Abstract
Listeria monocytogenes is a bacterial pathogen that can invade the central nervous system (CNS), causing meningoencephalitis and brain abscesses. The diagnosis of CNS listeriosis, based on the isolation of the bacteria in the cerebrospinal fluid (CSF), can be difficult because of previous antibiotic treatment and a low number of bacteria in the CSF. To improve the sensitivity of microbiological diagnosis, we have developed a real-time PCR assay for detecting and quantifying L. monocytogenes DNA in the CSF. The designed primers specifically amplify the L. monocytogenes hly gene, which encodes listeriolysin O, a pore-forming cytolysin. The PCR assay for the hly gene (PCR-hly) provides reproducible quantitative results over a wide dynamic range of concentrations and was highly sensitive while detecting a single gene copy/ml. By assaying a large panel of bacterial species, including species secreting pore-forming cytolysin, we determined the specificity of the PCR-hly, which exclusively detects the L. monocytogenes DNA. We then analyzed 214 CSF samples from patients suspected of having CNS listeriosis. PCR-hly was positive in all cases in which L. monocytogenes was isolated by culture. Positive PCR-hly of the CSF was also obtained for five additional, clinically confirmed cases of CNS listeriosis for which bacterial cultures were negative presumably due to previous treatment with antibiotics. As a complement to classical bacteriological CSF culture, our designed real-time PCR-hly assay proved to be valuable by enhancing the rapidity and the accuracy of the diagnosis of CNS infection by L. monocytogenes. In addition, the quantitative results provided may, in some instances, be useful for the follow-up of patients under treatment.
INTRODUCTION
Listeria monocytogenes is a facultative intracellular food-borne pathogen, widely found in the environment, which causes meningitis and meningoencephalitis (22, 35). Despite adequate antibiotic treatment, the overall mortality of central nervous system (CNS) infection is still high (25 to 30%) and neurological sequelae are frequent (22, 35). The apparent inefficacy of antibiotic therapy can be explained by the virulence of this facultative intracellular bacteria and because in many instances listeriosis occurs in immunocompromised hosts (22, 44, 46). Thus, the early initiation of an adequate antibiotic treatment is essential for a good outcome. However, an adequate therapy might be delayed, since the current first-line treatment for CNS infections relies on expanded-spectrum cephalosporins, which are inactive against L. monocytogenes (44, 46).
CSF cellular abnormalities are usually not characteristic, and diagnosis based on the detection of L. monocytogenes growth in cerebrospinal fluid (CSF) requires usually 24 to 72 h, with additional time needed for confirming the identification. L. monocytogenes culturing in the CSF, although specific, has an unacceptably low level of sensitivity because of a low number of bacteria within the CSF, low volumes of CSF, and/or previous treatment by an inadequate antibiotic regimen (35). Furthermore, encephalitis or brain abscesses may occur without any meningitis, suggesting that L. monocytogenes can cross the blood-brain barrier at the level of brain parenchyma (29, 35).
To enhance the diagnosis accuracy, a serodiagnosis assay based on the detection of serum antibodies directed against listeriolysin O (LLO) has been proposed (4, 16). However, it allows only a retrospective diagnosis for listeriosis and lacks specificity, since a false-positive result can be due to cross-reactions with hemolysins from other Gram-positive bacteria. In patients with severe immune deficiency, the serodiagnosis has somewhat lower sensitivity (4, 6, 16, 18).
Nucleic acid amplification testing by real-time PCR assay is a rapid diagnostic procedure, and it has been used successfully to diagnose a wide range of central nervous system infections (9, 14, 45). Several molecular methods have been developed in the food industry (12, 23, 26, 37, 43), based on the amplification of several specific genes of L. monocytogenes (iap, hly, prfA, actA) (27, 33, 36, 41). Some of these assays are sensitive enough to be valuable for the detection and quantification of L. monocytogenes in the environment and food products (23, 27, 33, 37, 41, 43). However, the results are highly dependent on the method used, the chosen target to be amplified, and the complexity of the food matrix product. Although rapid, specific, and sensitive detection of L. monocytogenes is important for medical diagnosis, only a few works have described the application of these tests for the diagnosis of CNS listeriosis in animals (25, 28, 40; K. A. Grant, N. M. Murphy, J. Hunter, I. Nwafor, and J. McLauchlin, presented at the XVIth International Symposium on Problems of Listeriosis, Savannah, GA, 20 to 23 March 2007) or in humans (25, 28; K. A. Grant et al., presented at the XVIth International Symposium on Problems of Listeriosis, Savannah, GA, 20 to 23 March 2007). Moreover, none of them have been subjected to prospective clinical evaluation for the diagnosis and follow-up of human listeriosis.
Our objective was first to design a highly specific and sensitive real-time PCR assay for the detection of L. monocytogenes by targeting the amplification of the hly gene (PCR-hly) and then to evaluate this PCR-hly assay for the detection of L. monocytogenes in CSF collected from patients suspected of having CNS listeriosis.
MATERIALS AND METHODS
Bacterial strains.
Reference bacteria and clinical isolates used in this study are listed in Table 1. All bacteria were grown under aerobic or anaerobic conditions, at 37°C, on the appropriate culture medium: brain heart infusion (BHI) (Becton Dickinson, Le Pont-de-Claix, France) agar (for Listeria spp.), blood agar (Streptococcus spp., Staphylococcus spp., Clostridium spp., Bacillus spp.), chocolate agar (Haemophilus influenzae, Neisseria meningitidis), and Lowenstein-Jensen (Mycobacterium tuberculosis) (bioMérieux, Marcy L'Étoile, France).
