Abstract
Bacteria of the genus Dehalococcoides are important members of bioremediation communities because of their ability to detoxify chloroethenes to the benign end product ethene. Genome-enabled studies conducted with Dehalococcoides ethenogenes 195 have revealed that two ATP-binding cassette (ABC)-type amino acid transporters are expressed during its exponential growth stages. In light of previous findings that Casamino Acids enhanced its dechlorination activity, we hypothesized that strain 195 is capable of importing amino acids from its environment to facilitate dechlorination and growth. To test this hypothesis, we applied isotopomer-based dilution analysis with 13C-labeled acetate to differentiate the amino acids that were taken up by strain 195 from those synthesized de novo and to determine the physiological changes caused by the significantly incorporated amino acids. Our results showed that glutamate/glutamine and aspartate/asparagine were almost exclusively synthesized by strain 195, even when provided in excess in the medium. In contrast, phenylalanine, isoleucine, leucine, and methionine were identified as the four most highly incorporated amino acids, at levels >30% of respective proteinogenic amino acids. When either phenylalanine or all four highly incorporated amino acids were added to the defined mineral medium, the growth rates, dechlorination activities, and yields of strain 195 were enhanced to levels similar to those observed with supplementation with 20 amino acids. However, genes for the putative ABC-type amino acids transporters and phenylalanine biosynthesis exhibited insignificant regulation in response to the imported amino acids. This study also demonstrates that using isotopomer-based metabolite analysis can be an efficient strategy for optimizing nutritional conditions for slow-growing microorganisms.
INTRODUCTION
Bacteria of the genus Dehalococcoides play a key role in bioremediation of chlorinated ethenes due to their ability to completely transform these toxic groundwater contaminants into benign ethene (4, 10, 24). However, bioremediation applications are limited by several physiological characteristics of Dehalococcoides that make these bacteria challenging to stimulate in the environment or to cultivate in the laboratory, including long doubling times (0.8 to 2.4 days), low cell yields (0.24 to 4.9 g of protein/mol of Cl− released) (8), and specific requirements for a variety of exogenously supplied compounds (e.g., hydrogen as an electron donor, acetate as a carbon source, and cobalamin as a cofactor) (9, 24, 35). Further, it has been reported that generating large quantities of active Dehalococcoides in a timely manner can represent a significant challenge (36, 41). Recent genome sequencing and annotation of several Dehalococcoides isolates have enabled novel molecular and biochemical approaches to improve our understanding of their physiology (16, 17, 20, 39), which in turn can lead to the identification of biological mechanisms that limit bioremediation applications using Dehalococcoides species.
Genome annotations of four sequenced Dehalococcoides strains reveal that each strain possesses nine genes encoding two putative ATP binding cassette (ABC)-type amino acid uptake carriers (19, 25, 35) (www.img.jgi.doe.gov) (see Table S1 in the supplemental material). For strain 195, one of the transporters encoded by six genes (DET0938, DET0941 to DET0945), is annotated as a high-affinity branched-chain amino acid transporter (35), which might be expressed to uptake leucine, isoleucine, and valine across the cytoplasmic membrane of cells. The other transporter encoded by three genes (DET0417 to DET0419) is likely a polar amino acid transporter (35). However, the putative substrates of this transporter are still indeterminate, in that each subunit of the DET0417 to DET0419 transporter seemingly has various substrates (see Table S1 in the supplemental material), and functional annotation based on sequence is often inexact. Nevertheless, a previous transcriptomics study with strain 195 demonstrated the expression of these genes during exponential growth stages (16), suggesting a potential role in the uptake of exogenous amino acids. Maymo-Gatell et al. (24) reported that the addition of Casamino Acids (a complex acid hydrolysate of casein including most amino acids except tryptophan [27]) resulted in the enhancement of strain 195 dechlorination. A recent paper reported that, in contrast to the initial sequence annotation of strain 195, this strain synthesizes all 20 common amino acids for biomass protein production with acetate and CO2 as carbon sources (39). Taken together, previous studies raised two questions about the utilization of exogenous amino acids by Dehalococcoides spp.: (i) whether the presence and expression of the ABC transporters lead to the phenotypic function of importing amino acids and (ii) whether any of the imported amino acids plays an important role in facilitating robust and sustainable Dehalococcoides growth.
Interestingly, a very recent study using a constraint-based model demonstrated an in silico enhancement of biomass yield of Dehalococcoides as a result of unlimited uptake of exogenous amino acids (15). The extensive computational model was largely based on the genome sequences and associated annotations of Dehalococcoides spp., which may or may not accurately reflect the specific phenotypic functionalities related to the import and incorporation of amino acids. The study described here used isotopomer-based metabolite analysis to quantitatively differentiate the amino acids that were preferentially incorporated from those that were de novo synthesized by D. ethenogenes strain 195. Further, physiological changes of strain 195 caused by the imported amino acids were quantified. Reverse transcription-quantitative PCR (RT-qPCR) was used to assess whether the genes involved in biosynthesis and transport of the favorable amino acids were differentially expressed in response to exogenous amino acid amendment.
