Abstract
Members of candidate division OP11 are widely distributed in terrestrial and marine ecosystems, yet little information regarding their metabolic capabilities and ecological role within such habitats is currently available. Here, we report on the microfluidic isolation, multiple-displacement-amplification, pyrosequencing, and genomic analysis of a single cell (ZG1) belonging to candidate division OP11. Genome analysis of the ∼270-kb partial genome assembly obtained showed that it had no particular similarity to a specific phylum. Four hundred twenty-three open reading frames were identified, 46% of which had no function prediction. In-depth analysis revealed a heterotrophic lifestyle, with genes encoding endoglucanase, amylopullulanase, and laccase enzymes, suggesting a capacity for utilization of cellulose, starch, and, potentially, lignin, respectively. Genes encoding several glycolysis enzymes as well as formate utilization were identified, but no evidence for an electron transport chain was found. The presence of genes encoding various components of lipopolysaccharide biosynthesis indicates a Gram-negative bacterial cell wall. The partial genome also provides evidence for antibiotic resistance (β-lactamase, aminoglycoside phosphotransferase), as well as antibiotic production (bacteriocin) and extracellular bactericidal peptidases. Multiple mechanisms for stress response were identified, as were elements of type I and type IV secretion systems. Finally, housekeeping genes identified within the partial genome were used to demonstrate the OP11 affiliation of multiple hitherto unclassified genomic fragments from multiple database-deposited metagenomic data sets. These results provide the first glimpse into the lifestyle of a member of a ubiquitous, yet poorly understood bacterial candidate division.
INTRODUCTION
Culture-independent surveys conducted within the last 2 decades have convincingly demonstrated the presence of a large number of yet-uncultured microbial lineages (16, 33). Members of yet-uncultured bacterial lineages at the phylum (division) level have been termed candidate divisions (CDs) (32). While many of such lineages are globally distributed in marine and terrestrial habitats, little is currently known regarding their metabolic capabilities and ecological role within various ecosystems.
One of the fascinating challenges currently facing microbial ecologists is to decipher the physiological properties, energy conservation pathways, and ecological significance of yet-uncultured CDs. Environmental genomic approaches offer the opportunity for culture-independent, in silico interrogation of genomic fragments from yet-uncultured CDs. Further, information regarding the physiological properties and metabolic capabilities could also guide future targeted enrichment and isolation efforts. However, the majority of CDs often constitute a minor fraction within microbial communities and are typically encountered within highly diverse ecosystems (20, 54, 63). Therefore, metagenomic surveys of microbial communities rarely yield large, genetically informative genomic fragments that could confidently be assigned to CDs, a situation aggravated recently by the wide utilization of shorter-read-length sequencing technologies in metagenomic surveys (30, 38).
Targeted metagenomic environmental surveys using fosmid and bacterial artificial chromosome library construction and screening have proved useful in deciphering interesting insights into the genomics of novel CDs, in spite of the relatively small size of the insert being sequenced (55). Recently, the coupling of single-cell-separation and multiple-displacement-amplification (MDA) approaches allowed the assembly of larger genomic fractions from a single uncultured bacterial cell (14). We have previously reported the utilization of a 28-channel microfluidic laser tweezer device for the sorting of individual cells of ammonia-oxidizing archaea (6) and the automation of MDA reactions in nanoliter volumes (42, 43). The present device utilizes 32 microfabricated channels for cell sorting and amplification. The features of this single-cell genome amplification platform strongly suppress the principal modes of contamination. Sorting operations are carried out inside the sealed microfluidic device, preventing extrinsic contamination. The use of optical tweezers minimizes the sorting volume to the volume of the cell itself, preventing the introduction of sample-borne contaminants to the reaction. Finally, the small amplification volumes concentrate the single-cell template with respect to reagent-borne contaminants (7).
Here, we focus on one of the widely distributed, yet-uncultured candidate divisions: CD OP11. 16S rRNA clones belonging to OP11 were first detected in obsidian pool sediments in Yellowstone National Park (33). The subsequent identification of OP11 16S rRNA-affiliated sequences from multiple habitats led to the recognition of the polyphyletic nature of this group. Sequences originally described as belonging to OP11 are now classified as members of CDs OP11, OD1, and SR1 (27). OP11 has been observed in several marine (27, 54) and terrestrial (1, 16, 33) habitats, including soil (20). We have previously documented the presence of OP11 as a minor constituent within the highly diverse bacterial community encountered at the anoxic source of Zodletone Spring, an anaerobic sulfide and sulfur-rich spring in southwestern Oklahoma (19, 63). Subsequent single-cell isolation and MDA of cells from Zodletone Spring source sediments, followed by 16S rRNA gene sequencing of individual sorts, led to the identification of a single OP11 sort (here referred to as ZG1). We describe in this study the main genomic features of this single-cell partial genome, with emphasis on metabolic capabilities, cell structure, and various traits allowing the survival of this microorganism in such a highly diverse and competitive habitat.
MATERIALS AND METHODS
Sampling.
