers contributed a single protocol.
Aspergillus PCR performance when testing serum specimens.
Of the 29 different protocols used to test the serum panel, 24 were able to reproducibly (100%) detect the designated threshold of 10 genomes/ml (Table 2). Despite detecting the designated threshold, protocols 19 and 24 achieved 66.7% and 0% positivity, respectively, when testing the 50-genome/ml sample. Protocols 8 and 9 were able to detect the threshold in two of three replicates, although protocol 9 was able to reproducibly detect the sample containing 5 genomes/ml. Protocols 15 and 16 detected the threshold in one of two replicates. Protocol 11 was unable to detect the designated threshold; most samples were determined to be inhibitory or as extraction failures. Most protocols (21/24) attaining the threshold could also reproducibly detect the sample with 5 genomes/ml, although at this concentration protocols 14, 17, and 19 generated positivity rates of 50% and 33.3%, respectively. Eleven protocols (1, 3, 4, 6, 7, 10, 12, 13, 19, 21, and 25) were able to detect the 1-genome/ml sample. At this concentration, not every 1-ml aliquot will contain DNA (Table 1), and logistical regression analysis generated a probability of 63.21% that an individual 1-ml aliquot of the 1-genome/ml specimen would actually contain target, and this would be accentuated if volumes less than 1 ml were used for the initial DNA extraction. Protocols 10 and 21 generated a 100% (4/4) false-positivity rate with Cq values almost identical to those for the samples containing ≤10 genomes/ml, casting doubt on the validity of the “true”-positive results. Additional false positivity was noted for protocols 9, 15 (1/4), 19 (2/6), 23, and 27 (1/2).
In determining overall serum PCR performance, assays 10 and 21 (extensive false positivity) and 11 (commercial method designed to process WB and unsuitable for serum testing) were considered unsatisfactory, leaving 26 protocols for analysis. The overall sensitivity and specificity for Aspergillus PCR, including all burdens were 86.1% (95% CI, 82.1 to 89.5%) and 93.6% (95% CI, 86.6% to 97.6%), respectively. Positive and negative likelihood ratios and the diagnostic odds ratio (DOR) were 13.5, 0.15, and 90.9, respectively.
PCR performance and relationship with fungal burden.
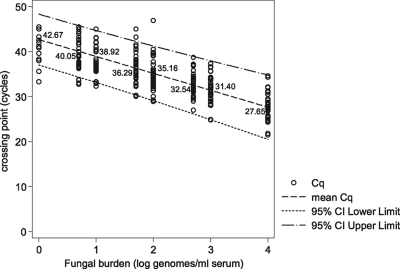
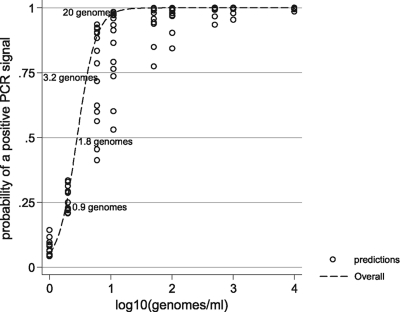
Predictably, there was a correlation between available fungal burden and PCR positivity (basic linear model: slope, −3.756; z value, −27.99; P < 0.0001). The mean PCR efficiency calculated from the slope coefficient of the regression line was 84.6% (95% CI, 77.4 to 93.3) (Fig. 1). Positivity rates and mean Cq values at the individual burdens are shown in Table 1 and Fig. 1 and 2. One hundred percent positivity was attained when testing the sample containing the equivalent of 500 genomes/ml, although at 100, 50, and 10 genomes/ml, positivity remained above 90%, and the difference in positivity rates between the four samples was not statistically significant (Table 1). The positivity rate for detecting burdens at or greater than the predesignated detection limit of 10 genomes/ml was 97.1% (95% CI, 94.5 to 99.6%). At 5 and 1 genome/ml, the positivity rates were 82.6% and 25.5%, respectively. The latter result was significantly lower than the positivity rates for every other burden tested (difference between 5 and 1 genome/ml, 57.1%; 95% CI, 37.8 to 70.3; P = 0.0001). Logistical regression analysis determined that a sample containing a burden of 1.82 genomes/ml would be associated with a 50% positivity rate, and the positivity rate when testing samples containing 1 genome/ml would be 27.59% (Fig. 2), as predicted by the Poisson distribution for a mean of 1 genome/ml.