MATERIALS AND METHODS
Chemicals.
Trichloroethene (TCE), cis-dichloroethene (cDCE), vinyl chloride (VC), and ethene were purchased from Fisher Scientific Co. (Pittsburgh, PA), Acros Chemical Co. (Pittsburgh, PA), Fluka Chemical Co. (Ronkonkoma, NY), and Alltech Associate, Inc. (Deerfield, IL), respectively. [U-13C]glucose (fully labeled, >98% purity) and [1-13C]sodium acetate (>99% purity) were purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA). The 20 amino acids were purchased from Sigma-Aldrich, Inc. (St. Louis, MO). All gases, including medical air, nitrogen, hydrogen, and hydrogen-CO2 mixture, were obtained from Praxair Inc (Oakland, CA).
Bacterial strain and culture conditions.
D. ethenogenes strain 195 was grown in batch cultures at 34°C in 160-ml glass serum bottles containing 100 ml of defined mineral salt medium and a H2/CO2 headspace (80/20 [vol/vol]) as described previously (39). The medium was amended with a modified Wolin vitamin solution containing a 50-μg/liter final B12 concentration (9). Sodium acetate (2 mM) and liquid TCE (7 μl) were added to the culture medium as the carbon source and electron acceptor, respectively. To identify the amino acids that strain 195 took up from the medium, tracer experiments were conducted using [1-13C]sodium acetate as the carbon source, with unlabeled 20 common amino acids (20 mg/liter each) as supplements. In order to observe amino acid uptake by fully acclimatized cultures and reduce the effect of unlabeled carbon from the inoculum (3%), strain 195 was subcultured five times in the same experimental conditions before being harvested for isotopomer-based metabolite analysis. Because of the presence of at least one uptake system that was found or predicted for each amino acid (23), Escherichia coli K-12 MG1655 was used as a positive control in order to verify the analytical methods. Strain MG1655 was cultured aerobically with [U-13C]glucose (2 g/liter) in M63 minimal medium (34), with or without amendment of 20 amino acids (50 mg/liter each). Strain MG1655 was grown from a 0.5% inoculum and harvested in the late exponential growth phase (optical density at 600 nm = 1).
In order to reveal the long-term growth effects of amino acid amendments, strain 195 was grown with different amino acid amendments over five subcultures (with similar initial cell densities) prior to quantifying dechlorination and growth rates for comparison. Four cultivation conditions were compared: AA20, amended with all 20 common amino acids (20 mg of each/liter); AA4, amended with phenylalanine, leucine, isoleucine, and methionine (20 mg of each/liter); AA1, amended with 20 mg of phenylalanine/liter; and AA0, an unamended culture. All subcultures were inoculated with similar initial cell densities in order to avoid the effect of various initial cell concentrations.
Analytical methods.
For isotopomer-based metabolite analysis, approximately 2 liters of liquid culture (∼7.7 × 107 cells/ml) from 20 individual 125-ml bottles were aseptically harvested by centrifugation (22,000 × g, 4°C, 15 min) for each experimental condition. Cell pellets were washed with sterile MilliQ water three times and stored at −80°C before use. The preparation and isotopomeric analysis of proteinogenic amino acids were performed as previously described (31, 38). In brief, biomass was hydrolyzed in 6 M HCl at 100°C for 24 h. During the hydrolysis, tryptophan and cysteine are degraded, while asparagine and glutamine are converted into aspartate and glutamate, respectively. The amino acid solution was dried under air flush overnight. Amino acid samples were derivatized in tetrahydrofuran (THF) and N-(tert-butyl dimethylsilyl)-N-methyl-trifluoroacetamide (Sigma-Aldrich) at 70°C for 1 h. A gas chromatograph (Hewlett-Packard, model 6890; Agilent Technologies, Palo Alto, CA) equipped with a DB5-MS column (J&W Scientific, Folsom, CA) and a mass spectrometer (model 5975; Agilent Technologies) was used for isotopomer analysis. Two types of charged fragments were clearly detected by gas chromatography-mass spectrometry for the derivatized amino acids: [M-57]+, which contains the entire amino acid, and [M-159]+, which contains the amino acid without the first carbon (α carboxyl group) (39). The [M-57]+ peaks in leucine, isoleucine, and proline overlap with other peaks, so the [M-159]+ group was used to obtain the isotopomer labeling information of those amino acids. The final isotopomer labeling fractions were indicated as follows: M0 (unlabeled fraction), M1 (singly labeled fraction), M2 (fraction with two labeled carbons), M3 (fraction with labeled three carbons), and so forth.
For determining the compositions of the proteinogenic amino acids of strain 195, approximately 100 mg (wet weight) of cell pellets (n = 2) was harvested from different amino acid amendment conditions by centrifugation as specified above for isotopomer-based metabolite analysis. The analyses were performed by the Molecular Structure Facility, University of California, Davis (http://msf.ucdavis.edu/), using a Beckman 6300 amino acid analyzer (Beckman Coulter, Inc., Fullerton, CA).