Sediment samples were collected from Zodletone Spring, an anoxic sulfide and sulfur-rich spring in southwestern Oklahoma. The hydrological and geochemical characteristics of Zodletone Spring, as well as its bacterial diversity and sulfur transformations, have been described previously (18, 53). Samples were obtained from the anoxic spring source (July 2009) and immediately mixed with the same volume of absolute ethanol for sample fixation. We avoided sample fixation with paraformaldehyde to overcome any downstream effects on microfluidic single-cell isolation and the subsequent multiple-displacement amplification. Fixed samples were stored on ice until they were brought back to the lab, where they were incubated at 4°C overnight. Samples were then centrifuged at high speed (14,000 × g for 10 min) to remove the ethanol. Cells were extracted from the sediment as described before (13). Briefly, the cell-containing sediments were vigorously shaken with phosphate-buffered saline (PBS; 154 mM NaCl, 1.69 mM KH2PO4, 5 mM Na2HPO4) containing 100 mM sodium pyrophosphate for 10 min to loosen the cells, followed by several steps of centrifugation with a gradual increase in centrifugation speed to remove as many sediment particles as possible. The sediment-free cells were finally suspended in PBS and used for microfluidic single-cell isolation.
Microfluidic device and cell sorting.
Thirty-two-channel microfluidic devices were produced by the Stanford microfluidics foundry (Fig. 1). These devices are similar to those previously described (6). The chip was pretreated for 10 min with 0.2% Pluronic F-127 polyol in 1× PBS, before it was filled with 1× PBS containing 0.01% Tween 20 and 0.01% Pluronic F-127 to reduce cell adhesion. Bovine serum albumin (BSA) was added to the treated cells to a final concentration of 0.1 mg/ml. Individual cells were separated from the bulk sample using the laser trap and through two valves in an air-lock configuration, opening one valve at a time to allow the trapped cell but not fluid flow to pass through. Each trapped cell was moved about 1 mm from the bulk sample to the reaction chamber using the laser trap.
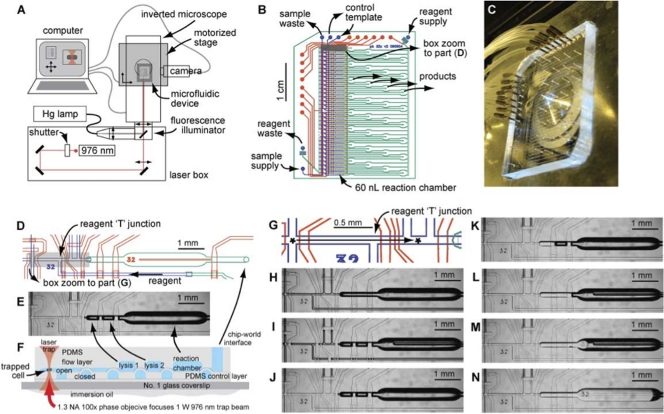
Fig. 1.
Optofluidic apparatus for isolating individual OP11 cells and amplifying their genomic contents. (A) Computer-controlled microscope fitted for fluorescence imaging and laser trapping. (B) Plan view of the two-layer 32-channel microfluidic chip used in this study. Control lines (25-μm depth, square profile, bottom layer) are shown in red, flow lines (10-μm depth, rounded profile, top layer) are shown in blue, and large channels/chambers (60-μm depth, rounded profile, top layer) are shown in green. (C) Photograph of the microfluidic chip with tubing to power control lines attached. (D) Plan view zoom of the box in panel B showing a single sorting/amplification channel. Cell suspension flows vertically in the blue channel at the left. Reagents are supplied to the indicated T junction from a supply line dedicated to 1 of the 32 reaction channels. Each reagent solution is flushed to the left from the T junction to backwash the blue channel, before being applied for the single-cell reaction by redirection to the right of the T junction. (E) Plan view micrograph of the chip region shown in panel D. (F) Elevation view (cross-sectional) schematic indicating components visible in panel E and layup of the microfluidic device. PDMS, polydimethylsiloxane. (G) Plan view zoom of the box in panel D showing the path by which cells are sorted using the optical trap. Cells traverse about 1.5 mm of channel containing clean buffer across two valves, which are opened sequentially to allow cells to pass. (H to N) Micrographs depicting device and MDA reaction setup. (H) Bare device with air-filled channels; (I) device with control lines filled with water (low-contrast channels) and pressurized (valves closed, visible where control channels cross air-filled flow lines); (J) device with reagent and sample lines prefilled with buffer (high-contrast channels, air; low-contrast channels, buffer); sorting takes place with this device configuration; (K) lysis chamber 1 (3.5-nl capacity) after reagent flush and dead-end fill; (L) lysis chamber 2 (3.5-nl capacity) after reagent flush and dead-end fill; (M) reaction chamber (60-nl capacity) initial filling by dead-end method after reagent flush; (N) reaction chamber with nearly complete dead-end fill.
Single-cell amplification.
