Abstract
Botulinum neurotoxin serotype E (BoNT/E) outbreaks in the Great Lakes region cause large annual avian mortality events, with an estimated 17,000 bird deaths reported in 2007 alone. During an outbreak investigation, blood collected from bird carcasses is tested for the presence of BoNT/E using the mouse lethality assay. While sensitive, this method is labor-intensive and low throughput and can take up to 7 days to complete. We developed a rapid and sensitive in vitro assay, the BoTest Matrix E assay, that combines immunoprecipitation with high-affinity endopeptidase activity detection by Förster resonance energy transfer (FRET) to rapidly quantify BoNT/E activity in avian blood with detection limits comparable to those of the mouse lethality assay. On the basis of the analysis of archived blood samples (n = 87) collected from bird carcasses during avian mortality investigations, BoTest Matrix E detected picomolar quantities of BoNT/E following a 2-h incubation and femtomolar quantities of BoNT/E following extended incubation (24 h) with 100% diagnostic specificity and 91% diagnostic sensitivity.
INTRODUCTION
Bacteria of the genus Clostridium produce seven distinct but structurally similar serotypes of botulinum neurotoxins (BoNT) designated A to G (for reviews, see references 10, 27, 31, and 43). BoNTs are zinc-dependent endopeptidases composed of a heavy chain responsible for neuron-specific receptor binding and cell entry and a catalytic light chain responsible for synaptic protein cleavage (36, 40). BoNTs cause botulism by inactivation of the neuromuscular junction. Upon entry into neurons, the neurotoxins specifically disrupt the protein machinery responsible for fusion of synaptic vesicles with the plasma membrane, thereby inhibiting neurotransmitter release into the postsynaptic junction (6, 7, 36, 39–41). In humans, progression of intoxication and eventual mortality are caused by respiratory failure due to flaccid paralysis of the diaphragm and intercostal muscles (44). In water birds, intoxication and subsequent paralysis result in loss of muscle tone and limber neck, with subsequent death from drowning due to the inability of the bird to maintain its head above water (34). BoNTs are the most lethal substances known, and depending on the serotype, the estimated human lethal dose ranges from 1 to 3 ng toxin per kg body weight (22, 31).
Intoxication by BoNT serotype C (BoNT/C) and BoNT/E is a significant contributor to avian mortality worldwide (14, 19, 21, 29, 45, 46). BoNT/E mortality events were first documented among fish-eating birds of the North American Great Lakes in the early 1960s, and outbreaks reemerged during the fall of 1999, with highest mortality documented among common loons, mergansers, long-tailed ducks, and gulls (34). Within the Great Lakes region, an estimated 17,000 birds perished from BoNT/E intoxication in 2007 alone, and total mortality of approximately 50,000 birds has been documented for the years from 1999 to 2009 (U.S. Geological Survey [USGS], National Wildlife Health Center [NWHC], unpublished data).
The environmental conditions that contribute to large-scale BoNT/E-related avian mortality events are not well understood. Unlike BoNT/C outbreaks that occur among filter-feeding and dabbling ducks, BoNT/E intoxication primarily impacts fish-eating birds, suggesting that fish are an important source for toxin delivery (34). This is supported by the isolation of Clostridium botulinum from fish muscle and alimentary canal tissues as well as detection of the BoNT/E gene in fish liver and intestine samples collected within the Great Lakes Basin (8, 16, 47). Additional studies have identified the BoNT/E gene in sediment samples collected from Lake Erie, suggesting that sediment ingestion is a route for bacterial infection or toxin accumulation in bottom-feeding fish or bottom-dwelling invertebrates such as filter-feeding dreissenid mussels (32, 33).
Diagnosis of avian botulinum in moribund or dead animals is based upon the identification of catalytically active BoNT in blood, together with the lack of clinical signs suggestive of other infectious diseases (34). Additionally, absence of pathology indicative of clostridial infection in wild birds indicates that food-borne intoxication and not infection is the mechanism of disease (34). The mouse lethality assay is recognized to be the “gold standard” for identifying catalytically active BoNT. This assay is performed by injecting test samples into pairs of mice, including one protected with BoNT antitoxin, and recording the time of death over 1 or more days (10). The mouse lethality assay is highly sensitive, with detection limits ranging from 5 to 10 pg (10, 27). However, the assay is low throughput and expensive and requires animal care equipment and training to complete. Large-scale sample screening using the mouse lethality assay is prohibitively expensive, and as the test requires the use of live animals, ethical questions arise. Thus, testing is often restricted to a limited number of samples collected postmortem during avian botulism outbreaks. Little information regarding the presence and concentration of BoNT in suspect food-web components is available, and rapid, high-throughput assays are necessary to investigate the reservoirs, drivers, and ecological pathways of large-scale avian mortality events.
Rapid diagnostic assays for the detection of both BoNTs and BoNT-producing bacteria have been developed, but because of various shortcomings, they have not been widely applied for ecological studies. PCR-based assays have been used to identify BoNT types C and E genes in fish and in lake-bed sediment samples (16, 30, 32), suggesting the presence of C. botulinum, but PCR does not indicate whether catalytically active BoNT is present. Other in vitro assays for detection of BoNTs use fluorescent substrates or mass spectrometry coupled with immunological techniques (5, 9, 13, 15, 17, 20, 23, 35). Shortcomings of these methods include poor sensitivity, an inability to detect BoNT in complex sample matrices, and expensive equipment requirements (13, 37).
Here we report a rapid and sensitive in vitro assay, the BoTest Matrix E assay, for the detection of BoNT/E in blood and serum. This assay combines immunologic isolation methods with a high-affinity endopeptidase reporter that uses Förster resonance energy transfer (FRET) technology to measure BoNT activity. The reporter was constructed using a portion of one of the natural targets for BoNT, synaptosome-associated protein-25 (SNAP-25), modified for specific measurement of catalytically active BoNT/E. Testing of archived avian blood samples with the activity-based BoNT/E assay demonstrated that the BoTest Matrix E assay possesses mouse lethality assay sensitivity and consistently detected and quantified BoNT/E in avian blood on the basis of changes in fluorescence emission. The BoTest Matrix E assay is also formatted for high throughput and can be completed in 24 h or less.