Table 1.
Bacterial strains tested by PCR-hly
| Bacteriuma | Referenceb | Result of PCR-hlyc |
|---|---|---|
| Listeriae | ||
| L. monocytogenes EGD-e 1/2a (LLO) | Sequenced strain EGD-e (20) | Positive |
| L. monocytogenes LO28 1/2c (LLO) | Sequenced strain L028 | Positive |
| L. monocytogenes 1/2a (LLO) | Serovar reference strain CLIP 74902 | Positive |
| L. monocytogenes 1/2b (LLO) | Serovar reference strain CLIP 74903 | Positive |
| L. monocytogenes 1/2c (LLO) | Serovar reference strain CLIP 74904 | Positive |
| L. monocytogenes 4b (LLO) | Serovar reference strain CLIP 74910 | Positive |
| L. monocytogenes 4b (LLO) | Clinical strain | Positive |
| L. ivanovii 5 (IVO) | Reference strain CIP 12229, CIP 7842 | Negative |
| L. seeligerii 1/2b (LSO) | Reference strain CIP 73021 | Negative |
| L. innocua 6a | Sequenced strain, CIP 11262 (20) | Negative |
| Gram-positive bacteria secreting a pore-forming cytolysin | ||
| Streptococcus pneumoniae (PLY) | Clinical strain | Negative |
| S. pyogenes (SLO) | Clinical strain | Negative |
| S. dysgalactiae subsp. dysgalactiae (SLO) | Clinical strain | Negative |
| S. dysgalactiae subsp. equisimilis (SLO) | Clinical strain | Negative |
| Clostridium perfringens (PFO) | Clinical strain | Negative |
| Bacillus cereus (CLY) | Clinical strain | Negative |
| Other bacterial strains frequently associated with community or nosocomial CNS infections | ||
| Neisseria meningitidis | Sequenced strain Z2491 | Negative |
| Streptococcus agalactiae (group B) | Sequenced strain Nem316 | Negative |
| Haemophilus influenzae (group B) | Clinical strain | Negative |
| Escherichia coli antigen K1 | Clinical strain | Negative |
| Staphylococcus aureus | Clinical strain | Negative |
| Staphylococcus epidermidis (Ica negative)d | CIP 68.21 | Negative |
| Staphylococcus epidermidis (Ica positive)d | CIP 105.777 | Negative |
| Pseudomonas aeruginosa | Clinical strain | Negative |
| Mycobacterium tuberculosis | Clinical strain | Negative |
When the bacterium secreted a thiol-activated pore-forming cytolysin, its abbreviation is indicated in parentheses: CLY, cereolysin; IVO, ivanolysin O; LLO, listeriolysin O; LSO, seeligerilysin O; PFO, perfringolysin; PLY, pneumolysin; SLO, streptolysin O.
CIP, Collection of Institut Pasteur; CLIP, Collection of Listeria Institut Pasteur.
Negative, hly gene was not detected after 45 CT.
Strains of S. epidermidis lacking (CIP 68.21) or carrying (CIP 105.777) the Ica operon, which is essential for biofilm production.
Real-time PCR for the hly gene (PCR-hly). (i) Probe and primer design for PCR-hly.
The target DNA consisted of a well-conserved region of the single gene hly encoding listeriolysin O, a thiol-activated pore-forming cytolysin (31). The primers were designed with the Primer Express Software, version 1.5 (Perkin-Elmer, Foster City, CA). The design of the primer was conducted after alignment of the sequences of various cytolysins published in GenBank with the hly gene of L. monocytogenes: the pneumolysin (PLY) of Streptococcus pneumoniae, the perfringolysin of Clostridium perfringens (PFO), the streptolysin of Streptococcus pyogenes (LSO), the ivanolysin O of Listeria ivanovii (IVO), and the seeligeriolysin O of Listeria seeligeri (LSO) (NCBI sequence navigator). The forward primer LmH.172F (5′-TT TCA TCC ATG GCA CCA CC-3′) and the reverse primer LmH.242R (5′-ATC CGC GTG TTT CTT TTC GA-3′) were used to amplify a 71-bp fragment. The amplicon was detected with a TaqMan internal oligonucleotide probe, LmH.199T (FAM-5′-CGC CTG CAA GTC CTA AGA CGC CA-3′-TAMRA), labeled with the reporter fluorescent dye 5-carboxyfluorescein (FAM) on the 5′ end and with a quencher molecule 6-carboxytetramethylrhodamine (TAMRA) covalently coupled on the 3′ end. Its hybridization temperature is 10°C below that of the primer pair. This oligonucleotide probe was unphosphorylated at the 3′ end to prevent probe elongation by the Taq DNA polymerase. The primer pairs and fluorescent probe were synthesized by Invitrogen (Cergy Pontoise, France).
(ii) DNA extraction for PCR.