Repli-G midi-MDA reagents (Qiagen, Valencia, CA) were used to amplify individual cells in 60-nl volumes on the device. First, cells contained in 0.75 nl PBS with 0.02% Tween 20 were flushed into the first lysis chamber with 3 nl Repli-G DLB buffer (supplemented with 0.1 M dithiothreitol) to complete lysis and denature the genomic DNA. Then, ∼50 nl of the reaction mix (43-μl aliquots were prepared from 29 μl Repli-G reaction buffer, 10 μl of 20 mM H2O with 0.6% Tween, 2 μl Repli-G enzyme, and 2 μl Repli-G stop solution) was added to each of the 32 reactions. The chip was then transferred to a hot plate set to 32°C and incubated overnight. The reaction volume was recovered by fitting the recovery ports on the chip with plastic pipette tips (P10 size) and by flushing the products into the pipette tips with the Tris solution plumbed into the reagent port at 8 lb/in2.
Phylogenetic identification of sorted cells.
DNA from MDA reactions of single-cell genomes was used in a PCR to amplify the 16S rRNA gene using 16S rRNA bacterial primers 27f (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1391r (5′-GACGGGCGGTGTGTRCA-3′). The PCRs were run in a 50-μl final volume containing 2 μl of a 1:10 dilution of the MDA reaction mixture as a template. The reaction mixture and PCR conditions utilized were described previously (20). PCR products were purified using an Invitrogen PureLink PCR purification kit (Invitrogen, Carlsbad, CA). Purified PCR products were sequenced at the Oklahoma State University core sequencing facility using the reverse primer.
Sequencing library creation and quantification.
Four microliters of the first-round reaction product identified as OP11 was reamplified using a Repli-G midi kit (Qiagen, Valencia, CA). This template solution was denatured by the addition of 3.5 μl buffer DLB for 5 min at room temperature and neutralized by addition of 3.5 μl stop solution. A reaction mix consisting of 29 μl reaction buffer, 10 μl water, and 1 μl enzyme was prepared on ice and then added to the denatured template. Reaction mixtures were incubated at 30°C for 12 h and then diluted 10-fold in 10 mM Tris with 0.02% Tween 20 and stored at −60°C.
A shotgun library was prepared from approximately 5 μg of the second-round MDA product according to the Roche/454 protocol for titanium shotgun libraries with the following modifications. Custom bar-coded adaptor oligonucleotides (Integrated DNA Technologies, Inc.) were used to enable pooling of multiple libraries in a single-emulsion PCR and picotiter plate region during sequencing. To obtain double-stranded DNA sequencing libraries and shorten the library preparation process, the library immobilization, fill-in, and single-stranded library isolation steps were omitted.
Sequencing libraries were quantified using digital PCR (dPCR) as previously reported (60), with the exceptions that 48.770 digital arrays (Fluidigm Corp., San Francisco, CA) were used for the microfluidic digital PCR step and that amplification primers complementary to the titanium adaptor sequences were used. Briefly, serial dilutions of the sequencing libraries were made in 10 mM Tris buffer with 0.02% Tween 20. Forty-eight sample preparations were combined according to the Fluidigm dPCR protocol with a reaction buffer containing thermostable DNA polymerase, deoxynucleoside triphosphates, GE sample-loading reagent (Fluidigm Corp., San Francisco, CA), and the primers and probe necessary to carry out the universal TaqMan (Applied Biosystems, Carlsbad, CA) amplification/detection scheme. The samples were loaded in the chip and run on a Biomark thermocycler (Fluidigm Corp., San Francisco, CA) for 45 cycles. Sample analysis was carried out using the default parameters for dPCR analysis using Fluidigm analysis software. The quantitated library was diluted to 2 × 106 molecules per microliter in 10 mM Tris with 0.02% Tween 20 and aliquoted for storage at −60°C. No normalization using duplex-specific exonuclease was attempted in this study (50).
Shotgun sequencing and assembly.
DNA pyrosequencing of the shotgun library was carried out on a Roche 454 Genome Sequencer FLX instrument using titanium chemistry. The OP11 library was sequenced in three runs of the instrument on beads prepared in emulsion PCRs with average DNA/bead ratios of 0.1:1, 0.3:1, and 0.5:1. A total of 77,493 reads (27,709,166 bases) were obtained. The G+C content of the single-cell reads formed a major peak centered near 42% GC. The dispersion of the read G+C content was typical of a single microbial genome sampled at the same average read length.
For assembly, reads from the pyrosequencing runs were separated by sample and trimmed using the sfffile tool (Roche), permitting one error in each 10-bp bar code, and assembled with the Newbler (version 2.3) program using default parameters, except for specifying the expected read depth at 100×.
Nucleotide content analysis.
We analyzed the nucleotide content of the OP11 contigs to assess the divergence from the nucleotide content profiles of sequenced microbes and check for contigs with possible outlying nucleotide content profiles that might represent contaminants. Three complete reference genomes were used for comparison: those of Escherichia coli O157:H7; another proteobacterium, Psychrobacter cryohalolentis K5; and the euryarchaeon Methanosarcina acetivorans. A custom Matlab code was used to calculate the frequency of pentanucleotides for overlapping 4,000-base samples from each organism and to carry out principal component analysis (PCA) of the pentanucleotide content of each organism. In this analysis, ZG1 contigs smaller than 4,000 bases were concatenated and sampled as described. In a separate analysis, the data sets were sampled as 200-mers, and the resulting PCA gave no evidence that short contigs constituted nucleotide frequency outliers (data not shown). To quantify the differences in pentanucleotide content between the 4 different microorganisms, distances between each PCA point representing a sequence belonging to one organism and every other PCA point representing a sequence belonging to each of the other three microorganisms were calculated and used to construct a histogram for each microorganism.