Fig. 1.
Crossing point (Cq) values for Aspergillus PCR when testing DNA extracted from serum spiked with various fungal burdens as determined by linear regression. The linear equation for the mean Cq values is y = −3.76 log(x) + 42.67, where log(x) = log10 genomes/ml. The 95% CI for the slope was −3.49 to −4.02.
Fig. 2.
Probability of PCR positivity at various fungal burdens as determined by logistic regression.
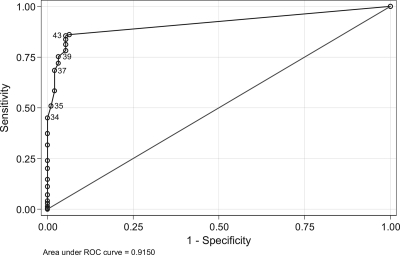
ROC analysis indicated that a Cq threshold of ≤43 cycles ensured a good diagnostic accuracy (DOR = 105) with a sensitivity and specificity of 85.6% and 94.7%, respectively, and using this threshold, 87.4% of samples were correctly classified (Fig. 3). Using a Cq threshold of ≤34 cycles generated a specificity of 100%.
Fig. 3.
Receiver operator curve for Aspergillus PCR methods when testing serum samples. PCR positivity was determined according to a variable Cq threshold. (Representative Cq thresholds are highlighted.) The area under the ROC was 0.9150 (95% CI, 0.89184 to 0.93806).
Linear regression analysis.
Bivariate linear regression determined a statistically significant positive association between PCR sensitivity and larger sample volumes, use of an internal control, and targeting the ITS region for PCR amplification. Separating the PCR performance on the basis of sample volume used (≥0.5 ml or <0.5 ml) generated sensitivities of 88.6% and 82.5% (difference, 6.1%; 95% CI, 14.9 to −1.0%; P = 0.1157), respectively. When only low burdens (≤10 genomes/ml) were included, sensitivities of 76.9% and 54.7% for ≥0.5-ml and <0.5-ml starting sample volumes, respectively, were generated. The difference in sensitivity at this clinically typical level was significant (difference, 21.8%; 95% CI, 5.6 to 37.1%; P = 0.0091).
A negative effect on the sensitivity of the PCR assay was detected for large elution volumes and for using mitochondrial targets for PCR (Table 4). When using elution volumes of ≥100 μl, 40% of methods generated a positive PCR result, on at least one occasion, for burdens of ≤5 genomes/ml, compared to 69.0% for methods eluting DNA in <100-μl volumes, the difference in positivity being 29.0% (95% CI, −3.4 to 55.1%; P = 0.1424). More significantly when detecting samples containing ≤5 genomes/ml, Cq values occurred earlier for DNA extraction methods eluting DNA in <100-μl volumes. The mean Cq for <100-μl elution volumes was 37.8 cycles, compared to 41.7 cycles for methods eluting DNA in volumes of ≥100 μl (difference, 3.9 cycles; 95% CI, 1.9 to 5.8 cycles; P = 0.0002).
Table 4.
Bivariate linear regression analysis between Cq and the additional covariates, excluding log10 genomes/mla
| Covariate | z value | P value |
|---|---|---|
| Sample vol | −2.27 | 0.023 |
| DNA extraction system | ||
| Roche | −0.11 | 0.909 |
| Qiagen | −1.15 | 0.250 |
| Other | 1.50 | 0.133 |
| Elution vol | 2.96 | 0.003 |
| PCR amplification system | ||
| Roche | −0.64 | 0.521 |
| ABI | 0.53 | 0.598 |
| Other | 0.25 | 0.803 |
| Template vol | −1.07 | 0.286 |
| PCR vol | 0.97 | 0.334 |
| PCR target | ||
| 18S rRNA gene | −0.28 | 0.779 |
| ITS region | −2.48 | 0.013 |
| 28S rRNA gene | 0.39 | 0.698 |
| Mitochondrial | 2.57 | 0.010 |
| Internal control | −2.18 | 0.029 |
Significant associations are highlighted in boldface. Note that a negative z value predicts an earlier Cq, and thus a higher probability of PCR positivity, whereas a positive value has the opposite effect.