Before the extraction of the bacterial DNA of each indicated strain (Table 1), the bacteria, grown overnight at 37°C in the appropriate culture broth (BHI) and blood culture bottles (BacT/Alert; bioMérieux), were collected, centrifuged at 5,000 × g for 10 min at 4°C, and then washed twice and resuspended in saline isotonic solution and frozen at −80°C in 1-ml aliquots. Titration of these aliquots was systematically performed by serial 10-fold dilutions on appropriate medium agar plates. These aliquots were thawed and diluted to obtain samples artificially infected at a calibrated inoculum (1 × 106 CFU/ml) for each indicated strain. Portions (200 μl) were used for DNA extraction by the QIAmp DNA minikit (catalog no. 51306; Qiagen S.A., Courtaboeuf, France) according to the manufacturer's recommendations. PCR-hly was then performed in triplicate for each strain. DNA was also extracted from CSF samples, previously stored at −20°C when necessary, by use of the MagNA Pure compact system (Roche Diagnostics).
(iii) Quantitative real-time PCR conditions.
PCRs were performed in a total volume of 50 μl containing 1× TaqMan universal PCR master mix (Applied Biosystems), 500 nM each primer, 100 nM probe, and 15 μl of the DNA extract. All of the amplification steps were carried out in a Thermocycler coupled to the ABI Prism 7700 sequence detector system (Applied Biosystems) with the following protocol: first heating (95°C for 10 min), followed by 45 amplification cycles, including denaturation (95°C for 15 s) and hybridization combined with elongation (60°C for 1 min). The DNA of an indicated amount of L. monocytogenes EGD-e was extracted and used to generate a standard curve. Quantitative results of the real-time PCR assay were expressed as a fractional cycle number, and then we determined the cycle threshold (CT) value, which marked the cycle when the fluorescence of a given sample significantly exceeded the baseline signal. In the absence of a signal beyond 45 cycles, the detection of the hly gene is considered negative. PCR inhibitors were systematically checked, using an external control of exogenous extracted DNA of Mycoplasma pneumoniae amplified under the same conditions. Negative and positive (L. monocytogenes DNA) controls were also processed in parallel in each series.
Study population: patients and CSF collection.
From January 2000 to December 2006, all of the CSF samples collected from patients hospitalized in our institution or other hospitals and suspected of having CNS listeriosis were analyzed by PCR-hly. DNA was extracted from at least 200 μl of the sampled CSF (as indicated above). CSF specimens were stored at −80°C before the PCR-hly was performed routinely once a week in our laboratory. In parallel, standard bacterial cultures of the CSF were performed by each laboratory on blood agar (bioMérieux) incubated 2 to 5 days under aerobic and anaerobic atmospheres and on chocolate agar (bioMérieux) incubated 2 to 5 days under a 5% CO2 atmosphere associated with an enrichment step in BHI broth (Difco). Bacterial cultures were done within 48 h and were subcultured under the same conditions for 2 to 5 days. Serology was performed for the detection of anti-LLO antibodies as a retrospective diagnosis (4). A definitive diagnosis of CNS listeriosis was retained when the patient had a neuromeningeal syndrome and/or a rhombencephalitis associated with (i) an L. monocytogenes isolate from CSF, (ii) an L. monocytogenes isolate from blood culture, and/or (iii) conversion of the anti-LLO serology when standard microbiological methods failed to isolate L. monocytogenes (4, 19).
Patients with clinically and microbiologically documented listeriosis but no sign of CNS infections (maternal-neonatal infection, isolated bacteremia, cutaneous focal infections) and from whom CSF was systematically collected were included in this study. Finally, normal CSF and those recovered for other known etiological types of meningitis (inflammatory, viral, bacterial, and nonlisterial) were used as control samples (for characteristics of each group, see Table S2 in the supplemental material).
Statistical analysis.
The differences in means of threshold cycle numbers were analyzed using a nonparametric Kruskal-Wallis test. P values of <0.05 were considered to be statistically significant. Statistical analyses were performed using Stata 9 (Statacorp, College Station, TX).
RESULTS
Development of real-time PCR-hly. (i) Sensitivity of the PCR-hly.
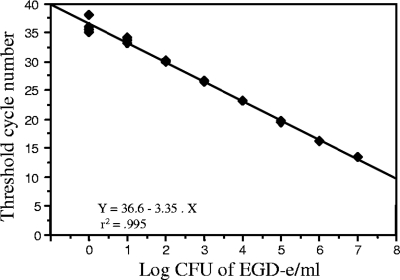
We first studied the sensitivity of the PCR-hly to detect an amount of pure DNA extracted from L. monocytogenes strain EGD-e with predetermined bacterial concentrations covering six logs ranging from 1 × 107 to 10 CFU/ml. As few as one copy per reaction could be detected between the 36th and 37th cycles. The calculated coefficients of correlation of the resulting standard curves of L. monocytogenes (r2 = 0.995) demonstrated the linearity of the quantification over a range of seven logs. Similar results were obtained using L. monocytogenes LO28 (serovar 1/2C) and a clinical strain (serovar 4b) (data not shown). PCR-hly performed in quadruplicate for the same extract is highly repeatable (Fig. 1). We further controlled the reproducibility of the quantitative PCR-hly on five aliquots of L. monocytogenes EGD-e suspension extracted independently and showed the same analytical sensitivities.
Fig. 1.
Result of real-time quantitative PCR-hly assay. Plot of the individual CT values from a four-time repetition assay against the input target quantity of L. monocytogenes EGD-e expressed in log10 CFU/ml (common log scale) showing the linearity of the quantification over a range of seven logs from dilutions of 1 × 100 to 1 × 107 CFU/ml. The computer calculated correlation coefficient is 0.995.
(ii) Specificity of PCR-hly.