Genome annotation.
Two systems (the integrated microbial genome [IMG] and Artemis/blastp systems) were used for the partial genome annotation (see Table S1 in the supplemental material). First, contigs were uploaded on the IMG system (44) for automated gene calling and annotation. In addition, contigs were also uploaded on Artemis, release 12.0 (52), for open reading frame (ORF) prediction. ORFs longer than 50 amino acids were then compared (35) against the sequences in the nonredundant (nr) protein database with an E-value cutoff of 0.001 using the blastp system. The two systems annotated 241 ORFs similarly, including 49 conserved hypothetical proteins. Thirteen ORFs (3.1%) were annotated by the blastp system but not the IMG system, and 139 ORFs (32.9%) were annotated by the IMG system but not the blastp system; 93% of these were hypothetical proteins. The remaining 7% of ORFs were annotated differently by the two systems. Those ORFs were individually compared to all the genes in the IMG system using the blastp system with an E-value cutoff of 10−5. Those genes with similarity to a database entry were annotated as their first blastp hit (4.5% of all ORFs). The remaining 2.4% of genes had no sequence similarity to any of the genes in the IMG system database and hence were annotated as hypothetical proteins unique to the ZG1 genome. The final annotation has been manually curated in the IMG system (taxon object identification [ID] number 2503283010) and provided in Table S2 in the supplemental material. The gene ID numbers from the IMG system final annotation are utilized throughout this report.
Genomic analysis.
For pathways analysis, we referred to the KEGG database (36). In cases where more specific details were required (e.g., subsets of major pathways), we used the MetaCyc database (12). For proteins that were not part of a pathway (e.g., domain- or repeat-containing proteins), we used the InterPro (2) and the pfam (21) databases. tRNAs were predicted using the tRNA-scan SE search server (41). Carbohydrate-active enzymes were classified according to the CAZY database (9). To identify all the potential proteases and peptidases, the ORFs were compared to the sequences in the Merops database (http://merops.sanger.ac.uk/) (49) using the blastp system (35). For identification of repeats, we used the tandem repeat finder (5), inverted repeat finder (58), CRISPR finder online program (http://crispr.u-psud.fr/Server/), and IS finder (http://www-is.biotoul.fr/). To identify potential noncoding RNAs (ncRNAs), all intergenic regions (IGRs) were identified and extracted. IGRs were defined as any sequence longer than 50 bp that was not annotated by the IMG system and was not part of an ORF with an E-value of <0.001 with the blastp system. All IGRs were compared to the sequences in the NCBI refseq genomic database using the blastn system (35). IGRs were also searched against the Rfam database (22) to locate ncRNAs and cis-regulatory structured RNAs.
Mining for OP11 sequences in metagenomic data sets.
We used the 16S rRNA gene sequence within the ZG1 partial genome to query published and database-deposited metagenomes in the IMG/M system database and NCBI environmental samples (env_nt) database using the blastn system. For each of the first 100 hits, the corresponding 16S rRNA gene was extracted and classified using the Greengenes database (15).
Next, we attempted to identify metagenomic fragments belonging to members of candidate division OP11 in metagenomic data sets available via the IMG/M system, as well as in the global ocean survey (GOS) data set (GenBank accession number AACY000000000), using a fragment-recruiting process (see Table S3 in the supplemental material for a list of all metagenomic data sets used in this study). To ensure that the fragments identified truly belong to OP11, we opted for a stringent approach rather than the high-throughput, less stringent approach previously described (51, 62). Only housekeeping genes rather than all genes identified in the assembly were utilized in the process, since the hypothetical and conserved hypothetical proteins encountered could not be confidently considered specific gene biomarkers for OP11 and since some of the genes identified in ZG1 might be plasmid mediated and/or might have recently been acquired via horizontal gene transfer. Genes utilized were those for ribosomal protein L31, ribosomal protein L9, ribosomal protein S2, ribosomal protein S6, ribosomal protein S18, ribosomal protein S21, dephospho coenzyme A (CoA) kinase, arginyl-tRNA synthetase, seryl-tRNA synthetase, prolyl-tRNA synthetase, leucyl-tRNA synthetase, glycyl-tRNA synthetase, cysteinyl-tRNA synthetase, homoserine kinase, nucleotide diphosphate kinase, thymidylate kinase, thymidylate synthase, deoxycytidylate deaminase, preprotein translocase subunit SecA, preprotein translocase subunit YidC, DNA-directed RNA polymerase (beta subunit/140-kDa subunit), and DNA-directed RNA polymerase (beta′ subunit/160-kDa subunit).