MATERIALS AND METHODS
Mouse lethality assay.
Clotted blood was removed from hearts collected from bird carcasses during the investigation of wildlife avian mortality events from August 2004 to October 2010 and assayed for BoNT using the mouse lethality assay (2), in accordance with USGS, NWHC, Institutional Animal Care and Use Protocol EP090924. Blood samples (n = 87) were collected from 18 bird species: American coot (n = 1), American white pelican (n = 5), California gull (n = 1), Caspian tern (n = 2), common loon (n = 17), double-crested cormorant (n = 11), great blue heron (n = 1), herring gull (n = 2), horned grebe (n = 6), long-tailed duck (n = 4), mallard duck (n = 16), northern pintail duck (n = 1), red-breasted merganser (n = 1), red-necked grebe (n = 5), red-throated loon (n = 1), ring-billed gull (n = 9), ring-necked duck (n = 1), and white-winged scoter (n = 3). All BoNT-containing samples were maintained in a laboratory at the NWHC registered with the Select Agent Program. Pairs of mice, consisting of antitoxin-protected and -unprotected animals, were observed for a total of 5 days for the development of clinical signs and/or patterns of mortality indicative of botulinum toxicosis.
Generation of FRET-based reporters.
Construction of the plasmid encoding the BoTest A/E reporter, pBoTest A/E, is described elsewhere (37). The BoNT/E-specific reporter plasmid (pBoTest E) was constructed by amplifying pBoTest A/E using phosphorylated primers 5′-Pi-CCA ACC AAG AGG CAA CAA AGA TGC-3′ and 5′-Pi-CTT CAT CAA TTC TGG TTT TGT TGG AG-3′ (where Pi indicates phosphorylated primer), designed to introduce an R198E mutation to deactivate the BoNT/A cleavage site (12). The PCR product was then gel purified and religated, using T4 DNA ligase (Promega, Madison, WI), to create plasmid pBoTest E. The BoNT null reporter plasmid (pBoTest KO) was constructed by amplifying pBoTest E using primers 5′-CAG AAT CGC CAG ATT GTC AGG TTC ATG GAG AAG-3′ and 5′-CTT CTC CAT GAA CCT GAC AAT CTG GCG ATT CTG-3′ and QuikChange (Stratagene, La Jolla, CA) site-directed mutagenesis according to the manufacturer's protocol. The resulting plasmid, pBoTest KO, encodes a protein reporter containing D179V, I181F, and R198E mutations that cannot be cleaved by BoNT/A, BoNT/C, or BoNT/E (12, 24).
Protein expression and purification.
All FRET reporter constructs were expressed and purified by double-affinity chromatography as described previously (37). The resulting reporters were quantified by bicinchoninic acid (Pierce, Rockford, IL), divided into single-use aliquots, and stored at −80°C. The construct pET28a-HcR/E was provided by J. T. Barbieri (Medical College of Wisconsin—Milwaukee) and purified as described previously (4). Purified heavy-chain receptor binding domain of BoNT/E (HcR/E) was dialyzed against 10 mM Tris-HCl [pH 7.6], 200 mM NaCl, 1 mM dithiothreitol (DTT), and 40% glycerol.
Antibody production, purification, and modification.
Chicken anti-HcR/E IgY was produced by Genetel Laboratories (Madison, WI) using purified HcR/E protein to inoculate hens and to affinity purify crude IgY. BoNT/E IgG was purified from ascites or serum using an N′ab antibody purification kit (Thermo Scientific, Rochester, NY) according to the manufacturer's specifications. When specified, purified antibodies were biotinylated using an EZ-Link solid-phase biotinylation kit (Pierce) according to the manufacturer's specification. BcMag magnetic beads (BioClone Inc., San Diego, CA) were coated with antibody according to the manufacturer's specifications. Antibody-coated beads were then blocked for 1 h with 1% casein (wt/vol) in phosphate-buffered saline (PBS). Following the blocking reaction, beads were washed and resuspended to 6 mg/ml in PBS-Tween 20 (PBS-T; 0.1%) and stored at 4°C.
BoNT/E pull-down/activity assay.
Twelve micrograms antibody-coated beads was incubated with 200 μl avian blood in black 96-well microtiter plates (Thermo Scientific) for 2 h at 25°C with shaking (700 rpm), unless otherwise indicated. BoNT/E-bound beads were then washed with 200 μl PBS-T per well and separated with a 96-well plate magnet (V&P Scientific, San Diego, CA), and the wash solution was removed by pipette. Washing steps were repeated a total of 5 times. After the final wash, 100 μl of a 250 nM solution of the indicated BoTest reporter (diluted in 50 mM HEPES-NaOH, pH 7.1, 5 mM NaCl, 0.1% Tween 20, 5 mM DTT, 10 μM ZnCl2) and a 10× concentration of EDTA-free complete protease inhibitor cocktail (Roche, Madison, WI), diluted according to the manufacturer's protocol, were added to each well containing BoNT/E-bound beads, and the plate was incubated at 37°C with shaking (700 rpm). Standard curves were prepared by diluting trypsinized BoNT/E (Metabiologics, Madison, WI) into PBS-T (final concentration range, 0.1 pM to 1 nM [15 pg/ml to 150 ng/ml]). BoNT/E dilutions were immunoprecipitated with antibody-conjugated beads as described above, and fluorescence was measured by exciting the reporter at 434 nm and collecting emission readings at 470 nm and 526 nm using either a Varioskan or an Ascent Fluoroscan microplate reader (Thermo Scientific). FRET emission ratio values were calculated by dividing the relative fluorescence unit (RFU) value at 526 nm by the RFU value at 470 nm.
BoNT/E sandwich enzyme-linked immunosorbent assay (ELISA).