Ethenes in culture headspace were measured using a gas chromatograph (Hewlett-Packard, model 5890; Agilent Technologies) with a 30-m J&W capillary column and a flame ionization detector as described previously (9, 38).
Quantification of cell numbers with different amino acid amendments.
To compare the growth rates of strain 195 under different amino acid amended conditions, 2-ml cultures were periodically sampled from three replicate culture bottles for DNA extraction. Cells were pelleted by centrifugation (14,000 rpm and 15 min at 4°C) and stored at −20°C until extraction. Genomic DNA (gDNA) was isolated by using a DNeasy Blood & Tissue kit (Qiagen, Inc., Valencia, CA) according to the manufacturer's instructions. Since each strain 195 cell contains only one copy of the tceA gene (35), cell numbers were represented as tceA gene copies per ml of culture medium. tceA gene copies were quantified from triplicate qPCRs performed on each of the triplicate DNA samples by using a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA). Briefly, each qPCR mixture contained 2 μl of sample gDNA or a 5-fold serially diluted standard, 1× Fast SYBR green Master Mix (Applied Biosystems), and 0.5 μM concentrations of each forward and reverse primer (see Table S2 in the supplemental material). Standard curves for qPCR analysis of tceA genes were generated from known concentrations of strain 195 gDNA with linear regression R2 values of 99.9% and amplification efficiencies of 99.5 to 105.3%.
RNA extraction, reverse transcription, and qPCR.
When ca. 67% of added TCE was dechlorinated, 100 ml of active strain 195 cells (mid-exponential-phase cultures) were collected for RNA extraction from each of triplicate culture bottles representing AA0, AA4, and AA20. Cell samples were collected by filtration through sterile 0.2-μm-pore-size membrane filters, flash-frozen in liquid nitrogen, and stored at −80°C for up to 2 weeks before RNA extraction (16). Total RNA was prepared from frozen cells using an RNeasy minikit (Qiagen) according to the manufacturer's directions. Contaminating DNA in RNA samples was removed with three consecutive rigorous DNase I treatments using a DNA-free kit (Ambion, Austin, TX) according to the manufacturer's directions. Purified RNA was stored at −80°C prior to further use. RNA integrity was checked on a 2100 Bioanalyzer RNA 6000 Nano LabChip (Agilent Technologies), and RNA quantification was determined using a Ribogreen assay on a NanoDrop 3300 Fluorospectrometer (Thermo Scientific, Wilmington, DE). The total mass of purified RNA extracted from the 100-ml cultures ranged from 1.7 to 3.4 μg. cDNA was synthesized in 40-μl reaction mixes containing 50-ng RNA templates, 0.5 μM concentrations of random hexamer, and 50 U of reverse transcriptase by using a TaqMan reverse transcription reagent kit (Applied Biosystems) as described previously (21).
cDNA samples from the reverse transcriptions were diluted 5-fold with nuclease-free water and quantified in three replicate qPCRs using Applied Biosystems SYBR green fast mix and primers specific to strain 195's genes of interest (see Table S2 in the supplemental material) as previously described for quantifying tceA gene copies. Relative standard curves for qPCR analysis were generated from 5- or 10-fold serially diluted pooled cDNA with linear regression R2 values of >99.0% and amplification efficiencies within a range of 100% ± 10% (see Table S2 in the supplemental material). Three optimal reference genes—16S rRNA gene, DET0435 (prs), and DET1606 (recA)—representing the most stable reference genes from a set of tested candidate reference genes were selected by the GeNorm software for normalization (42). REST 2009 software (30) was used to normalize the genes of interest to the geometric mean of the three reference genes, to calculate the relative expression ratios between AA4, AA20, and control (AA0) samples, and to identify statistically significant differences in transcript levels.
Statistical significance for the gene expression data of the branched-chain amino acid transporter (DET0941, DET0942, and DET0944), tceA (DET0079) and pheA (DET1547) were tested in three conditions (AA0, AA4, and AA20) using pairwise comparison, in order to evaluate their response to the presence of the putative substrates. The P values obtained from these tests were then adjusted to correct for multiple comparisons by applying a sequentially rejective procedure using Holm tests (7). The polar amino acid transport genes (DET0417 to DET0419) were assessed for only two experimental conditions (AA0 and AA20), since AA4 probably does not contain the putative substrates for this transporter. P values of <0.05 were considered statistically different for DET0417 to DET0419. Only genes exhibiting differential expression of >2-fold, i.e., log2(relative expression ratio) > 2, were considered biologically significant (16).
RESULTS
Incorporation of amino acids by strain 195.
In order to determine which amino acids were imported and incorporated by strain 195 during growth, experiments were performed using 13C-labeled acetate as the carbon source with amendment of 20 unlabeled amino acids, and isotopomer analysis was applied to quantify dilution of labeled amino acids within cells. Since strain 195 possesses all pathways needed for complete de novo amino acid biosynthesis (39), the rationale behind this experiment was to evaluate whether strain 195 prefers to import some amino acids from the medium rather than synthesizing them de novo.