In addition to the selective utilization of ZG1 housekeeping genes in fragment recruiting, a three-tiered approach for phylogenetic inference was implemented. First, we used an E-value cutoff corresponding to each protein's blastp first hit E value against the nr database (blastp system first hits in the nr database were always genes from sequenced bacterial genomes belonging to different microbial phyla). Proteins with E values lower than those of the closest genomic hits were extracted from the IMG/M system database and aligned to a set of corresponding gene products from 44 other complete genomes available in the IMG system database (44), including TM7, the closest OP11 relative with available partial genomes (for a list, see Table S3 in the supplemental material), using the ClustalX program, version 2.0 (40). We then constructed phylogenetic trees (parsimony, distance, neighbor joining) using the PAUP program (version 4.01b10; Sinauer Associates, Sunderland, MA) to determine whether the metagenome-derived sequences are monophyletic, with reproducible bootstrap support, to the ZG1 gene product but not to the TM7 gene product or the product of any other lineage. Finally, the metagenomic gene sequences (75 total) that were closer to the ZG1 genome than any of the other 44 genomes (lower E values) and that formed a monophyletic branch with ZG1 genes were finally compared to all genome sequences within the IMG system database (including ZG1 genome) using the blastp system. Only sequences with ZG1 genes as their first hit with the highest score, percent identity, and lowest E value compared to those for sequences of all other complete genomes were considered to be of OP11 origin. For those putative OP11 sequences, the corresponding scaffolds were extracted from the IMG/M system database to study the annotation of neighboring genes in the genomic fragment.
Nucleotide sequence accession number.
The sequence and annotation of the ZG1 partial assembly are available at the IMG system. The taxon object ID number is 2503283010.
RESULTS
Single-cell sorting and phylogenetic affiliation of ZG1.
A single cell belonging to OP11 (ZG1) was obtained during a general cell sorting effort with Zodletone Spring source sediments. The sorted cell was a small coccus less than 0.5 μm in diameter (Fig. 2A). To rule out gross contamination of the MDA-amplified DNA from the targeted cell prior to DNA pyrosequencing, the PCR product obtained using 16S rRNA general bacterial primer pair 27f and 1391r was cloned and 12 clones were sequenced. All clones gave identical 16S rRNA OP11-affiliated sequences.
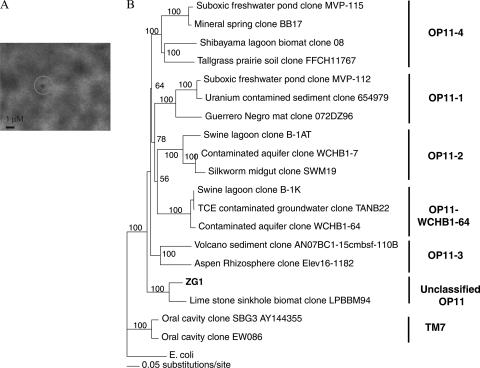
Fig. 2.
(A) Phase-contrast micrograph of candidate division OP11 ZG1 cell. (B) 16S rRNA gene tree highlighting the phylogenetic affiliation of the ZG1 cell. Bootstrap values (expressed as percentages) are based on 1,000 replicates and are shown for branches with more than 50% support. The evolutionary distance-based tree was constructed with the neighbor-joining algorithm with the Jukes-Cantor corrections. TCE, trichloroethylene.
Using the Greengenes phylogenetic framework, OP11 is divided into 5 different classes (OP11-1 to OP11-4 and WCHB1-64), with multiple additional sequences within OP11 unaffiliated with any of these 5 classes. ZG1 did not belong to any of the 5 established OP11 lineages. The ZG1 closet relative was a clone from phreatic limestone sinkholes in northeastern Mexico (GenBank accession number FJ902078; Fig. 2B).
Genome assembly, estimated size, annotation, and general features.
For a total of 77,493 reads, 27,709,166 bases were obtained from a single pyrosequencing run (average read length, 357 bp). Assembly resulted in a total of 269,392 bp of nonredundant sequences in 170 contigs, ranging in size from 112 to 11,464 bp (average, 1,589 bp; N50, 3,715 bp for contigs of >500 bp). The average depth of coverage in our assembly is 95.4, with a standard deviation of 184. Only 22 out of a core gene list of 182 housekeeping genes (45) were encountered in ZG1. By extrapolation, a crude genome size estimate of 2.25 Mbp could be suggested. The relatively small proportion of genome obtained in spite of the high coverage (95.4×) suggests that MDA bias is responsible for the incomplete assembly obtained.
Nucleotide content analysis.
We analyzed the pentanucleotide content of the ZG1 contigs to assess the divergence from the nucleotide content profile of sequenced microbes and check for contigs with possible outlying nucleotide content profiles that might represent contaminants. Nucleotide content analysis was conducted to test the putative occurrence of contamination in the ZG1 assembly. The results (Fig. 3), presented as a PCA (Fig. 3A), show that the ZG1 sequences form a cohesive cluster distinct from a bacterium (Psychrobacter cryohalolentis K5) and an archaeon (Methanosarcina acetivorans) with nearly identical GC contents (42%). We quantified the pentanucleotide content distinction among the cluster of sequences sampled from each organism by plotting histograms of the distances between points in the 3-dimensional PCA space. The ZG1 cluster is the most distinct within this set, exhibiting a greater separation from both P. cryohalolentis and M. acetivorans than that found between any of the others and its closest neighbor in the PCA space. This is evident in the scarcity of distances measured to be less than 10 units between ZG1 PCA points and points from the other organisms (Fig. 3B). This indicates the highly unique nature of the ZG1 sequence set and provides strong evidence that this sequence set is not contaminated with sequences from another organism.