Ninety-six-well Maxisorp microtiter plates (Thermo Scientific) were coated with 100 ng/well anti-HcR/E IgY capture antibody in PBS overnight at 4°C. Plates were blocked for 1 h with 1% (wt/vol) casein in PBS and then incubated with 50 μl sample per well for 2 h at 25°C with shaking (700 rpm). Following incubation with samples, plates were washed 5 times with 200 μl PBS-T per well. Plates were then incubated with 100 ng/well biotinylated burro anti-BoNT/E (Centers for Disease Control and Prevention, Atlanta, GA) detection antibody for 2 h at 25°C with shaking. Plates were washed again 3 times and incubated with streptavidin-conjugated poly-horseradish peroxidase (Thermo Scientific) diluted 1:5,000 in 1% (wt/vol) casein in PBS for 1 h at 25°C. After a final wash, wells were incubated with 100 μl per well of 3,3′,5,5′-tetramethylbenzidine (TMB; Thermo Scientific) for 30 min. Colorimetric reactions were stopped by adding 100 μl 0.19 M H2SO4 per well, and the absorbance was measured at 450 nm using a Varioskan or Ascent Fluoroscan microplate reader (Thermo Scientific).
Data analysis.
For all line graphs, data shown are averages from triplicate reactions, with bars indicating standard deviations (SDs). Half-maximal (50%) effective concentrations (EC50) were calculated using Prism (version 4.0) software (GraphPad, La Jolla, CA) from data fitted with the sigmoidal dose-response equation Y = bottom + [(top − bottom)/1 + 10(logEC50−x) · Hillslope)], where Y is the response that starts at a top (no acitivity) and goes to a bottom (assay saturation) and x is the logarithm of the concentration. Limits of detection (LODs) were calculated by determining the minimal BoNT concentration producing a signal >3 SDs below control values in the absence of BoNT (n = 6). Limits of quantitation (LOQs) were calculated by determining the minimal BoNT concentration producing a signal >10 SDs below control values in the absence of BoNT (n = 6) (37). The quantitative range was determined to be >10 SDs from the values in the presence and absence of BoNT that generated a maximal response. Coefficients of variation (CVs) were determined by comparing emission ratios from an immunoprecipitated blood sample containing 50 pM BoNT/E to those on a BoNT/E standard curve generated from immunoprecipitated samples. Percent CVs were calculated by dividing the SD of a single run (intraassay) or 3 independent runs (interassay) by the average of the run or the average of the 3 runs, respectively.
Diagnostic sensitivity was calculated using the equation Bpos/(Bpos + Bfneg), where Bpos is the number of samples that tested positive using both the BoTest Matrix E and lethality assays, and Bfneg is the number of false-negative samples (samples that tested negative for BoNT/E by BoTest Matrix E but positive for BoNT/E by the mouse assay). Diagnostic specificity was calculated using the equation Bneg/(Bneg + Bfpos), where Bneg is the number of samples that tested negative using both BoTest Matrix E and the mouse lethality assay, and Bfpos is the number of false-positive samples (samples that tested positive for BoNT/E by BoTest Matrix E but negative for BoNT/E by the mouse assay). Results for samples determined to be positive by both assays but that also cleaved the BoTest KO reporter were identified to be equivocal and were excluded from further analysis.
RESULTS
Ruge et al. recently reported the use of fluorogenic substrates, called the BoTest reporters, to develop high-throughput and sensitive assays to detect BoNT endopeptidase activity (37). For the detection of BoNT/A and BoNT/E, the BoTest A/E reporter is composed of SNAP-25 residues 141 to 206 fused to an N-terminal cyan fluorescent protein (CFP) and a C-terminal yellow fluorescent protein (YFP) (13, 37). In the absence of BoNT, excitation of the CFP moiety of the BoTest A/E reporter results in FRET to the YFP moiety: FRET results in YFP fluorescence emission while quenching CFP emission (Fig. 1A). In the presence of BoNT, the substrate is cleaved, causing separation of the fluorescent proteins and loss of FRET (YFP emission decreases and CFP emission increases). By calculating the ratio of YFP emission to CFP emission, BoNT activity can be quantified.
Fig. 1.
BoTest reporter design and characterization. (A) Schematic representation of the BoTest reporters. BoTest reporters consist of the BoNT recognition sequence of SNAP-25 flanked by CFP and YFP moieties. The BoTest E reporter has a single point mutation at the BoNT/A P1′ cleavage site. The BoTest KO reporter construct has three mutations at the BoNT/A cleavage site (P1′) and BoNT/E cleavage sites (P2 and P1′, respectively). The BoTest A/E (B), BoTest E (C), or BoTest KO (D) reporter was incubated at 25°C with the indicated concentrations of BoNT/A, BoNT/C, or BoNT/E. CFP and YFP (FRET) emissions (Em) were collected after 4 h, and the ratios of the emissions were plotted as a function of BoNT concentration. Trypsin (100 μg/ml) was used as a positive control.
Development of BoNT/E-specific and control BoTest reporters.
We reengineered the SNAP-25 moiety of the BoTest A/E reporter to create a BoNT/E-specific assay. BoNT/A cleavage of the BoTest reporter was eliminated by introducing a single arginine-to-glutamate substitution at P1′ residue 198 of the SNAP-25 BoNT/A cleavage site (Fig. 1A) (12, 24). This amino acid substitution yielded a FRET reporter that was sensitive to cleavage by BoNT/E but completely resistant to cleavage by 10 nM BoNT/A (Fig. 1C). The R198E mutation had no effect on BoNT/E-dependent cleavage compared to the BoTest A/E reporter, with EC50s of 7.3 and 8.1 pM BoNT/E, respectively (Fig. 1B and C). The resulting FRET reporter, BoTest E, was serotype specific for BoNT/E.
To identify potential sources of nonspecific proteolytic activity and to control for false-positive results, we developed a knockout reporter that is refractory to BoNT-dependent cleavage but remains sensitive to nonspecific proteases. This FRET reporter, BoTest KO, was created by introducing two amino acid substitutions, D179V and I181F, previously shown to be important for BoNT/E cleavage of SNAP-25 (11), in addition to the R198E mutation required to eliminate BoNT/A cleavage of SNAP-25 (Fig. 1C). The BoTest KO reporter was refractory to cleavage by 1 nM BoNT/A, BoNT/C, and BoNT/E but was still sensitive to proteolysis by trypsin (Fig. 1D).
Effects of matrix molecules on BoTest assay.