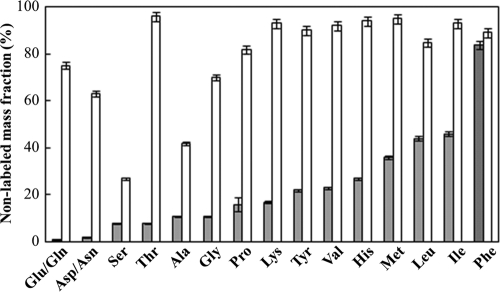
In order to validate this isotopomer dilution method, we first applied it to E. coli strain K-12 MG1655, a strain known to be capable of both complete de novo amino acid synthesis and amino acid uptake from medium (3, 23, 40), as a positive control. When this strain was grown with [U-13C]glucose in the presence of 20 unlabeled amino acids, isotopomer results (mass fraction M0) showed 30 to 98% dilution of all tested amino acids in the exponential growth biomass (Fig. 1). This result confirmed the isotopomer dilution method for differentiating amino acid uptake from the medium from de novo synthesis. In addition to tryptophan and cysteine that are degraded during protein hydrolysis, Fig. 1 does not contain labeling information for arginine, since the mass spectra for arginine fragments (either [M-57]+ or [M-159]+) are not accurate due to peak overlap and rearrangement (2).
Fig. 1.
Mass fraction of unlabeled proteinogenic amino acids (M0 fraction) of D. ethenogenes strain 195 (▩) and E. coli K-12 MG1655 (□).
When strain 195 was grown in medium with labeled acetate and unlabeled amino acids, a total of 13 amino acids exhibited dilutions greater than 5% (Fig. 1). Eight of these are hydrophobic in nature, while the other five are hydrophilic with polar side chains (see Table S3 in the supplemental material). Only background level dilution rates (<5%) were detected for glutamate, glutamine, aspartate, and asparagine, indicating that they were not significantly imported by strain 195. The dilutions of serine and glycine were relatively low (∼10%), indicating that they are also mostly synthesized de novo (from 3-P-glycerate or C1 metabolism) (29). Four amino acids exhibited >35% dilution (Fig. 1), with phenylalanine (Phe) exhibiting the highest dilution of over 80%. Isoleucine (Ile), leucine (Leu), and methionine (Met) were the next highest imported amino acids, with 46 to 37% dilution. Although tyrosine shares a similar biosynthesis pathway to phenylalanine, it exhibited a lower labeling dilution (∼20%), indicating that tyrosine was mainly synthesized de novo.
Effects of amino acids on physiological traits of D. ethenogenes strain 195.
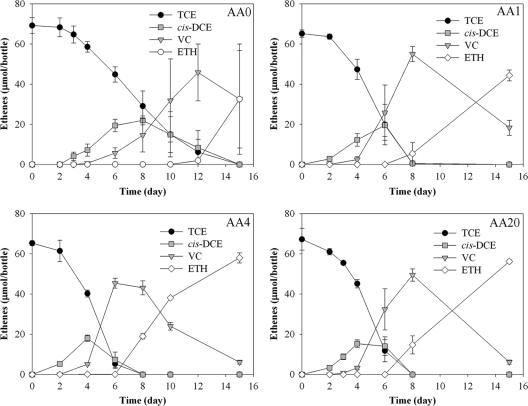
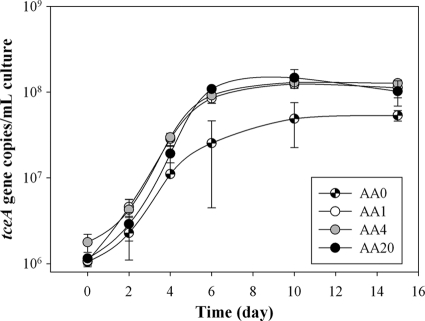
The high dilution of Phe, Leu, Ile, and Met indicated that strain 195 was importing these amino acids under the experimental conditions. In order to assess the effects of the amendments on strain 195 physiology, a comparison of growth and TCE degradation was performed for strain 195 amended with different amino acid combinations, including no amino acids (AA0), Phe only (AA1), Phe, Iso, Leu, and Met (AA4), and all 20 common amino acids (AA20). TCE dechlorination patterns demonstrated that the AA0 culture dechlorinated ∼65 μmol TCE after 15 days, while the AA1, AA4, and AA20 cultures depleted the TCE within 10 days (Fig. 2). The experiment also showed that AA1 generated less ethene than AA4 and AA20, and thus more vinyl chloride remained in the end. Furthermore, all three amino acid-amended cultures grew faster than the unamended culture AA0 (Fig. 3). During the exponential phase, the amino acid-amended cultures exhibited shorter doubling times than did the unamended culture (Table 1). Amino acid-amended cultures reached their highest cell number before day 8, while the unamended culture reached its highest cell number after day 10. In addition, the cell yields of the amino acid amended cultures were two to three times that of the unamended culture (Table 1). All amino acid amended cultures reproducibly exhibited very similar growth and degradation profiles, suggesting that the incorporated amino acids, especially phenylalanine, play an important role in enhancing biomass growth.