Fig. 3.
Pentanucleotide content of ZG1 versus that of sequenced bacteria and archaea. (A) PCA of pentanucleotide content of ZG1, P. cryohalolentis K5, E. coli O157:H7, and M. acetivorans. (B to E) Distance histograms of sequences from each organism present in panel A to sequences from the other three organisms. Units are normalized variance and can be compared within and across panels B to E.
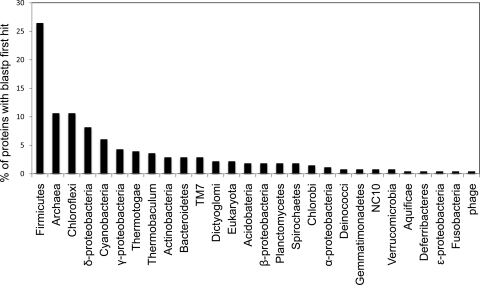
Table 1 summarizes the general genomic features of ZG1. A total of 427 genes were identified, with 423 protein-coding genes and 4 tRNA genes corresponding to codons for alanine (2 anticodons), proline, and arginine (for detailed annotation results obtained using the IMG and blastp systems, see Table S1 in the supplemental material). Almost 45.4% of the protein-coding genes had no function prediction (27.6% of those were conserved hypothetical proteins with similarities in the databases, and 72.4% were unique to the ZG1 genome with no similarities in the databases). This value is relatively high compared to that for an average genome from well-characterized phyla (e.g., 14.73% for Escherichia coli K-12 and 27.62% for Bacillus licheniformis ATCC 14580). Indeed, such a high proportion of hypothetical proteins has been reported in only a few (but not all) CD genomes, e.g., 54.6% for CD TM7 (43) and 48% for CD WWE1 (48). ZG1 also has the lowest percentage of protein-coding genes assigned to clusters of orthologous groups (COGs; 41%), and protein families (pfams; 49%), even compared to genomes of other CDs (e.g., 53% and 58% for TM7 [43], 63% and 67% for WWE1 [48], and 80% and 81% for TG1 [31]). Annotated proteins encoded by the ZG1 genome show no particular similarity to genomes within a specific phylum (Fig. 4), with the higher percentage of the Firmicutes and Proteobacteria first hits probably being a reflection of the larger proportion of published genomes belonging to these two phyla. No CRISPR elements were encountered in the partial genome of ZG1. Finally, housekeeping protein trees confirm the unique phylogenomic position of ZG1 compared to other bacterial phyla (see Fig. S1 in the supplemental material).
Table 1.
ZG1 genome general features
| Feature | Value |
|---|---|
| Total no. of bases | 269,392 |
| No. (%) of bases coding DNA | 227,142 (84) |
| % G+C content | 42.16 |
| No. of ORFs | 423 |
| No. of tRNA genes | 4 |
| No. of ORFs with function prediction | 230 |
| No. of ORFs without function prediction | 193 |
| No. of ORFs with COGs | 172 |
| No. of ORFs with pfams | 206 |
| No. of ORFs with signal peptide | 94 |
| No. of ORFs with transmembrane domains | 113 |
| Avg length of ORFs (no. of amino acids) | 187 |
Fig. 4.
Phylogenetic affiliation of ZG1 first hits for all genes identified within the partial ZG1 genome.
Genomic analysis of various pathways/processes within ZG1 partial genome.
The ZG1 partial genome was analyzed for clues regarding various metabolic and regulatory pathways. In spite of the relatively small assembly, various valuable insights regarding the cell wall structure, metabolic capabilities, secretory pathways, and defense mechanisms of ZG1 were gained. Important features pertaining to the lifestyle of ZG1 are described below. A more complete description of the genomic features of ZG1 is provided as supplementary text (see Text S1 in the supplemental material).
(i) Catabolism: evidence for plant polymer degradation and sugar fermentation capabilities.
Multiple ZG1 genes suggesting its ability to metabolize multiple plant polymers, as well as glucose (and possibly galactose), via fermentation were identified. The ZG1 genome encodes a cellulase A enzyme (IMG system gene ID number 2503336224), also known as glycoside hydrolase family 5 (GH5) (9). Cellulase A is an endoglucanase that hydrolyzes β-1→4 internal bonds to disrupt the crystalline structure of cellulose and expose individual cellulose polysaccharide chains. The cellulase A gene identified contained only a catalytic module and not a cellulosomal dockerin or carbohydrate-binding modules. This suggests free extracellular production, similar to what has typically been observed in cellulolytic Gram-negative bacteria.
The ZG1 partial genome also encodes a GH57 amylopullulanase enzyme (IMG system gene ID number 2503335832) (9), members of which are known to hydrolyze both the α-1→4 (amylase activity) and the branch α-1→6 (pullulanase activity) glucosidic linkage of starch. The end products of starch metabolism would be glucose, maltose, and maltotriose (64). The enzyme maltose-O-acetyltransferase is encoded by the genome and potentially indicates maltose-utilizing capabilities.