Quantitative measurements of BoNT activity in complex matrices, including biological and environmental samples, are complicated by the presence of substances such as albumin and NaCl that are known to interfere with either BoNT function or fluorescent reporter performance. Human blood contains albumin and NaCl at concentrations of approximately 3 to 5 g/dl and 100 mM, respectively (1, 18). Increasing amounts of human serum-derived albumin (HSA) significantly decreased BoNT/E-dependent reporter cleavage, with a BoNT/E EC50 shift from 8.0 pM at 0% HSA to 45.9 pM at 10% HSA (Fig. 2A). The dynamic range of the assay also decreased with increasing amounts of albumin, suggesting that albumin had an adverse effect on the fluorescent reporter.
Fig. 2.
Effects of albumin and sodium chloride on BoTest reporter performance. The BoTest E reporter was incubated at 25°C with the indicated concentrations of BoNT/E in 100 μl of 1× BoTest reaction buffer supplemented with 0 to 10% (wt/vol) HSA (A) or 5 to 150 mM NaCl (B). The fluorescence emissions were collected after 4 h, and the ratios of the emissions were plotted as a function of BoNT/E concentration.
NaCl caused a dose-dependent decrease in the BoNT/E EC50 for the BoTest E reporter (Fig. 2B). Addition of 100 mM NaCl, equivalent to concentrations in human blood, shifted the BoNT/E EC50 of the BoTest E assay to 45.2 pM from the 11.8 pM with 5 mM NaCl. As with albumin, the dynamic range was also affected by the addition of NaCl, indicating that removal of interfering substances will be a necessary component of a fluorescence-based assay to be used for the analyses of complex matrices such as blood or environmental samples.
Development of immunoprecipitation methods.
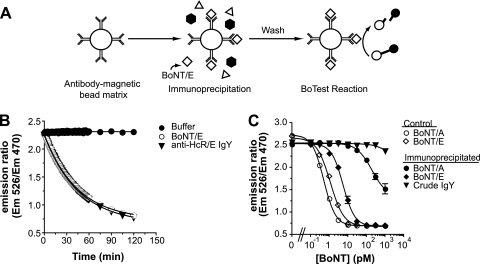
We used BoNT/E-specific antibodies coupled to magnetic beads as the immunoprecipitation reagent to capture BoNT/E and separate it from molecules that might interfere with BoNT activity determinations in vitro. Following immunoprecipitation or pull-down, captured BoNT catalytic activity can then be quantified using BoTest reporters (Fig. 3A). We screened 19 different monoclonal (n = 15) and polyclonal (n = 4) antibody preparations in an attempt to find antibodies with the affinity and specificity required to detect picomolar levels of BoNT/E. Most antibody preparations failed to efficiently immunoprecipitate less than 1 nM BoNT/E and were considered inappropriate for assay development (data not shown). Desired assay performance was achieved using a polyclonal antibody against the heavy-chain receptor binding domain of BoNT/E (HcR/E) (4). This IgY preparation was generated by immunizing chickens against HcR/E and did not interfere with BoNT/E-dependent cleavage of the BoTest E reporter (Fig. 3B). The anti-HcR/E IgY was then conjugated to magnetic beads and tested for its ability to capture and isolate BoNT/E. The anti-HcR/E IgY-coated beads effectively immunoprecipitated BoNT/E, as determined by measuring the proteolytic activity of the captured BoNT/E using the BoTest A/E reporter (Fig. 3C). The BoNT/E activity that immunoprecipitated with crude IgY (from nonimmunized hens)-coated beads did not fall within the calculated limits of detection of the assay. The BoNT/E EC50 for the assay using the anti-BoNT/E beads was 5.6 pM, less than a 0.5 log difference from the EC50 of the BoNT/E control assay (1.2 pM), the theoretical sensitivity limit for the assay.
Fig. 3.
Development of the BoTest Matrix E assay. (A) Schematic representation of the BoTest Matrix E design. Antibodies are conjugated to magnetic beads and used to immunoprecipitate BoNT/E from complex samples. Interfering substances are then washed away, and captured BoNT/E activity is quantified with the BoTest reporter. (B) Antibody interference. BoNT/E (100 pM) was incubated with or without 10 μg/ml anti-HcRE IgY antibody for 30 min at 25°C. BoTest E reporter was then added and fluorescence emissions were measured at 30-s intervals for 60 min and then at 15-min intervals for a total of 120 min. The emission ratio was plotted as a function of time. (C) Anti-HcRE IgY specificity. BoNT/A or BoNT/E was incubated with either anti-HcRE IgY- or preimmune crude IgY-coated magnetic beads for 2 h at 25°C at the indicated BoNT concentrations (closed symbols). Beads were then washed and incubated with BoTest A/E. Reactions for immunoprecipitated samples were compared to control BoNT/A and BoNT/E holotoxin reactions, where BoNT was directly incubated with the BoTest reporters without first being subject to immunoprecipitation (open symbols). After 4 h, fluorescence emissions were collected and the ratios of the emissions were plotted as a function of BoNT concentration.
We assessed the specificity of anti-HcR/E IgY using antibody-coated beads incubated with BoNT/A, followed by activity determination with BoTest A/E (Fig. 3C). The anti-HcR/E IgY-coated beads captured BoNT/A at concentrations of >300 pM, based upon detection using the BoTest A/E reporter. Nonspecific capture of BoNT/A by anti-HcR/E IgY, however, was circumvented using the BoNT/E-serotype specific reporter, BoTest E. Anti-HcR/E IgY-coupled bead-based immunoprecipitation followed by activity determination with the BoTest E serotype-specific reporter is referred to here as the BoTest Matrix E assay.
Performance specifications of BoTest Matrix E assay.
We tested the ability of the BoTest Matrix E assay to measure toxin activity in human serum and whole chicken blood spiked with BoNT/E at various concentrations. Using the BoTest Matrix E assay, BoNT/E was immunoprecipitated from PBS-T, serum, and blood (Fig. 4A). A slight shift in pull-down efficiency was seen with the blood and serum samples. Overall, however, assay sensitivity was largely unaffected by sample composition, indicating the suitability of the assay for use with avian blood samples.
Fig. 4.