Multivariate analysis confirmed the negative association between sensitivity and elution volume (P = 0.003) and PCR amplifying the mitochondrial targets (P = 0.011).
DISCUSSION
The testing of serum is less technically demanding than the testing of WB (7), since lysis of red and white cells is not required. The perceived target in serum is free circulating DNA (DNAemia), released from the organism through the actions of the host's immune defenses, antifungal therapy, autolysis, and possibly exponential growth (6, 9). Any cell-associated DNA sources in serum will be limited, being trapped during blood clot formation and/or removed during blood fractionation (12). If the target in serum is free DNA, then the extraction process is straightforward, although it is necessary to remove any serum components (proteins, electrolytes, waste products, drugs, etc.) that may interfere with the PCR amplification process.
Theoretically, targeting free circulating DNA through serum testing requires less standardization as commercial extraction methods can be employed. The commercial DNA extraction systems evaluated in the study provided comparable levels of performance. Nevertheless, each extraction system is directly associated with other variables such as sample input and elution volume, and it is essential to determine the analytical performance thresholds of any protocols utilizing commercial kits, highlighting methodologies that provide inadequate performance and should not be used in clinical practice. In this study, only five protocols (8, 9, 11, 15, and 16) failed to reproducibly detect the designated detection threshold of 10 genomes/ml serum (Table 2). Method 11 had been designed to test WB samples, providing excellent performance when testing this specimen type (11). Being fully automated, it could not be adjusted for serum testing; consequently, results were poor, and it should not be used for Aspergillus PCR testing of serum. Each protocol is constructed of many individual, yet linked, steps that will influence further steps downstream, and it is important to identify the critical step or steps that impact performance. For example, methods 8 and 9 use extraction platforms similar to methods 4 to 6 and 22, yet the latter methods provide better performance. It would be easy to conclude that the extraction platform provided inconsistent interlaboratory performance. However, detailed examination of the protocols showed that methods 8 and 9 only extracted 0.2 ml of the available specimen, compared to 1.0 ml for methods 4 to 6 and 22, limiting the amount of DNA target for extraction and negatively affecting final performance. In this study, a positive association between PCR positivity and the volume of starting material was shown.
False positivity was not a major issue, although two methods, 10 and 21, consistently generated false-positive results, suggesting a contamination issue. Both DNA extraction systems in these protocols had previously been associated with Aspergillus DNA contamination (4) (EAPCRI, group communication), but within this study, other centers had also used these systems without contamination, highlighting the transient, possibly batch-variable nature of this problem. Alternatively, the contamination issue may be associated with the individual center or other parts of these individual processes, and fungal DNA contamination of certain PCR reagents has been noted (5).
A PCR positivity threshold of 43 cycles should be used to provide the greatest degree of diagnostic accuracy, and at this threshold, 87.4% of results were correctly classified. This is in keeping with a previous report where the authors also concluded that clinically the burden of DNA in serum was very low (fg/μl), to such a degree that positive PCR signals became inconsistent (8).
The overall sensitivities and and specificities for Aspergillus PCR testing of serum were 86.1% and 93.6%, respectively. This compares favorably with the same performance parameters for Aspergillus PCR methods testing WB (11). When testing WB using protocols compliant with the EAPCRI recommendations provided to participants, the sensitivity and specificity were 88.7% and 91.6%, respectively. The probability of obtaining positive PCR results at specific fungal burdens is higher for serum than WB PCR, which is probably a consequence of the additional processing required when testing WB (results not shown). As the initial evaluation of existing serum PCR methods provides performance that is comparable to that of the agreed WB PCR protocol, it is not necessary to attempt to enhance performance through the evaluation of further serum panels with associated procedural recommendations. Indeed, our initial hypothesis that serum PCR methods would require less technical standardization appears to be correct. Nevertheless, mixed-model linear analysis did highlight several important parameters that form the basis of the EAPCRI recommendations for Aspergillus PCR when testing serum samples, as highlighted in Table 5.