Surprisingly, the ZG1 partial genome also encodes a multicopper polyphenol oxidoreductase (laccase) (IMG system gene ID number 2503336172). Laccases are known to be involved in aerobic (oxygen-dependent) metabolism of lignin and are mainly present in fungi and aerobic bacteria (37). While their presence in a microorganism from an anaerobic habitat is peculiar, the facultative nature of ZG1 cannot be ruled out, and so this phenol oxidase activity of that enzyme might be involved in the digestion of lignin by OP11 in the presence of oxygen. It is interesting to note that ZG1 laccase belongs to the DUF152 family. This is a recently described family of laccases related to conserved hypothetical proteins harboring the domain DUF152. The first laccase activity within members of this family was reported after the enzyme was mined from a metagenomic library from an anaerobic rumen ecosystem (4). Therefore, it seems that these DUF152 structures appear to be the hallmark of laccases from anoxygenic/hypoxic habitats.
The ZG1 partial genome encodes only two of the reversible enzymes of the Embden-Meyerhof pathway, DhnA-type fructose-1,6-bisphosphate aldolase (class I; IMG system gene ID number 2503336037) and phosphoglycerate mutase (IMG system gene ID number 2503335845), both of which are also involved in gluconeogenesis. None of the irreversible glycolytic enzymes are present in the partial genome. However, one of the irreversible gluconeogenic enzymes, fructose-1,6-bisphosphatase class II (GlpX type; IMG system gene ID number 2503336038), is also encoded by the partial genome. Since the genome encodes two carbohydrolases (a cellulase and an amylopullulanase for degrading cellulose and starch, respectively), with glucose being the ultimate end product of both enzymes, we suggest that ZG1 should be able to use glucose as a carbon and energy source. Hence, we believe that ZG1 can utilize glucose via glycolysis. In addition to glucose, genes suggesting galactose utilization capabilities were identified: a predicted kinase related to galactokinase and mevalokinase (IMG system gene ID number 2503336213) and a gene encoding UDP glucose-4-epimerase (IMG system gene ID number 2503336175) (GalE of the galactose metabolism pathway) (36).
No pyruvate-metabolizing enzyme was identified. However, the ZG1 partial genome encodes a cytoplasmic formate:hydrogen lyase α subunit (IMG system gene ID number 2503336078). Formate is most probably produced from pyruvate (end product of glycolysis) via pyruvate:formate lyase, suggesting a possible mixed-acid fermentation pathway for glucose utilization.
(ii) Cell wall machinery: evidence for a Gram-negative bacterial cell wall.
Several enzymes involved in biosynthesis of the cell membrane (glycerophospholipid, lipoprotein, glycolipids, hopanoids) and cell wall (peptidoglycan, lipoprotein, glycoprotein) are encoded within the ZG1 partial genome assembly (see Text S1 in the supplemental material). Of specific interest are genes encoding lipopolysaccharide (LPS) synthesis enzymes that were encountered, since their presence suggests that ZG1 has a Gram-negative bacterial cell wall. LPS is formed of three parts: a hydrophobic region known as lipid A anchored to the outer envelope of the cell, a polysaccharide core region, and a distal oligosaccharide repeat known as the O antigen. Of the five enzymes required for the biosynthesis of the ADP d-glycero-β-d-manno-heptose (56), one of the building blocks of the inner core of lipopolysaccharide, three are present in ZG1: sedoheptulose-7-phosphate isomerase (SIS domain), which converts sedoheptulose-7-phosphate to d-glycero-d-manno-heptose-7-phosphate; d-glycero-d-manno-heptose-7-phosphate kinase, which adds a 1-phosphate group to form the 1,7-bisphosphate; and the d-glycero-d-manno-heptose-1-phosphate adenosyltransferase that adds an ADP to yield ADP d-glycero-β-d-manno-heptose. The genes for the last two are most probably encoded by an operon since they have an overlap of 9 bp.
Enzymes involved in O-antigen biosynthesis that were encountered in ZG1 include several glycosyltransferases (lipid carrier-phosphate-sugar-phosphotransferases) that transfer glycosyl moieties from nucleoside diphosphate to the lipid carrier phosphate. These include undecaprenyl-phosphate galactosyl phosphotransferase, dolichol-phosphate mannosyltransferase, and dolichol-phosphate glucose synthase. In addition, the two enzymes that transfer the repeating sugar unit to the growing O-antigen chain (O-antigen polymerase) and that transfer the completed O antigen from its lipid carrier to the lipid A core (lipid A core–O-antigen ligase) are also present in the genome. None of the genes involved in the biosynthesis of lipid A were encountered in the partial genome of ZG1.
(iii) Secretory pathways.
Evidence for multiple secretory pathways in ZG1 was identified. These include pathways involved in translocation of molecules across the plasma membrane to the periplasm (e.g., Sec-dependent and TAT-dependent pathways [see Text S1 in the supplemental material]). Surprisingly, in addition to these two general pathways, the ZG1 genome also encodes an LPXTG-site transpeptidase (a sortase belonging to peptidase family C60 [49]). Sortases are integral membrane proteins that recognize proteins with the LPXTG signal and covalently attach them to the cell surface through the pentaglycine bridge of peptidoglycans of Gram-positive bacteria (59). Those enzymes were originally thought to be restricted to the cell wall of Gram-positive bacteria since the peptidoglycan layer does not have a cross-linking pentaglycine bridge in Gram-negative organisms and is not the outermost layer of the cell. The function of sortases in the cell walls of Gram-negative bacteria is not fully understood. However, sortases have previously been identified in few Gram-negative bacteria: according to the pfam database (3), 1,464 sortases are from Gram-positive bacteria, as opposed to only 113 from Gram-negative bacteria and 5 in the Archaea.