Assessment of sensitivity of BoNT/E assay BoTest Matrix E. (A) BoNT/E was diluted in either PBS-T (0.1% Tween 20), human serum, or whole chicken blood and then immunoprecipitated with anti-HcRE IgY-conjugated beads for 2 h at 25°C. Beads were then incubated with the BoTest E reporter at 37°C for 2 h before collecting fluorescence emissions. (B) The sensitivity of the BoTest Matrix E activity assay is dependent on the incubation time of bead-captured BoNT/E with the BoTest reporters. The indicated concentrations of BoNT/E were immunoprecipitated with anti-HcRE IgY-coated beads for 2 h at 25°C. Beads were then washed and incubated with the BoTest E reporter. Fluorescence emissions were collected at the indicated time points, and the ratios of the emissions were plotted as a function of BoNT concentration. (C) Performance parameters of the BoTest Matrix E assay. EC50s, LODs, and LOQs were calculated as described in Materials and Methods. Percent coefficients of variation (CV) are shown.
We investigated time-dependent toxin activity and assay sensitivity using BoNT/E holotoxin immunoprecipitated from PBS-T, followed by activity determination with the BoTest E reporter at multiple time points (Fig. 4B). Assay BoNT/E EC50s decreased over time from 157.6 pM at 2 h to 3.6 pM at 24 h. The dilution curve at 2 h indicated a quantitative range of ∼26 to 1,000 pM with an LOD of 6.4 pM, whereas the 24-h reading had a quantitative range of ∼1.4 to 10 pM with an LOD of 500 fM (Fig. 4C). Thus, assay performance was dependent on the time that captured BoNT/E was incubated with the BoTest E reporter, demonstrating the ability to tune the assay's sensitivity using reaction time. Assay precision was quantified using samples composed of 50 pM BoNT/E suspended in chicken blood compared to a standard curve composed of BoNT/E diluted into PBS-T. Intra- and interassay CVs were 9.6% and 2.4%, respectively (Fig. 4C).
Qualification of BoTest Matrix E assay with avian blood samples.
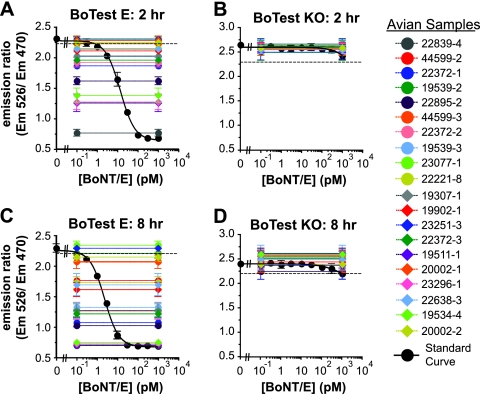
The NWHC investigates suspected wildlife avian BoNT intoxication events in the United States. We used the BoTest Matrix E activity assay to test archived avian blood samples previously determined by the NWHC to contain either BoNT/C or BoNT/E using the mouse lethality assay. The BoNT/E activity in a subset (n = 20) of the total avian blood samples was quantified using a BoNT/E holotoxin standard, as shown in Fig. 5. Readings from the standard curve were fitted with the sigmoidal dose-response equation described in Materials and Methods, to which emission ratios taken from blood samples were compared and solved for concentration (Fig. 5A and C). To rule out nonspecific proteolytic cleavage of the reporter and to confirm the presence of BoNT/E in the samples, the activities of immunoprecipitates were also tested using BoTest KO (Fig. 5B and D). At the 2-h reading, 11 of the 20 total samples had significantly cleaved the BoTest E reporter (Fig. 5A). When measurements were taken after 8 h of incubation, 18 of the 20 immunoprecipitated samples had cleaved the BoTest E reporter and were thus considered positive for BoNT/E (Fig. 5C). None of the immunoprecipitated avian blood samples significantly cleaved the BoTest KO reporter at either time point, indicating that the quantified protease activity was specific to BoNT/E (Fig. 5B and D).
Fig. 5.
Detection of BoNT/E in avian blood samples using the BoTest Matrix E assay. Avian blood samples were tested for BoNT/E using the BoTest Matrix E assay as described in Materials and Methods. Immunoprecipitated BoNT/E was quantified using the BoTest E reporter (A and C) or the BoTest KO reporter (B and D) to control for nonspecific proteases. The resulting emission ratio for each sample was compared to a standard curve consisting of known amounts of BoNT/E (black circles). Representative data are shown from readings following a 2-h (A and B) or 8-h (C and D) incubation of capture BoNT/E with the reporters. LODs were calculated as described in Materials and Methods and indicated with dashed lines. Samples with emission ratio values below calculated LOD values were considered positive for BoNT/E.
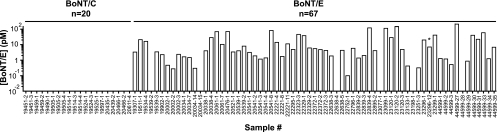
A total sample set of 67 mouse lethality assay-confirmed BoNT/E-positive samples from 18 different avian species was tested for BoNT/E using the BoTest Matrix E assay (Fig. 6). An additional set of 20 mouse lethality assay-confirmed BoNT/C-positive samples was used as a BoNT/E-negative control. The BoTest Matrix E assay confirmed the positive test results previously generated by the mouse lethality assay for 61 of the 67 BoNT/E-positive samples. One sample (23296-12) that was identified to contain BoNT/E by the mouse assay cleaved both the BoTest E and BoTest KO reporters and was thus identified to be equivocal and eliminated from further analyses. All BoNT/C-containing blood samples (n = 20) tested negative for BoNT/E using the BoTest Matrix E assay. The diagnostic sensitivity and specificity were determined by comparison of the BoTest Matrix E assay results to those of the mouse lethality assay. The BoTest Matrix E assay performed with an overall diagnostic sensitivity of 91.0% and specificity of 100%, with the BoTest Matrix E assay providing positive results for BoNT/E for 61 samples, negative results for 26 samples, false-negative results for 6 samples, and the equivocal result for 1 sample. None of the samples had false-positive results.
Fig. 6.
Quantification of BoNT/E activity in avian blood samples. Samples confirmed to contain BoNT/C (n = 20) and BoNT/E (n = 67) by the mouse lethality assay were tested using the BoTest Matrix E assay as described in the legend to Fig. 5. The resulting emission ratio value for each sample was compared to the standard curve and solved for an apparent concentration of BoNT/E (pM) as described in Materials and Methods. The asterisk denotes a sample that cleaved both the BoTest E and BoTest KO reporters and was thus determined to be equivocal and excluded from further analysis.