The specificity of designed primers to amplify exclusively the hly gene of L. monocytogenes was tested in a variety of different Listeria species, including L. seeligerii, L. ivanovii, and L. innocua, and a large panel of other Gram-positive or Gram-negative bacteria, as shown in Table 1.
All of the L. monocytogenes species tested gave a positive amplification. In contrast, the PCR-hly failed to amplify any product of L. monocytogenes EGD-e with a deletion of the hly gene (EGD Δhly) (5), demonstrating that the primers are specifically directed to the hly gene (data not shown). We then tested the species in the Listeria genus that are able to secrete a hortolog for the hly gene: L. seeligerii and L. ivanovii, secreting seeligerolysin (LSO) and ivanolysin (ILO), respectively. The PCR-hly failed to amplify any product in these bacteria as it did with L. innocua, a nonhemolytic and nonvirulent species used as a negative control (Table 1). As expected, PCR-hly failed to amplify any PCR product with bacterial pathogens that also secrete a pore-forming cytolysin, such as perfringolysin of Clostridium perfringens (PFO), pneumolysin of Streptococcus pneumoniae (PLY), or streptolysin of Streptococcus pyogenes and S. dysgalactiae (SLO), or with bacterial pathogens from other genera usually encountered in meningitis (Table 1). In order to decipher whether the PCR-hly results are influenced by the biological abnormalities of CSF samples, we tested 55 clinical CSF specimens collected from patients hospitalized in our institution and for which the diagnosis was known (nonlisterial meningitis, including viral etiologies and systematic samples of noninfectious meningitis) (see Table S1 in the supplemental material). With all the tested samples, no PCR inhibition was detected and the PCR-hly amplification never gave a false-positive result.
Altogether, the results showed no cross-amplification between L. monocytogenes and other Listeria species or members of other bacterial genera when CSF was biologically abnormal. We thus clearly demonstrated that the designed PCR-hly is highly specific to amplifying the hly gene of L. monocytogenes virulent strains.
Prospective clinical evaluation. (i) Detection of L. monocytogenes in cerebrospinal fluid (CSF).
On the basis of these preliminary in vitro results, we aimed at prospectively evaluating the performance of the real-time quantitative PCR-hly carried out with DNA extracted from CSF samples collected prospectively over a 7-year period from patients suspected of having CNS listeriosis. We analyzed 313 CSF samples (corresponding to 304 patients) originating from Necker-Enfants Malades hospital (18%) and from other French hospitals (82%). Although negative after PCR amplification, 99 of the 313 CSF samples were not considered for the final evaluation of our PCR-hly assay, because they lacked at least one of the diagnostic criteria for CNS listeriosis (see Materials and Methods).
The median age and the male-to-female sex ratio of the population studied were 40 years (range, 1 day to 93 years) and 1.3, respectively. Nineteen percent were children (<15 years), with a median age of 2.7 years (range, 0 to 13.4 years), and 81% were adults, with a median age of 48 years (range, 16.1 to 93.0 years). Among the 205 patients included in the study (i.e., 214 eligible CSF samples), we identified 30 cases of listeriosis for which we analyzed 39 CSF: in six cases, CSF was collected twice, and in one case it was collected four times for controlling infection under antibiotic treatment.
Clinical presentations are presented in Table 2: (i) CNS listeriosis (n = 24) and (ii) non-CNS listeriosis, including maternal-neonatal infections (n = 3), isolated bacteremia with sepsis (n = 2), and cutaneous focal infection (n = 1). For all of the other patients (n = 175), the diagnosis of listeriosis was ruled out according to the criteria previously defined or because other bacterial etiologies were finally identified (n = 13) (Table 2).
Table 2.
Standard microbiological test and PCR-hly results for the first collected CSF sample
| PCR-hly result | No. with result for sample typea |
||||
|---|---|---|---|---|---|
| Listeriosis |
Non-Listeria meningitisb (n = 175) |
||||
| CNS infection (n = 24) |
Non-CNS infection (n = 6)c | ||||
| Cult+ | Cult− | Cult+ | Cult− | ||
| Positive | 9 | 5 | 0 | 0 | 0 |
| Negative | 0 | 10 | 6 | 13d | 162 |
Shown are the numbers of results according to clinical presentation. Cult+ or Cult−, positive or negative bacterial culture of the first collected CSF sample, respectively.
CNS listeriosis was ruled out according to criteria defined in Materials and Methods.
All samples from non-CNS infection were Cult−.
Bacteria (no. of bacteria) isolated in non-Listeria meningitis: Streptococcus agalactiae (3), Escherichia coli (2), Cryptococcus neoformans (2), Staphylococcus epidermidis (2), Steptococcus pneumoniae (2), Neisseria meningitidis (1), and Streptococcus pyogenes (1).
CNS infections were divided into two distinct groups according to the characteristics of the CSF sent to our laboratory for analysis, notably the delay between the time when the CSF was collected and the onset of the disease. We observed 14 cases (12 adults and 2 neonates) for which the CSF was early collected within the first two days and 10 cases for which the CSF was collected between 5 and 10 days after the onset of the disease. In the 10 latter cases, all patients were already under active antibiotic treatment; however, clinical presentation, cellular and biochemical disorders of the CSF, and LLO serology conversion were strongly suggestive for the diagnosis of CNS listeriosis. Thus, the diagnosis of CNS listeriosis was retained although L. monocytogenes could not be isolated from the CSF collected or from other clinical specimens.