Fig. 2.
Dechlorination profiles by D. ethenogenes strain 195 amended with 0 (AA0), 1 (AA1), 4 (AA4), or 20 (AA20) amino acids after five subcultures. The data shown are the mean values from triplicate biotic experiments, with error bars representing one standard deviation.
Fig. 3.
Growth curves of D. ethenogenes strain 195 amended with 0 (AA0), 1 (AA1), 4 (AA4), or 20 (AA20) amino acids after five subcultures. The data shown are the mean values from triplicate biotic experiments, with error bars representing one standard deviation.
Table 1.
Doubling times and cell yields of D. ethenogenes strain 195 using TCE and amended with 0 (AA0), 1 (AA1), 4 (AA4), or 20 (AA20) amino acids after five subcultures
| Culture | Mean ± SDa |
|
|---|---|---|
| Doubling time (days)b | Cell yield (107/μmol of Cl− released)c | |
| AA0 | 1.3 ± 0.3 | 3.8 ± 0.6 |
| AA1 | 0.95 ± 0.04 | 7.8 ± 0.3 |
| AA4 | 0.91 ± 0.16 | 10.0 ± 0.6 |
| AA20 | 0.76 ± 0.08 | 7.5 ± 2.5 |
F tests were performed to determine statistically significant differences (P < 0.05) in variances of the four conditions.
The doubling times were calculated from cell numbers during exponential growth on days 2 and 6 (tceA copies).
The Cl− released was computed using only metabolic dechlorination from TCE to VC as described by Duhamel and Edwards (6).
To test the hypothesis that variations in availability of amino acids for import and/or variations in growth rates may affect amino acid ratios within cells (32), the proteinogenic amino acids in strain 195 were analyzed in AA0, AA4, and AA20. In each condition, overall amino acid composition of stain 195 exhibited negligible differences (see Table S4 in the supplemental material). Interestingly, we also found that the most abundant proteinogenic amino acids (i.e., alanine, aspartate/asparagine, glycine, and glutamate/glutamine) did not correspond to the amino acids that were most preferentially imported from the medium (phenylalanine, isoleucine, leucine, and methionine).
Transcript levels of the genes involved in ABC transporters and phenylalanine synthesis.
RT-qPCR was used to investigate the transcriptional response of the putative ABC-type transporter genes to the presence of the putative substrates. Here, both ABC transporter genes exhibited no significant response to the presence of putative amino acid substrates in either the AA4 or the AA20 culture (Table 2). Although DET0944, the gene encoding the substrate-binding protein for the branched-chain amino acid transporter, exhibited downregulation that was significant according to the P value, the fold change in relative expression ratio was not significant.
Table 2.
Differential transcript levels of genes involved in ABC-type amino acid transporters and phenylalanine synthesisa
| Gene | Relative expression ratiob |
Pc |
Annotated functions | ||
|---|---|---|---|---|---|
| AA4 | AA20 | AA4 | AA20 | ||
| DET0417 | NAd | 0.65 | NA | 0.68 | ATP binding protein of the polar amino acid transporter |
| DET0418 | NA | 0.73 | NA | 0.63 | Permease subunit of the polar amino acid transporter |
| DET0419 | NA | 0.64 | NA | 0.28 | Substrate binding protein of the polar amino acid transporter |
| DET0941 | 0.69 | 0.45 | 0.16 | 0.05 | ATP binding protein of the branched-chain amino acid transporter |
| DET0942 | 0.73 | 0.52 | 0.22 | 0.10 | Permease subunits of the branched-chain amino acid transporter |
| DET0944 | 0.82 | 0.84 | 0.05* | 0.01* | Substrate binding protein of the branched-chain amino acid transporter |
| pheA (DET1547) | 0.86 | 0.78 | 0.42 | 0.21 | Prephenate dehydratase |
| tceA (DET0079) | 0.78 | 0.49 | 0.41 | 0.20 | Reductive dehalogenase |
All measurements are averages of triplicate qPCR results from triplicate biological samples.
Relative expression ratios were generated from REST 2009 software using the mathematical model described by Pfaffl et al. (32).
P values were generated by using REST 2009 software. *, Statistically significant P values in pairwise comparison that fulfilled the criterion in the Holm test.
NA, not applicable.
Since a major fraction of phenylalanine (>80%) was imported by strain 195, the potential regulation of imported phenylalanine on its biosynthesis pathway was investigated. The gene pheA (DET1547) that encodes the enzyme transforming prephenate to phenylpyruvate (see Fig. S1 in the supplemental material) contains an ACT domain. The ACT domain, derived from aspartokinase, chorismate mutase and TyrA (prephenate dehydrogenase), has a putative role as a regulatory module involved in a feedback inhibition regulated by the presence of phenylalanine on the catalytic enzyme(s) (22). The transcript levels of this gene showed no significant differences between amino acid amended cultures and control AA0, indicating the absence of transcriptional feedback inhibition to phenylalanine synthesis by imported amino acids (Table 2). Similarly, the reductive dehalogenase gene tceA was not regulated under AA4 and AA20 conditions.