We also investigated the correlation between BoNT/E activity determined by BoTest Matrix E assay and the BoNT/E protein concentration based upon a sandwich ELISA (Fig. 7A) using a subset of samples (n = 14). High BoTest Matrix E activity relative to BoNT/E concentration would indicate activity from substances other than BoNT/E, such as nonspecific proteases. Conversely, low activity determination relative to total BoNT/E concentration would indicate a loss of BoNT/E function.
Fig. 7.
BoNT/E concentration correlates with toxin activity in avian blood samples. (A) BoNT/E protein concentration was measured by sandwich ELISA as described in Materials and Methods. Avian blood samples (colored circles) were compared to a standard curve of BoNT/E at the indicated concentrations (black circles). LOQs are indicated with dashed lines. (B) Avian blood samples containing BoNT/E protein concentrations within the LOQ as determined by sandwich ELISA (A) were plotted against BoNT/E activity measurements as determined by the BoTest Matrix E assay (Fig. 6). The slope of the linear regression was 1.224 ± 0.07 (R2 = 0.972).
Avian blood sample values that fell within the quantitative range for both the BoTest Matrix E activity assay and the sandwich ELISA (n = 11) were plotted (Fig. 7B) and fit by linear regression. We found a linear correlation between BoNT/E activity and BoNT/E concentration with a slope of 1.2 (R2 = 0.972; Fig. 7B), demonstrating that the activity quantified by the BoTest Matrix E assay correlates with total BoNT/E contained in the avian blood samples. The BoTest Matrix E assay is ∼100-fold more sensitive than the sandwich ELISA, which limited the number of samples that could be compared by both assays.
DISCUSSION
Development of a sensitive, in vitro, high-throughput diagnostic test to detect BoNTs is imperative to facilitate ecological studies to better understand and manage botulism outbreaks in free-ranging wildlife populations and to enhance public health surveillance capabilities. The current gold standard test for detecting BoNT is a mouse bioassay which is sensitive but requires animal-handling facilities, takes up to 7 days to complete, and is highly variable when used to quantify BoNT activity (CVs, 20 to 40% [42]). The inaccuracy of the mouse lethality assay for quantitation of toxin can be addressed by using large numbers of animals (>100) per sample analyzed (38, 42), but this is expensive and presents animal welfare concerns.
Ruge et al. recently reported the use of fluorogenic substrates to develop high-throughput and sensitive assays to detect BoNT (37). These substrates, called the BoTest reporters, provide femtomolar-level BoNT endopeptidase quantification in a time-tunable manner. However, substances such as albumins and salts found in complex matrixes such as blood and serum interfere with BoNT activity in vitro, limiting the utility of BoTest reporters as stand-alone reagents (Fig. 2). To circumvent problems with assay interference, we coupled immunoprecipitation purification techniques with BoTest reporter activity measurements. The resulting BoTest Matrix E assay isolates and concentrates BoNT/E from interfering molecules and enables subsequent high-sensitivity measurement of BoNT/E activity. BoTest Matrix E detected femtomolar quantities (less than 30 pg) of BoNT/E in avian blood in less than 24 h, similar to the detection limits of the mouse lethality assay (Fig. 4). In addition, the BoTest Matrix E assay, when used along with a control substrate (BoTest KO), distinguished between BoNT/E endopeptidase activity and the activity of nonspecific proteases commonly found in biological samples and reduced the potential number of false-positive samples (Fig. 5).
The BoTest Matrix E assay was able to detect and quantify BoNT/E in avian blood samples from 18 different species containing variable toxin burdens (Fig. 6) under various postmortem conditions, ranging from freshly euthanized birds to partially scavenged carcasses that had been dead for 1 to 3 days. In addition, these archived samples had been frozen at −70°C for up to 6 years prior to BoTest Matrix E assay testing. In spite of this high variability among samples, the BoTest Matrix E assay accurately identified BoNT/E in archived avian blood samples with high specificity (100%) and sensitivity (91%) compared to the mouse lethality assay, demonstrating assay robustness and utility (Table 1).
Of the five blood samples identified to have false-negative results by the BoTest Matrix E assay, three samples originated from birds that had been euthanized and thus may have harbored sublethal levels of BoNT, below the detection limits of the BoTest Matrix E assay. Studies directly comparing limits of detection for the mouse lethality and the BoTest Matrix E assays would further define in vitro assay performance capabilities. Lastly, adherence to more stringent collection protocols that limit postmortem decay of carcasses may further reduce assay variability.
Environmental factors that contribute to large-scale BoNT/E outbreaks are not well understood. BoNT/E outbreaks in the Great Lakes region typically occur between June and December and correlate with low mean annual water levels and increased surface water temperatures (25). It has been hypothesized that BoNT/E is mobilized through Great Lakes aquatic food webs consisting of exotic species (19, 25, 32, 33), providing a potential link between sediment-inhabiting clostridia and the intoxication of fish-eating birds. Using the BoTest Matrix E assay to conduct enhanced epidemiological investigations of avian BoNT/E outbreaks, including the analysis of aquatic food web components, would aid in our understanding of toxin mobilization pathways and provide critical insights for the management and conservation of bird species in the Great Lakes region.
The BoTest Matrix E assay also has potential application for human health diagnostics. In humans, BoNT/E intoxication can result from ingestion of improperly prepared fish products and home-canned foods (8, 47). In the United States, approximately 25% of all reported food-borne botulism outbreaks are attributed to BoNT/E (43). In Canada and Alaska, approximately 90% of food-borne botulism cases are attributed to BoNT/E (28), with an incidence rate of ∼1 per 2,000 people in some areas of northern Canada (26), where the consumption of fish and other meats aged according to native customs is prevalent (3). Rapid diagnostic tests such as BoTest Matrix E using equipment commonly found in biochemistry, toxicology, and diagnostic laboratories provide the potential to decentralize botulism testing, reduce the time to diagnosis, and speed initiation of medical countermeasures (43).