(ii) Result of standard microbiological tests performed on CSF.
Standard bacterial culture of the first collected CSF yielded a L. monocytogenes isolate in nine cases (64%) (Table 2). Among these nine cases, three were positive only after using the enrichment method (patients 1, 6, and 9; see Table S2 in the supplemental material).
Nine additional CSF samples (corresponding to 7 patients) were collected several days after the start of antibiotic therapy for controlling treatment efficacy. Among these nine CSF samples, only two specimens from patient 5 (see Table S2 in the supplemental material) were positive in culture after using the enrichment method.
Direct examination of CSF retrieved evocative Gram-positive bacilli in only four cases. All of these cases corresponded to positive culture. This highlights the low number of bacteria present in the CSF specimens, possibly due to an antibiotic treatment started before CSF collection.
(iii) Result of quantitative PCR-hly.
PCR-hly did not provide a false-negative result in any of the analyzed cases when L. monocytogenes was isolated by standard culture of CSF. There was no positive PCR-hly result for patients with listeriosis without CNS involvement (maternal-neonatal infections, bacteremia, and cutaneous focal infection). PCR-hly remained negative in all the cases for which the diagnosis of listeriosis was definitely excluded (Table 2).
However, PCR-hly was positive in five additional CSF samples for which L. monocytogenes was not retrieved by culture even after enrichment technique (patients 2, 3, 7, 8, and 14; see Table S2 in the supplemental material), thus completing the diagnosis of CNS listeriosis. In all cases, discrepancies between culture and PCR could be explained by the administration of antibiotics started between 1 and 5 days before CSF collection.
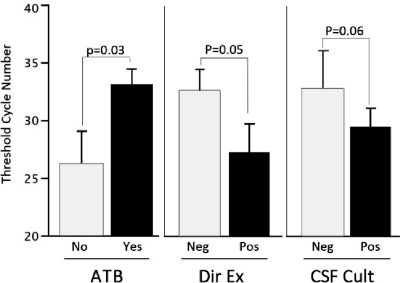
The analysis of the quantitative results of PCR-hly showed the presence of a significantly smaller amount of L. monocytogenes DNA in cases with previous administration of effective antibiotics (see Table S2 in the supplemental material). Similarly, the low bacterial inoculum in the CSF, evidenced by the small amount of bacterial DNA, made the standard bacterial techniques, like culture and the direct examination for observation of specific Gram-positive bacilli, less sensitive (Fig. 2).
Fig. 2.
Comparison of the quantitative results of PCR-hly according to the previous antibiotic regimen and the result of direct examination and culture performed on CSF. ATB, previous antibiotics were given (Yes) or not given (No) before CSF samples were taken. Dir Ex, direct examination of CSF retrieved (Pos) or did not retrieve (Neg) Gram-positive rods evocative of Listeria monocytogenes. CSF cult, standard culture of CSF yielded (Pos) or did not yield (Neg) Listeria monocytogenes, with or without after an enrichment step. The difference in means of threshold cycle numbers was analyzed using a Kruskal-Wallis test. P values of <0.05 were considered to be statistically significant.
Concerning CSF collected for control under antibiotic treatment, PCR-hly was positive for four specimens, including the two positive CSF in culture (collected 2 and 5 days after effective antibiotic treatment was initiated). In all cases, the CT values were lower than those obtained for the first collected CSF specimen.
DISCUSSION
The development of a molecular test for the detection of L. monocytogenes in clinical specimens was motivated by the need for a rapid and reliable test for the diagnosis of CNS listeriosis, complementary to the culture. Indeed, the rapid detection and identification of causative pathogens in bacterial meningitis are critical for a rapid adaptation of antibiotic treatment (7). Moreover, this test should be highly sensitive, because CNS listeriosis is often linked to the presence of a low number of bacteria in CSF.
The hly gene, part of the genome of L. monocytogenes, was chosen as a target for detection and quantification by real-time PCR of L. monocytogenes infection. It is a 1,590-bp single-copy gene coding for the thiol-activated pore-forming cytolysin listeriolysin O (LLO), a major and specific virulent factor for L. monocytogenes which is necessary for the invasiveness of the bacteria (31). Thus, the hly gene appeared to be a reliable and highly conserved (about 99%) candidate target, and it is already used for specific detection of L. monocytogenes in food products (23, 36, 37; K. A. Grant et al., presented at the XVIth International Symposium on Problems of Listeriosis, Savannah, GA, 20 to 23 March 2007). However, various sequences of the hly gene have been tested, leading to various results in terms of specificity (23, 36–38; K. A. Grant et al., presented at the XVIth International Symposium on Problems of Listeriosis, Savannah, GA, 20 to 23 March 2007). Despite important similarities between the hly gene and genes encoding proteins belonging to the same pore-forming cytolysin family, especially ivanolysin O (77% identity) (1, 31), the targeted region was found to be specific for L. monocytogenes. Moreover, since the PCR-hly does not cross with other bacteria usually responsible for meningitis, this test could be included in the scheme of multiplex PCR for identifying the major etiologies of bacterial meningitis (7, 9, 45).