Table 5.
Recommendations of the EAPCRI for Aspergillus PCR protocols when testing serum
| Result | Recommendation |
|---|---|
| A positive association between sensitivity and initial sample vol was noted (z value, −2.27; P = 0.023). | The use of larger sample volumes (≥0.5 ml) will improve sensitivity. |
| When detecting low fungal burdens (<10 genomes/ml), the difference in sensitivity using a starting volume of <0.5 ml serum compared to ≥0.5 ml is 21.8% (95% CI, 5.6 to 37.1; P = 0.0091). | To enhance detection of low fungal burdens (<10 genomes/ml), a minimum of 0.5 ml starting material should be used. |
| Twenty-eight of 29 protocols evaluated were able to reach the designated threshold of detection on at least one occasion, generating a threshold positivity rate of 91.5%. | Most commercially available nucleic acid extraction systems described in Table 2 can be used to extract Aspergillus DNA using the protocols as described by the manufacturer. However, all kits should be screened for contamination and a limit of detection determined prior to clinical use. |
| Method 11 (Table 2) was specifically designed to test whole blood and was not able to efficiently process serum samples. | Protocols specifically designed for testing alternative specimen types (e.g., whole blood) should not be used. |
| A negative association between sensitivity and elution vol was noted (z value, 2.96; P = 0.003). | The use of larger elution volumes (≥100 μl) will reduce sensitivity. |
| When detecting low fungal burdens (<10 genomes/ml), the difference in Cq for methods eluting in <100 μl (37.8 cycles) compared to ≥100 μl (41.7 cycles) volumes was 3.9 cycles (95% CI, 1.9 to 5.8 cycles; P = 0.0002) and results in a 29.0% reduction in positivity. | For optimal detection of low fungal burdens (<10 genomes/ml), nucleic acid should be eluted in vol of <100 μl. Elution in vol of ≥100 μl delays Cq values to the degree where real-time PCR is inconsistent and positivity will be affected. |
| A positive association between sensitivity and PCR targeting the ITS region was noted (z value, −2.48; P = 0.013). | The reaction kinetics for each PCR reaction will vary according to the design of specific oligonucleotides and the optimization of each particular PCR assay. Performance may not be directly associated with the target gene but exclusive to the individual assay itself. When designing a new assay, the MIQE guidelines (2a) should be followed. |
| A negative association between sensitivity and PCR targeting the mitochondrial regions was noted (z value, 2.57; P = 0.010). | The reaction kinetics for each PCR reaction will vary according to the design of specific oligonucleotides and the optimization of each particular PCR assay. Performance may not be directly associated with the target gene but exclusive to the individual assay itself. When designing a new assay, the MIQE guidelines (2a) should be followed. |
| A positive association between sensitivity and the use of an internal control PCR was noted (z value, −2.18; P = 0.029). | An internal control PCR should be used and incorporated at the start of the extraction procedure, monitoring both PCR inhibition and DNA extraction efficiency. Internal control Cq values should be representative of typical Aspergillus PCR-positive results (≥35 cycles), and human DNA targets should be avoided due to the possibility of variable target amounts within individual specimens generating varied Cq values. |
| Using a threshold of 43 cycles generates a DOR of 105, and 87.4% of results were correctly classified. | A PCR positivity threshold of 43 cycles provides the greatest degree of diagnostic accuracy. |
There were no significant associations between sensitivity and DNA template volume, PCR amplification platform, and final PCR volume, although PCR amplification of the ITS region was positively associated with sensitivity, and the opposite was true for the mitochondrial targets. This should be interpreted with caution as the reaction kinetics for each PCR will vary according to the design of specific oligonucleotides, and the optimization of each particular PCR assay and performance may not be directly associated with the target gene but exclusive to the individual assay.