Our goal in designing an in vitro diagnostic assay for detecting BoNT/E was to maintain mouse lethality assay sensitivity while improving throughput and speeding time to diagnosis. The BoTest Matrix E assay provides an accurate and reproducible in vitro method of quantifying toxin activity in complex biological samples that can be modified on the basis of user requirements to account for sample volume, composition, and toxin load. With sample-specific protocol optimization and validation, the BoTest Matrix E assay will be applicable to a wide range of environmental, food, and human health applications and provide insight into BoNT disease prevention and management.
ACKNOWLEDGMENTS
We thank Daniel Ruge (BioSentinel Pharmaceuticals Inc.) for preparation of the BoTest A/E reporter. We also thank T. Rocke (USGS) and S. Riley (USGS) for helpful comments during the preparation of the manuscript.
This project was funded by the U.S. Geological Survey Midwest Area (to NWHC), the Great Lakes Restoration Initiative (Template 73 to NWHC), the National Park Service Midwest Regional Office (interagency agreement F6620090039 to the University of Wisconsin—Madison), the Wisconsin Department of Commerce (contract FY09-19294-0602609 to BioSentinel Pharmaceuticals Inc.), and the U.S. Department of Defense (contract W81XWH-07-2-0045 to BioSentinel Pharmaceuticals Inc.). This research was conducted in accordance with Cooperative Research and Development Agreement C-06-349 between the U.S. Geological Survey and BioSentinel Pharmaceuticals Inc.
Use of trade, product, or firm names does not imply endorsement by the U.S. government.
T. Piazza, M. Dunning, F. Zeytin, and W. Tucker are employees or owners of BioSentinel Inc. BioSentinel currently manufactures some of the reagents presented in this report and intends to commercialize some or all of the methods presented here.
Footnotes
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Allison S. 2004. Fluid, electrolytes and nutrition. Clin. Med. 4:573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. AOAC International 2001. AOAC official method 977.26 (sec. 17.7.01). Clostridium botulinum and its toxins in foods. In Official methods of analysis, 17th ed AOAC International, Gaithersburg, MD [Google Scholar]
- 3. Austin J. W., Leclair D. 2011. Botulism in the north: a disease without borders. Clin. Infect. Dis. 52:593–594 [DOI] [PubMed] [Google Scholar]
- 4. Baldwin M. R., et al. 2005. Characterization of the antibody response to the receptor binding domain of botulinum neurotoxin serotypes A and E. Infect. Immun. 73:6998–7005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barr J. R., et al. 2005. Botulinum neurotoxin detection and differentiation by mass spectrometry. Emerg. Infect. Dis. 11:1578–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blasi J., et al. 1993. Botulinum neurotoxin A selectively cleaves the synaptic protein SNAP-25. Nature 365:160–163 [DOI] [PubMed] [Google Scholar]
- 7. Blasi J., et al. 1993. Botulinum neurotoxin C1 blocks neurotransmitter release by means of cleaving HPC-1/syntaxin. EMBO J. 12:4821–4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bott T. L., Deffner J. S., McCoy E., Foster E. M. 1966. Clostridium botulinum type E in fish from the Great Lakes. J. Bacteriol. 91:919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boyer A. E., et al. 2005. From the mouse to the mass spectrometer: detection and differentiation of the endoproteinase activities of botulinum neurotoxins A-G by mass spectrometry. Anal. Chem. 77:3916–3924 [DOI] [PubMed] [Google Scholar]
- 10. Cai S., Singh B. R., Sharma S. 2007. Botulism diagnostics: from clinical symptoms to in vitro assays. Crit. Rev. Microbiol. 33:109–125 [DOI] [PubMed] [Google Scholar]
- 11. Chen S., Barbieri J. T. 2007. Multiple pocket recognition of SNAP25 by botulinum neurotoxin serotype E. J. Biol. Chem. 282:25540–25547 [DOI] [PubMed] [Google Scholar]
- 12. Chen S., Barbieri J. T. 2006. Unique substrate recognition by botulinum neurotoxins serotypes A and E. J. Biol. Chem. 281:10906–10911 [DOI] [PubMed] [Google Scholar]
- 13. Dong M., Tepp W. H., Johnson E. A., Chapman E. R. 2004. Using fluorescent sensors to detect botulinum neurotoxin activity in vitro and in living cells. Proc. Natl. Acad. Sci. U. S. A. 101:14701–14706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galvin J. W., Hollier T. J., Bodinnar K. D., Bunn C. M. 1985. An outbreak of botulism in wild waterbirds in southern Australia. J. Wildl. Dis. 21:347–350 [DOI] [PubMed] [Google Scholar]
- 15. Gaunt P. S., Kalb S. R., Barr J. R. 2007. Detection of botulinum type E toxin in channel catfish with visceral toxicosis syndrome using catfish bioassay and Endopep mass spectrometry. J. Vet. Diagn. Invest. 19:349–354 [DOI] [PubMed] [Google Scholar]
- 16. Getchell R. G., et al. 2006. Quantitative polymerase chain reaction assay used to measure the prevalence of Clostridium botulinum type E in fish in the lower great lakes. J. Aquat. Anim. Health 18:39–50 [Google Scholar]
- 17. Gilmore M. A., et al. 2011. Depolarization after resonance energy transfer (DARET): a sensitive fluorescence-based assay for botulinum neurotoxin protease activity. Anal. Biochem. 413:36–42 [DOI] [PubMed] [Google Scholar]