DISCUSSION
Although strains of Dehalococcoides have been found in many different subsurface environments (11), researchers have observed that the growth of Dehalococcoides is usually more robust in complex microbial communities than in isolation (9). Additions of cofactors such as vitamin B12 and complex nutrient supplements have been found to enhance both the dechlorination and growth of Dehalococcoides (24). Identification of other important growth factors for the robust and sustained cultivation of Dehalococcoides could lead to improved understanding of the physiology of this organism and to potentially more successful bioremediation strategies (36, 41). Conventional cultivation-dependent screening techniques, such as a monofactorial analysis in batch culture experiments by stepwise omission of single amino acids, can be used for such identification as demonstrated by Holliger et al. (13) in their study with Dehalobacter restrictus.
In the present study, we used results of isotopomer dilution analysis to differentiate imported and synthesized amino acids by D. ethenogenes 195 and to further assay the physiological effects of these amino acids on this strain. Four amino acids—phenylalanine, isoleucine, leucine, and methionine—were successfully identified as effective nutrient factors for enhancing the growth yield and dechlorination rates of strain 195. By importing amino acids rather than synthesizing them de novo, strain 195 may save energy and alleviate bottlenecks associated with anabolic pathways that limit the growth rate of Dehalococcoides, as has been recently demonstrated in silico using a constraint-based model (15). Based on estimations made for E. coli using acetate as a sole carbon and energy source, it costs about 13 to 87 high-energy phosphate bonds for a cell to synthesize one amino acid (1). Our isotopomer dilution results revealed that strain 195 could import some energetically costly amino acids (e.g., 35 to 58 phosphate bonds per amino acid in E. coli), such as phenylalanine, isoleucine, leucine, and methionine (37). As a comparison, the energy cost is estimated to be 1 to 2 ATPs per amino acid for importing them from the growth medium (12, 26, 28).
In addition, aromatic precursors (e.g., chorismate) of phenylalanine are likely the same precursors of ubiquinones that are integrated in large amounts into Dehalococcoides membranes, possibly involved in protecting against free radicals during reductive dechlorination (43). Therefore, the availability of large amounts of exogenous phenylalanine might reduce the anabolic energy cost compared to biosynthesis of these high-energy consuming precursors. Interestingly, tyrosine shares some of the same precursor, prephenate, as phenylalanine (see Fig. S1 in the supplemental material), but tyrosine was mainly synthesized de novo by strain 195. Tyrosine synthesis is via prephenate dehydrogenase (EC 1.3.1.12; DET0460): prephenate + NAD+ → 4-hydroxyphenylpyruvate + CO2 + NADH, whereas phenylalanine synthesis occurs via prephenate dehydratase: prephenate → phenylpyruvate + H2O + CO2. Consequently, the biosynthesis of tyrosine requires NAD+ as a reactant while producing NADH, which is predicted to be an important limiting factor for Dehalococcoides growth (15). Therefore, Dehalococcoides cells may be able to grow more efficiently by allowing tyrosine synthesis to occur (generating NADH) while importing available phenylalanine from the medium in order to diminish competition for the prephenate precursor. Furthermore, although tryptophan is the most energy-intensive amino acid to synthesize (1, 37), isotopomer analysis using current protocols is unable to measure this amino acid. Nevertheless, comparison of TCE dechlorination and the growth of strain 195 in AA4 and AA20 demonstrates negligible differences, indicating that the amino acids beyond the four most preferentially imported ones (including tryptophan) provide little additional benefit to strain 195 under the conditions tested in the present study.
The amino acid dilution results indicated that strain 195 completely synthesized aspartate, asparagine, glutamate, and glutamine, even though these amino acids were available in the medium. This is distinctly different from fast-growing bacteria, e.g., E. coli strain K-12 MG1655, which showed significant import and utilization of all exogenous amino acids, as shown in Fig. 1. This difference might be associated with the environmental origins of these two bacteria, where E. coli likely evolved a metabolism for fast growth in nutrient-rich environments, such as the guts of animals (14), whereas Dehalococcoides is common in nutrient-scarce environments such as groundwater aquifers, where its metabolism may have evolved to rely heavily on de novo amino acid biosynthesis (5). In addition, a total of 13 amino acids exhibited dilutions from 5% to >80% demonstrated that strain 195 can only selectively import and incorporate certain amino acids with different efficiencies. This finding is different from an assumption in the previous modeling study that Dehalococcoides were capable of unselectively taking in all amino acids (15). The discrepancy between our results and the previous study indicates the understanding of Dehalococcoides physiology is still limited. Thus, the knowledge obtained here could be integrated into future in silico models of Dehalococcoides for a better simulation.