To conclude, the testing of serum by Aspergillus PCR can be performed using commercial nucleic acid extraction methods, providing standardization and quality control. The EAPCRI previously showed PCR amplification not to be rate limiting, and commercial extraction in combination with most amplification methods will provide acceptable analytical performance(11). The use of serum is less technical than testing of WB, reducing both time to result reporting and labor, and permits fully automated nucleic acid extraction, limiting performance variability. It allows the use of a single sample for galactomannan enzyme-linked immunosorbent assay (ELISA), β-d-glucan, and PCR analysis, thereby reducing costs if high-throughput screening of high-risk patients is required. As with WB PCR testing, a multicenter clinical trial is paramount to determine the true clinical validity and utility of serum PCR testing. More so, as the clinically relevant biological target circulating in blood has not been precisely determined, a multicenter comparison of WB PCR (targeting fungal cell-associated DNA) and serum PCR (targeting free circulating DNA) is required to determine the clinically relevant biological target and consequently the optimal clinical specimen for PCR testing.
ACKNOWLEDGMENTS
We acknowledge the European Aspergillus Initiative of the ISHAM.
The EAPCRI thanks Andreas Opitz (Institute of Transfusional medicine, Wuerzburg University Hospital, Wuerzburg, Germany) for drawing and screening the blood kindly donated by our volunteers.
The EAPCRI Steering Group consists of the following members: J. Peter Donnelly, chair of foundation, Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands; Juergen Loeffler, Secretary, Wuerzburg University, Wuerzburg, Germany; Rosemary A Barnes, treasurer, Cardiff University, Cardiff, United Kingdom.
The EAPCRI Laboratory Working Group consists of the following members: Juergen Loeffler, Laboratory Working Group—lead, Data Analysis Group, Wuerzburg University, Wuerzburg, Germany; P. Lewis White, panel development and distribution, Data Analysis Group, NPHS Microbiology, Cardiff, United Kingdom; Stephane Bretagne, Data Analysis Group, Henri Mondor Hospital, Creteil, France; Willem Melchers, EAPCRI website administrator, Radboud University, Nijmegen Medical Centre, Nijmegen, The Netherlands; Lena Klingspor, Karolinska University Hospital, Stockholm, Sweden; Niklas Finnstrom, Cepheid AB, Toulouse, France; Elaine McCulloch, External Quality Control Programme, Royal Hospital for Sick Children, Glasgow, United Kingdom; Craig Williams, External Quality Control Programme, Royal Hospital for Sick Children, Glasgow, United Kingdom; and Manuel Cuenca-Estrella, Sapnish National Centre for Microbiology, Instituto de Salud Carlos III, Madrid, Spain.
The EAPCRI Clinical Working Group consists of the following members: Rosemary A. Barnes, Clinical Working Group—lead, Cardiff University, Cardiff, United Kingdom; Catherine Cordonnier, Henri Mondor Hospital, Creteil, France; Johan Maertens, University Hospital Gasthuisberg, Leuven, Belgium; Lena Klingspor, Karolinska University Hospital, Stockholm, Sweden; Werner Heinz, Wuerzburg University, Wuerzburg, Germany; and Brian Jones, Glasgow Royal Infirmary, Glasgow, United Kingdom.
The EAPCRI Statistical Working Group consists of the following members: Carlo Mengoli, University of Padua, Padua, Italy; Mario Cruciani, University of Verona, Verona, Italy; Juergen Loeffler, Wuerzburg University, Wuerzburg, Germany; Rosemary A. Barnes, Cardiff University, Cardiff, United Kingdom; and J Peter Donnelly, Radboud University Nijmegen Medical Centre, Nijmegen, The Netherlands.