Some teams have described the use of amplification of the universal bacterial 16S rRNA gene for detecting L. monocytogenes in clinical samples (3, 8, 25, 39, 42). However, the broad-range PCR 16S rRNA gene presents some drawbacks, like less sensitivity, and the requirement for sequencing the amplicons provides a delaying response that does not match our objective of rapid etiological diagnosis (10, 32, 48). In order to enhance the rapidity of the diagnosis, we preferred real-time PCR, which appears to be highly sensitive, as the theoretical threshold detection was as low as 1 gene copy/ml (7, 9, 13, 14, 45). Turnaround time of this real-time PCR-hly is 2 h; thus, the expected time to result of this test, when performed on demand, could be less than 24 h. Rapid and accurate results are helpful for rapid adaption of the empirical antibiotic treatment, usually based on a high dose of expanded-spectrum cephalosporins inactive against L. monocytogenes or on adding or removing ampicillin (44).
Nevertheless, technical improvements should enhance the efficacy of DNA extraction and avoid inhibitors in order to optimize molecular detection of the hly gene in other clinical samples, like blood, placenta, and feces, but also in environmental and food samples (2, 30). The sensitivity of PCR-hly could be further improved by targeting genes with multiple copies in the L. monocytogenes genome.
According to the results of the prospective clinical evaluation, PCR-hly appears to be more sensitive than standard bacterial culture and direct examination. PCR-hly rescues the diagnosis of CNS listeriosis when culture remains sterile or the isolation of the bacterium has been delayed because of the need of an enrichment step. It should also be emphasized that discrepancies between culture and PCR corresponded to unusual clinical presentations like ventriculoperitoneal shunt infection (see patient 5 in Table S2 in the supplemental material) (32a), exclusive encephalitis presentation (patient 8, Table S2), or previous administration of antibiotics active against L. monocytogenes (patients 2, 4, 5, 7, 8, and 14, Table S2), as already described by Fayol et al. (15) (patient 14, Table S2). In all of these cases, the quantitative value of PCR-hly confirmed the low number of bacteria present in the CSF specimens.
Moreover, in the case of patient 5, the results of quantitative PCR-hly were used for demonstrating the persistence of infection from a medical device despite effective antibiotic therapy. Our results suggest a quantitative indication of the PCR-hly for monitoring treatment efficacy (32a).
For the 10 probable cases not microbiologically documented, the anamnesis and LLO serology conversion reported are not definitive evidence of evolutive CNS listeriosis despite evocative CSF anomalies. Moreover, the lack of sensitivity of both culture and PCR-hly is reinforced when CSF specimens are sampled late after the onset under effective antibiotic treatment. However, in at least two cases (patients 26 and 30; see Table S2 in the supplemental material), PCR-hly failed to confirm the diagnosis despite a high suspicion of CNS listeriosis based on the clinical presentation, the radiological findings evocative of a bacterial abscess localized in the rhombencephalic area by magnetic resonance imaging (MRI), the LLO serology conversion, and the favorable evolution under antibiotic treatment active against L. monocytogenes.The exclusive encephalitis presentation without meningitis constitutes a potential source of false-negative results. Indeed, the bacterium could be located exclusively in the brain parenchyma with little or no bacterial release in the CSF at early times (34, 35). In these cases, culture is, however, no more contributive. LLO serology conversion appears as an interesting alternative but only for retrospective diagnosis when culture and PCR remain noncontributive (4, 16, 18).
The analysis of other evolutive clinical presentations of listeriosis highlights the fact that L. monocytogenes is not systematically linked to CNS infection, although some physicians systematically include the search for possible subclinical meningitis in the investigation and management of such severe infections.
Until now, the definitive diagnosis of listeriosis has been exclusively based on the isolation of L. monocytogenes in clinical samples. Cases of listeriosis diagnosed by the detection of L. monocytogenes DNA are currently not included in the data on epidemiological surveillance because of the lack of prospective evaluation of molecular assays.
Although the trend shows an increasing number of cases of listeriosis in France and throughout the world (21), listeriosis remains a rare disease. During the period studied, between 200 and 250 cases of listeriosis were diagnosed yearly in France (i.e., an incidence of 3 to 4 cases per million inhabitants). Less than one-third of these cases consisted of CNS listeriosis, in adults or more rarely in newborns (data from the annual report of the French National Reference Centre for Listeria). Although it is the second most frequent cause of bacterial meningoencephalitis in France, behind Mycobacterium tuberculosis (34), meningoencephalitis due to L. monocytogenes is an even rarer clinical presentation. Only 12 cases were diagnosed in 2007 during a 1-year study collecting encephalitis cases in France (34). Thus, national recruitment of cases was an absolute requirement to conduct such studies but led inevitably to several missing data and inconclusive cases because of a loss of information. Despite some problems in methodology, this study is the first prospective evaluation of the diagnostic value of a molecular assay for the diagnosis of CNS listeriosis.
We are aware that molecular diagnostic testing must not replace culture, the only technique that allows antimicrobial susceptibility testing and epidemiological and microbiological surveillance of emerging strains, including investigation of clusters of cases (17, 21). However, altogether our results suggest that the definition of listeriosis should include cases diagnosed by molecular tests, as with other infectious diseases (7, 11, 14, 47, 48).
In conclusion, we have demonstrated that as a routine test, real-time PCR-hly provides rapid and reliable information allowing antibiotic therapy to be quickly adapted. This study also showed, in our studied population, that Listeria culture can be insensitive when the patient has been previously treated with antibiotics, whereas PCR is still highly sensitive. In our institution, we perform PCR-hly as a complementary test to standard culture for rapid or retrospective diagnosis of Listeria meningoencephalitis. We also used the quantitative results for the follow-up of a patient receiving antibiotic treatment until CSF sterilization.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all colleagues who sent CSF samples and clinical data.