The genome annotations of strain 195 and other sequenced Dehalococcoides strains predict that the two ABC-type amino acid transporters would likely uptake amino acids with different characteristics of hydrophobicity and polarity (see Table S1 in the supplemental material). Although data from the isotopomer analysis confirmed 195's ability to import amino acids with different chemical characteristics (see Table S3 in the supplemental material) (e.g., hydrophilic Thr, Lys, and His, together with hydrophobic Pro, Ala, Tyr, Val, Met, Leu, Ile, and Phe), the actual substrate(s) for the transporters have not yet been positively identified. For example, the predicted branched-chain transporter (DET0938, DET0941 to DET0944) was annotated to have a broad substrate specificity, and bioinformatics findings suggest that this transporter is similar to the LIV-I/LS system in E. coli (33), which catalyzes the translocation of not only Leu, Ile, and Val but also Phe and Tyr from the periplasm to the cytoplasm (18). An analogous broad specificity was observed in E. coli strain K-12 MG1655 (Fig. 1). However, these hypothetical substrates were not equally imported by strain 195, and a differential expression analysis revealed that the transporter genes were not regulated by the presence of their hypothetical substrates. The same was observed for the putative polar amino acid transporters of strain 195. It has been reported that transporters with broad substrate specificities tend to be constitutively expressed regardless of the presence of their substrates (23) and that may be the case in the present study.
In general, we demonstrate here that the specific amino acids that have stimulatory effects on slow-growing bacteria can be effectively identified by an isotopomer-based metabolite analysis. Moreover, although D. ethenogenes 195 exhibited no metabolic regulation for import of amino acids under the experimental conditions, exogenous amendment with one or more amino acids resulted in enhanced growth and dechlorination.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by the Superfund Basic Research Program under grant NIEHS ES04705, the Strategic Environmental Research and Development Program at U.S. Department of Energy (project ER-1587). This study was also partially supported by an NSF Career Grant (MCB0954016).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Akashi H., Gojobori T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 99:3695–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antoniewicz M. R., Kelleher J. K., Stephanopoulos G. 2007. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal. Chem. 79:7554–7559 [DOI] [PubMed] [Google Scholar]
- 3. Blattner F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 4. Cupples A. M., Spormann A. M., McCarty P. L. 2003. Growth of a Dehalococcoides-like microorganism on vinyl chloride and cis-dichloroethene as electron acceptors as determined by competitive PCR. Appl. Environ. Microbiol. 69:953–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Devlin J. F., Katic D., Barker J. F. 2004. In situ sequenced bioremediation of mixed contaminants in groundwater. J. Contam. Hydrol. 69:233–261 [DOI] [PubMed] [Google Scholar]
- 6. Duhamel M., Edwards E. A. 2007. Growth and yields of dechlorinators, acetogens, and methanogens during reductive dechlorination of chlorinated ethenes and dihaloelimination of 1,2-dichloroethane. Environ. Sci. Technol. 41:2303–2310 [DOI] [PubMed] [Google Scholar]
- 7. Glantz S. A. 2001. Primer of biostatistics, 5th ed McGraw-Hill, New York, NY [Google Scholar]
- 8. He J., Sung Y., Krajmalnik-Brown R., Ritalahti K. M., Löffler F. E. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichloroethene-respiring anaerobe. Environ. Microbiol. 7:1442–1450 [DOI] [PubMed] [Google Scholar]
- 9. He J. Z., Holmes V. F., Lee P. K. H., Alvarez-Cohen L. 2007. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl. Environ. Microbiol. 73:2847–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He J. Z., Ritalahti K. M., Yang K. L., Koenigsberg S. S., Löffler F. E. 2003. Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium. Nature 424:62–65 [DOI] [PubMed] [Google Scholar]
- 11. Hendrickson E. R., et al. 2002. Molecular analysis of Dehalococcoides 16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Higgins C. F., Linton K. J. 2004. The ATP switch model for ABC transporters. Nat. Struct. Mol. Biol. 11:918–926 [DOI] [PubMed] [Google Scholar]
- 13. Holliger C., et al. 1998. Dehalobacter restrictus gen. nov. and sp. nov., a strictly anaerobic bacterium that reductively dechlorinates tetra- and trichloroethene in an anaerobic respiration. Arch. Microbiol. 169:313–321 [DOI] [PubMed] [Google Scholar]
- 14. Hudault S., Guignot J., Servin A. L. 2001. Escherichia coli strains colonizing the gastrointestinal tract protect germfree mice against Salmonella typhimurium infection. Gut 49:47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Islam A. M., Edwards E. A., Mahadevan R. 2010. Characterizing the metabolism of Dehalococcoides with a constraint-based model. PLoS Comput. Biol. 6:e1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Johnson D. R., et al. 2008. Temporal transcriptomic microarray analysis of “Dehalococcoides ethenogenes” strain 195 during the transition into stationary phase. Appl. Environ. Microbiol. 74:2864–2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson D. R., Nemir A., Andersen G. L., Zinder S. H., Alvarez-Cohen L. 2009. Transcriptomic microarray analysis of corrinoid responsive genes in Dehalococcoides ethenogenes strain 195. FEMS Microbiol. Lett. 294:198–206 [DOI] [PubMed] [Google Scholar]