- 18. Gupta D., Lis C. 2010. Pretreatment serum albumin as a predictor of cancer survival: a systematic review of the epidemiological literature. Nutr. J. 9:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hannett G. E., Stone W. B., Davis S. W., Wroblewski D. 2011. Biodiversity of Clostridium botulinum type E associated with a large outbreak of botulism in wildlife from Lake Erie and Lake Ontario. Appl. Environ. Microbiol. 77:1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hines H. B., et al. 2008. Use of a recombinant fluorescent substrate with cleavage sites for all botulinum neurotoxins in high-throughput screening of natural product extracts for inhibitors of serotypes A, B, and E. Appl. Environ. Microbiol. 74:653–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoque M. A., Skerratt L. F., Rahman M. A., Beg A. B., Debnath N. C. 2010. Factors limiting traditional household duck production in Bangladesh. Trop. Anim. Health Prod. 42:1579–1587 [DOI] [PubMed] [Google Scholar]
- 22. Horowitz B. Z. 2005. Botulinum toxin. Crit. Care Clin. 21:825–839 [DOI] [PubMed] [Google Scholar]
- 23. Kalb S. R., et al. 2006. The use of Endopep-MS for the detection of botulinum toxins A, B, E, and F in serum and stool samples. Anal. Biochem. 351:84–92 [DOI] [PubMed] [Google Scholar]
- 24. Kumaran D., Rawat R., Ahmed S. A., Swaminathan S. 2008. Substrate binding mode and its implication on drug design for botulinum neurotoxin A. PLoS Pathog. 4:e1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lafrancois B. M., Riley S. C., Blehert D. S., Ballmann A. E. 2011. Links between type E botulism outbreaks, lake levels, and surface water temperatures in Lake Michigan, 1963-2008. J. Great Lakes Res. 37:86–91 [Google Scholar]
- 26. Leclair D., Pagotto F., Farber J. M., Cadieux B., Austin J. W. 2006. Comparison of DNA fingerprinting methods for use in investigation of type E botulism outbreaks in the Canadian Arctic. J. Clin. Microbiol. 44:1635–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lindstrom M., Korkeala H. 2006. Laboratory diagnostics of botulism. Clin. Microbiol. Rev. 19:298–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Loutfy M. R., Austin J. W., Blanchfield B., Fong I. W. 2003. An outbreak of foodborne botulism in Ontario. Can. J. Infect. Dis. 14:206–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neimanis A., et al. 2007. An outbreak of type C botulism in herring gulls (Larus argentatus) in southeastern Sweden. J. Wildl. Dis. 43:327–336 [DOI] [PubMed] [Google Scholar]
- 30. Nol P., Williamson J. L., Rocke T. E., Yuill T. M. 2004. Detection of Clostridium botulinum type C cells in the gastrointestinal tracts of Mozambique tilapia (Oreochromis mossambicus) by polymerase chain reaction. J. Wildl. Dis. 40:749–753 [DOI] [PubMed] [Google Scholar]
- 31. Peck M. W., Robert K. P. 2009. Biology and genomic analysis of Clostridium botulinum. Adv. Microb. Physiol. 55:183–265, 320 [DOI] [PubMed] [Google Scholar]
- 32. Pérez-Fuentetaja A., Clapsadl M. D., Einhouse D., Bowser P. R. G. R. G., Lee W. T. 2006. Influence of limnological conditions on Clostridium botulinum type E presence in eastern Lake Erie sediments (Great Lakes, USA). Hydrobiologia 563:189–200 [Google Scholar]
- 33. Pérez-Fuentetaja A., et al. 2011. Clostridium botulinum type E in Lake Erie: interannual differences and role of benthic invertebrates. J. Great Lakes Res. 37:238–244 [Google Scholar]
- 34. Rocke T. E., Friend M. 1999. Avian botulism. United States Geological Survey information and technology report 1999-2001. U.S. Geological Survey, Reston, VA [Google Scholar]
- 35. Ross J. A., et al. 2011. Characterization of Forster resonance energy transfer in a botulinum neurotoxin protease assay. Anal. Biochem. 413:43–49 [DOI] [PubMed] [Google Scholar]
- 36. Rossetto O., et al. 1994. SNARE motif and neurotoxins. Nature 372:415–416 [DOI] [PubMed] [Google Scholar]
- 37. Ruge D. R., et al. 2011. Detection of six serotypes of botulinum neurotoxin using fluorogenic reporters. Anal. Biochem. 411:200–209 [DOI] [PubMed] [Google Scholar]
- 38. Schantz E. J., Kautter D. A. 1978. Microbiological methods: standardized assay for Clostridium botulinum toxins. J. AOAC 61:96–99 [Google Scholar]
- 39. Schiavo G., Rossetto O., Benfenati F., Poulain B., Montecucco C. 1994. Tetanus and botulinum neurotoxins are zinc proteases specific for components of the neuroexocytosis apparatus. Ann. N. Y. Acad. Sci. 710:65–75 [DOI] [PubMed] [Google Scholar]
- 40. Schiavo G., et al. 1993. Identification of the nerve terminal targets of botulinum neurotoxin serotypes A, D, and E. J. Biol. Chem. 268:23784–23787 [PubMed] [Google Scholar]
- 41. Schiavo G., Shone C. C., Rossetto O., Alexander F. C., Montecucco C. 1993. Botulinum neurotoxin serotype F is a zinc endopeptidase specific for VAMP/synaptobrevin. J. Biol. Chem. 268:11516–11519 [PubMed] [Google Scholar]
- 42. Sesardic D., Leung T., Gaines Das R. 2003. Role for standards in assays of botulinum toxins: international collaborative study of three preparations of botulinum type A toxin. Biologicals 31:265–276 [DOI] [PubMed] [Google Scholar]
- 43. Shapiro R. L., Hatheway C., Swerdlow D. L. 1998. Botulism in the United States: a clinical and epidemiologic review. Ann. Intern. Med. 129:221–228 [DOI] [PubMed] [Google Scholar]
- 44. Sobel J. 2005. Botulism. Clin. Infect. Dis. 41:1167–1173 [DOI] [PubMed] [Google Scholar]
- 45. Woo G. H., et al. 2010. Outbreak of botulism (Clostridium botulinum type C) in wild waterfowl: Seoul, Korea. J. Wildl. Dis. 46:951–955 [DOI] [PubMed] [Google Scholar]
- 46. Work T. M., Klavitter J. L., Reynolds M. H., Blehert D. 2010. Avian botulism: a case study in translocated endangered Laysan ducks (Anas laysanensis) on Midway Atoll. J. Wildl. Dis. 46:499–506 [DOI] [PubMed] [Google Scholar]
- 47. Yule A. M., Austin J. W., Barker I. K., Cadieux B., Moccia R. D. 2006. Persistence of Clostridium botulinum neurotoxin type E in tissues from selected freshwater fish species: implications to public health. J. Food Prot. 69:1164–1167 [DOI] [PubMed] [Google Scholar]