We thank the following members of the participating laboratories: Vanda Chrenkova, Motol Hospital, Prague, Czech Republic; Malcolm Guiver, HPA Northwest, Manchester, United Kingdom; Catriona Halliday and Sue Sleiman, Westmead Hospital, Westmead, NSW, Australia; Chris Linton and Elizabeth Johnson, UK Mycology Reference Lab, HPA Southwest, Bristol, United Kingdom; Rebecca Gorton and Chris Kibbler, Royal Free Hospital, London, United Kingdom; Martina Lengerova, Central Molecular Biology, Gene Therapy and Haematology Clinic, Brno, Czech Republic; Eva Rosello Mayans, Vall d'Hebron University Hospital, Barcelona, Spain; Tom Rogers and Oliver Morton, Trinity College, Dublin, Ireland; Boualem Sendid, Lille University, Lille, France; Angie Caliendo, Emory University, Atlanta, GA; Tom Patterson, UTHCSA, San Antonio, TX; Katrien Lagrou, University Hospital Gasthuisberg, Leuven, Belgium; Mark Wilks, Barts and the London Hospital, London, United Kingdom; Adrian Moody, Myconostica, Ltd., Manchester, United Kingdom; Maria J. Buitrago and Leticia Bernal-Martinez, Spanish National Centre for Microbiology, Madrid, Spain; and Jaques Billes and Phillippe Hauser, Lausanne, Switzerland.
Footnotes
Published ahead of print on 21 September 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Bossuyt P. M., et al. 2004. Towards complete and accurate reporting of studies of diagnostic accuracy: the STARD initiative. Fam. Pract. 21:4–10 [DOI] [PubMed] [Google Scholar]
- 2. Bretagne S., et al. 1998. Comparison of serum galactomannan antigen detection and competitive polymerase chain reaction for diagnosing invasive aspergillosis. Clin. Infect. Dis. 26:1407–1412 [DOI] [PubMed] [Google Scholar]
- 2a. Bustin S. A., et al. 2009. The MIQE guidelines: minimum information for the publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 3. Costa C., et al. 2002. Real-time PCR coupled with automated DNA extraction and detection of galactomannan antigen in serum by enzyme-linked immunosorbent assay for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 40:2224–2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jaeger E. E., et al. 2000. Rapid detection and identification of Candida, Aspergillus, and Fusarium species in ocular samples using nested PCR. J. Clin. Microbiol. 38:2902–2908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Loeffler J., et al. 1999. Contaminations occurring in fungal PCR assays. J. Clin. Microbiol. 37:1200–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mennink-Kersten M. A., Ruegebrink D., Wasei N., Melchers W. J., Verweij P. E. 2006. In vitro release by Aspergillus fumigatus of galactofuranose antigens, 1,3-beta-D-glucan, and DNA, surrogate markers used for diagnosis of invasive aspergillosis. J. Clin. Microbiol. 44:1711–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Millon L., et al. 2011. Ribosomal and mitochondrial DNA target for real-time PCR diagnosis of invasive aspergillosis. J. Clin. Microbiol. 49:1058–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Millon L., et al. 2005. Use of real-time PCR to process the first galactomannan-positive serum sample in diagnosing invasive aspergillosis. J. Clin. Microbiol. 43:5097–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Morton C. O., et al. 2010. Dynamics of extracellular release of Aspergillus fumigatus DNA and galactomannan during growth in blood and serum. J. Med. Microbiol. 59:408–413 [DOI] [PubMed] [Google Scholar]
- 10. Pham A. S., et al. 2003. Diagnosis of invasive mold infection by real-time quantitative PCR. Am. J. Clin. Pathol. 119:38–44 [DOI] [PubMed] [Google Scholar]
- 11. White P. L., et al. 2010. Aspergillus PCR: one step closer towards standardization. J. Clin. Microbiol. 48:1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. White P. L., Barnes R. A. 2009. Aspergillus PCR, p. 373–390 In Latge J.-P., Steinbach W.J. (ed.), Aspergillus fumigatus and aspergillosis. ASM Press, Washington, DC [Google Scholar]
- 13. White P. L., Linton C. J., Perry M. D., Johnson E. M., Barnes R. A. 2006. The evolution and evaluation of a whole blood polymerase chain reaction assay for the detection of invasive aspergillosis in hematology patients in a routine clinical setting. Clin. Infect. Dis. 42:479–486 [DOI] [PubMed] [Google Scholar]
- 14. Williamson E. C., et al. 2000. Diagnosis of invasive aspergillosis in bone marrow transplant recipients by polymerase chain reaction. Br. J. Haematol. 108:132–139 [DOI] [PubMed] [Google Scholar]