- 18. Koyanagi T., Katayama T., Suzuki H., Kumagai H. 2004. Identification of the LIV-I/LS system as the third phenylalanine transporter in Escherichia coli K-12. J. Bacteriol. 186:343–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kube M., et al. 2005. Genome sequence of the chlorinated compound respiring bacterium Dehalococcoides species strain CBDB1. Nat. Biotechnol. 23:1269–1273 [DOI] [PubMed] [Google Scholar]
- 20. Lee P. K. H., He J., Zinder S. H., Alvarez-Cohen L. 2009. Evidence for nitrogen fixation by “Dehalococcoides ethenogenes” strain 195. Appl. Environ. Microbiol. 75:7551–7555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee P. K. H., Macbeth T. W., Sorenson K. S., Deeb R. A., Alvarez-Cohen L. 2008. Quantifying genes and transcripts to assess the in situ physiology of “Dehalococcoides” spp. in a trichloroethene-contaminated groundwater site. Appl. Environ. Microbiol. 74:2728–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liberles J. S., Thórólfsson M., Martínez A. 2005. Allosteric mechanisms in ACT domain containing enzymes involved in amino acid metabolism. Amino Acids 28:1–12 [DOI] [PubMed] [Google Scholar]
- 23. Marin K., Krämer R. 2007. Amino acid transport systems in biotechnologically relevant bacteria, p. 289–325 In Wendisch V. F. (ed.), Amino acid biosynthesis: pathways, regulation, and metabolic engineering. Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 24. Maymo-Gatell X., Chien Y. T., Gossett J. M., Zinder S. H. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568–1571 [DOI] [PubMed] [Google Scholar]
- 25. McMurdie P. J., et al. 2009. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet. 5:e1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mimmack M. L., et al. 1989. Energy coupling to periplasmic binding protein-dependent transport-systems: stoichiometry of ATP hydrolysis during transport in vivo. Proc. Natl. Acad. Sci. U. S. A. 86:8257–8261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mueller J. H., Johnson E. R. 1941. Acid hydrolysates of casein to replace peptone in the preparation of bacteriological media. J. Immunol. 40:33–38 [Google Scholar]
- 28. Patzlaff J. S., van der Heide T., Poolman B. 2003. The ATP/Substrate stoichiometry of the ATP-binding cassette (ABC) transporter OpuA. J. Biol. Chem. 278:29546–29551 [DOI] [PubMed] [Google Scholar]
- 29. Perrenoud A., Sauer U. 2005. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 187:3171–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pfaffl M. W., Horgan G. W., Dempfle L. 2002. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pingitore F., Tang Y., Kruppa G. H., Keasling J. D. 2007. Analysis of amino acid isotopomers using FT-ICR MS. Anal. Chem. 79:2483–2490 [DOI] [PubMed] [Google Scholar]
- 32. Pramanik J., Keasling J. D. 1998. Effect of Escherichia coli biomass composition on central metabolic fluxes predicted by a stoichiometric model. Biotechnol. Bioeng. 60:230–238 [DOI] [PubMed] [Google Scholar]
- 33. Saier M. H., Yen M. R., Noto K., Tamang D. G., Elkan C. 2009. The transporter classification database: recent advances. Nucleic Acids Res. 37:D274–D278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sambrook J., Fritsch E., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 35. Seshadri R., et al. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105–108 [DOI] [PubMed] [Google Scholar]
- 36. Simpkin T. J., Norris R. D. 2010. Engineering and implementation challenges for chlorinated solvent remediation, p. 109–143 In Stroo H. F., Ward C. H. (ed.), In situ remediation of chlorinated solvent plumes. Springer, New York, NY [Google Scholar]
- 37. Stephanopoulos G. N., Aristidou A. A., Nielsen J. P. 1998. Metabolic engineering principles and methodologies, vol. 75 Academic Press, Inc., San Diego, CA [Google Scholar]
- 38. Tang Y., et al. 2007. Pathway confirmation and flux analysis of central metabolic pathways in Desulfovibrio vulgaris Hildenborough using gas chromatography-mass spectrometry and Fourier transform-ion cyclotron resonance mass spectrometry. J. Bacteriol. 189:940–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang Y. J., et al. 2009. Investigation of carbon metabolism in “Dehalococcoides ethenogenes” strain 195 by use of isotopomer and transcriptomic analyses. J. Bacteriol. 191:5224–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tao H., Bausch C., Richmond C., Blattner F. R., Conway T. 1999. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J. Bacteriol. 181:6425–6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vainberg S., Condee C., Steffan R. 2009. Large-scale production of bacterial consortia for remediation of chlorinated solvent-contaminated groundwater. J. Ind. Microbiol. Biotechnol. 36:1189–1197 [DOI] [PubMed] [Google Scholar]
- 42. Vandesompele J., et al. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:0034.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. White D. C., et al. 2005. Phospholipid furan fatty acids and ubiquinone-8: lipid biomarkers that may protect Dehalococcoides strains from free radicals. Appl. Environ. Microbiol. 71:8426–8433 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.