This work was supported by the Institut Pasteur, the Institut de Veille Sanitaire, and the Assistance Publique Hôpitaux de Paris (APHP).
Footnotes
Supplemental material for this article may be found at http://jcm.asm.org/.
Published ahead of print on 14 September 2011.
REFERENCES
- 1. Alouf J. E. 2000. Cholesterol-binding cytolytic protein toxins. Int. J. Med. Microbiol. 290:351–356 [DOI] [PubMed] [Google Scholar]
- 2. Al-Soud W. A., Radstrom P. 2001. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 39:485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Backman A., Lantz P., Radstrom P., Olcen P. 1999. Evaluation of an extended diagnostic PCR assay for detection and verification of the common causes of bacterial meningitis in CSF and other biological samples. Mol. Cell. Probes 13:49–60 [DOI] [PubMed] [Google Scholar]
- 4. Berche P., et al. 1990. Detection of anti-listeriolysin O for serodiagnosis of human listeriosis. Lancet 335:624–627 [DOI] [PubMed] [Google Scholar]
- 5. Chakraborty T., et al. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chatzipanagiotou S., Hof H. 1988. Sera from patients with high titers of antibody to streptolysin O react with listeriolysin. J. Clin. Microbiol. 26:1066–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chiba N., et al. 2009. Rapid detection of eight causative pathogens for the diagnosis of bacterial meningitis by real-time PCR. J. Infect. Chemother. 15:92–98 [DOI] [PubMed] [Google Scholar]
- 8. Clarridge J. E., III 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin. Microbiol. Rev. 17:840–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Corless C. E., et al. 2001. Simultaneous detection of Neisseria meningitidis, Haemophilus influenzae, and Streptococcus pneumoniae in suspected cases of meningitis and septicemia using real-time PCR. J. Clin. Microbiol. 39:1553–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Corless C. E., et al. 2000. Contamination and sensitivity issues with a real-time universal 16S rRNA PCR. J. Clin. Microbiol. 38:1747–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Costa J. M., et al. 2000. Real-time PCR for diagnosis and follow-up of Toxoplasma reactivation after allogeneic stem cell transplantation using fluorescence resonance energy transfer hybridization probes. J. Clin. Microbiol. 38:2929–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Agostino M., et al. 2004. A validated PCR-based method to detect Listeria monocytogenes using raw milk as a food model—towards an international standard. J. Food Prot. 67:1646–1655 [DOI] [PubMed] [Google Scholar]
- 13. Doumith M., Buchrieser C., Glaser P., Jacquet C., Martin P. 2004. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 42:3819–3822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Espy M. J., et al. 2006. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin. Microbiol. Rev. 19:165–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fayol L., Beizig S., Le Monnier A., Lacroze V., Simeoni U. 2009. Neonatal meningitis due to Listeria monocytogenes after 3 weeks of maternal treatment during pregnancy. Arch. Pediatr. 16:353–356(In French.) [DOI] [PubMed] [Google Scholar]
- 16. Gaillard J. L., et al. 1992. Serological evidence for culture-negative listeriosis of central nervous system. Lancet 340:560. [DOI] [PubMed] [Google Scholar]
- 17. Gerner-Smidt P. K., et al. 2006. PulseNet USA: a five-year update. Foodborne Pathog. Dis. 3:9–19 [DOI] [PubMed] [Google Scholar]
- 18. Gholizadeh Y., Juvin M., Beretti J. L., Berche P., Gaillard J. L. 1997. Culture-negative listeriosis of the central nervous system diagnosed by detection of antibodies to listeriolysin O. Eur. J. Clin. Microbiol. Infect. Dis. 16:176–178 [DOI] [PubMed] [Google Scholar]
- 19. Gholizadeh Y., et al. 1996. Serodiagnosis of listeriosis based upon detection of antibodies against recombinant truncated forms of listeriolysin O. J. Clin. Microbiol. 34:1391–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Glaser P., et al. 2001. Comparative genomics of Listeria species. Science 294:849–852 [DOI] [PubMed] [Google Scholar]
- 21. Goulet V., Hedberg C., Le Monnier A., de Valk H. 2008. Increasing incidence of listeriosis in France and other European countries. Emerg. Infect. Dis. 14:734–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Goulet V., Marchetti P. 1996. Listeriosis in 225 non-pregnant patients in 1992: clinical aspects and outcome in relation to predisposing conditions. Scand. J. Infect. Dis. 28:367–374 [DOI] [PubMed] [Google Scholar]
- 23. Gouws P. A., Lidedemann I. 2005. Evalution of diagnostic PCR for the detection of Listeria monocytogenes in food products. Food Technol. Biotechnol. 43:201–205 [Google Scholar]
- 24. Reference deleted.
- 25. Greisen K., Loeffelholz M., Purohit A., Leong D. 1994. PCR primers and probes for the 16S rRNA gene of most species of pathogenic bacteria, including bacteria found in cerebrospinal fluid. J. Clin. Microbiol. 32:335–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guilbaud M., et al. 2005. Quantitative detection of Listeria monocytogenes in biofilms by real-time PCR. Appl. Environ. Microbiol. 71:2190–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hein I., et al. 2001. Detection and quantification of the iap gene of Listeria monocytogenes and Listeria innocua by a new real-time quantitative PCR assay. Res. Microbiol. 152:37–46 [DOI] [PubMed] [Google Scholar]
- 28. Jaton K., Sahli R., Bille J. 1992. Development of polymerase chain reaction assays for detection of Listeria monocytogenes in clinical cerebrospinal fluid samples. J. Clin. Microbiol. 30:1931–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Join-Lambert O. F., et al. 2005. Listeria monocytogenes-infected bone marrow myeloid cells promote bacterial invasion of the central nervous system. Cell Microbiol. 7:167–180 [DOI] [PubMed] [Google Scholar]
- 30. Jordan J. A., Durso M. B. 2005. Real-time polymerase chain reaction for detecting bacterial DNA directly from blood of neonates being evaluated for sepsis. J. Mol. Diagn. 7:575–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kayal S., Charbit A. 2006. Listeriolysin O: a key protein of Listeria monocytogenes with multiple functions. FEMS Microbiol. Rev. 30:514–529 [DOI] [PubMed] [Google Scholar]
- 32. Kotilainen P., et al. 1998. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J. Clin. Microbiol. 36:2205–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a. Le Monnier A., Blanot S., Abachin E., Beretti J.-L., Berche P., Kayal S. 2011. Listeria monocytogences: a rare complication of ventriculoperitoncal shunt in children. J. Clin Microbiol. 49:3924–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Longhi C., et al. 2003. Detection of Listeria monocytogenes in Italian-style soft cheeses. J. Appl. Microbiol. 94:879–885 [DOI] [PubMed] [Google Scholar]
- 34. Mailles A., Stahl J. P., Brouard C., Vaillant V. 2008. Epidemiology, improvement of etiological diagnosis, and outcome of infectious encephalitis in France in 2007: preliminary results of a national prospective study. Med. Mal. Infect. 38(Suppl. 2):S37–S38(In French.) [DOI] [PubMed] [Google Scholar]
- 35. Mylonakis E., Hohmann E. L., Calderwood S. B. 1998. Central nervous system infection with Listeria monocytogenes. 33 years' experience at a general hospital and review of 776 episodes from the literature. Medicine 77:313–336 [DOI] [PubMed] [Google Scholar]
- 36. Nogva H. K., Rudi K., Naterstad K., Holck A., Lillehaug D. 2000. Application of 5′-nuclease PCR for quantitative detection of Listeria monocytogenes in pure cultures, water, skim milk, and unpasteurized whole milk. Appl. Environ. Microbiol. 66:4266–4271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Norton D. M. 2002. Polymerase chain reaction-based methods for detection of Listeria monocytogenes: toward real-time screening for food and environmental samples. J. AOAC Int. 85:505–515 [PubMed] [Google Scholar]
- 38. Norton D. M., Batt C. A. 1999. Detection of viable Listeria monocytogenes with a 5′ nuclease PCR assay. Appl. Environ. Microbiol. 65:2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pandit L., Kumar S., Karunasagar I., Karunasagar I. 2005. Diagnosis of partially treated culture-negative bacterial meningitis using 16S rRNA universal primers and restriction endonuclease digestion. J. Med. Microbiol. 54:539–542 [DOI] [PubMed] [Google Scholar]
- 40. Peters M., Pohlenz J., Jaton K., Ninet B., Bille J. 1995. Studies of the detection of Listeria monocytogenes by culture and PCR in cerebrospinal fluid samples from ruminants with listeric encephalitis. Zentralbl. Veterinarmed. B 42:84–88 [DOI] [PubMed] [Google Scholar]
- 41. Rossmanith P., Krassnig M., Wagner M., Hein I. 2006. Detection of Listeria monocytogenes in food using a combined enrichment/real-time PCR method targeting the prfA gene. Res. Microbiol. 157:763–771 [DOI] [PubMed] [Google Scholar]
- 42. Rothman R., et al. 2010. Use of quantitative broad-based polymerase chain reaction for detection and identification of common bacterial pathogens in cerebrospinal fluid. Acad. Emerg. Med. 17:741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Somer L., Kashi Y. 2003. A PCR method based on 16S rRNA sequence for simultaneous detection of the genus Listeria and the species Listeria monocytogenes in food products. J. Food Prot. 66:1658–1665 [DOI] [PubMed] [Google Scholar]
- 44. Stahl J. P. 2009. Treatment of community acquired bacterial meningitis, after microbiological identification. Med. Mal. Infect. 39:513–520 (In French.) [DOI] [PubMed] [Google Scholar]
- 45. Taha M. K., Olcen P. 2004. Molecular genetic methods in diagnosis and direct characterization of acute bacterial central nervous system infections. APMIS 112:753–770 [DOI] [PubMed] [Google Scholar]
- 46. Troxler R., von Graevenitz A., Funke G., Wiedemann B., Stock I. 2000. Natural antibiotic susceptibility of Listeria species: L. grayi, L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri and L. welshimeri strains. Clin. Microbiol. Infect. 6:525–535 [DOI] [PubMed] [Google Scholar]
- 47. van Haeften R., et al. 2003. A quantitative LightCycler PCR to detect Streptococcus pneumoniae in blood and CSF. Diagn. Microbiol. Infect. Dis. 47:407–414 [DOI] [PubMed] [Google Scholar]
- 48. Welinder-Olsson C., et al. 2007. Comparison of broad-range bacterial PCR and culture of cerebrospinal fluid for diagnosis of community-acquired bacterial meningitis. Clin. Microbiol. Infect. 13:879–886 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.