The ZG1 partial genome also encodes multiple nucleases, proteases, and carbohydrolases that function extracellularly. Evidence for two extracellular protein secretory systems were identified in ZG1: a Sec-independent type I secretion system and a Sec-dependent type IV secretion system.
(a) Sec-independent type I secretory system (ABC transporter). The type I secretory system exports proteins directly from the cytoplasm to outside the cell without passing through the periplasm. The system is comprised of 3 secretory proteins: an inner membrane ATP-binding protein, an inner membrane auxiliary protein, and an outer membrane auxiliary protein. The secretory genes are usually linked to the genes for the protein to be exported (59). The only ABC transporter proteins encoded by the ZG1 partial genome are 2 copies of a peptidase family C39 (49). Peptidases belonging to this family are specialized in bacteriocin (nonlantibiotic) activation and transport. The enzyme is an integral membrane protein that serves the 2 functions, where it cleaves a Gly—Gly bond found in the leader peptide (inactive bacteriocin), and it also possesses an ATP-binding domain responsible for the transport of the activated bacteriocin. No evidence for the 2 auxiliary proteins was detected in the partial genome.
(b) Sec-dependent protein secretion via type IV secretory pathway. The type IV secretion system can be classified into three subfamilies: conjugation system, DNA uptake and release, and effector translocation (10). Conjugation systems are found in most Gram-negative and Gram-positive bacterial species. ZG1 encodes 2 copies of the conjugation system coupling protein possessing a DNA-binding domain (pfam TrwB_AAD_bind). Conjugation system coupling protein is the first of three substructures comprising the conjugation systems in Gram-negative bacteria (together with a transenvelope protein complex and a conjugative pilus) (10). In E. coli, this protein is involved in binding single-stranded DNA during conjugation and is encoded on plasmids (26). The 2 copies of this protein in ZG1 are encoded on separate contigs, and each spans the whole contig. There is a possibility that these contigs are pieces of plasmid rather than chromosomal DNA.
(iv) Antibiotic production and resistance in ZG1.
Analysis of the ZG1 partial genome assembly suggests the capability for bacteriocin and extracellular peptidase M23 production: ZG1 possesses a peptidase C39-coding gene that has been shown to be specialized in the activation and transport of the antibiotic bacteriocin (28). While none of the genes involved in biosynthesis of bacteriocin have been detected in the partial genome, the presence of the bacteriocin activation/export system strongly suggests its production by ZG1. The genome also encodes a metalloendopeptidase belonging to the M23B family and possessing a LysM domain. Peptidases of this family are secreted extracellularly to lyse the cell wall peptidoglycan of Gram-positive bacteria (49).
In addition to antibiotic and cell wall lysis-mediating peptidases, the ZG1 partial genome also encodes genes for resistance to β-lactam and aminoglycoside antibiotics. The ZG1 genome encodes a β-lactamase enzyme belonging to molecular class A. In Gram-negative bacteria, class A β-lactamases are usually encoded on plasmids (34). However, the contig harboring the β-lactamase gene in ZG1 also contains a housekeeping gene (the gene for arginyl-tRNA synthetase), making it highly unlikely that it is a part of a plasmid. β-Lactamases are known to be secreted in the periplasm to deactivate any β-lactam antibiotics that might cross the outer membrane before the antibiotic enters the cell (38). In addition, the genome encodes one copy of an aminoglycoside phosphotransferase known to inactivate aminoglycoside antibiotics via phosphorylation (47).
(v) Stress response in ZG1 partial genome.
Survival in a eutrophic, highly competitive environment such as Zodletone Spring requires the possession of efficient stress response systems. Indeed, ZG1 contains a rich repertoire and a moderately high proportion of transcription-associated genes (3.55%), a hallmark of the genomes of free-living, highly adaptable, and cosmopolitan microorganisms (11). For comparison, transcription-associated genes constitute 2.88 and 1.84% of the genomes of the intracellular microbes Rickettsia africae and the Wolbachia endosymbiont of Culex quinquefasciatus, respectively. Genes associated with the stress response in the ZG1 genome consist of heat shock proteins, DNA repair proteins, housecleaning enzymes, and toxin-antitoxin systems (see Text S1 in the supplemental material). In brief, heat shock proteins identified in ZG1 include a class III heat shock ATP-dependent LonA protease (peptidase S16 family) and an ATP-independent heat shock-induced protease (peptidase family S1/S6) (49). DNA repair proteins identified include a DNA photolyase, the UvrC subunit of UvrABC endonuclease, as well as the helper UvrD helicase, and a uracil DNA glycosylase. Housecleaning enzymes include two proteins of the NUDIX hydrolase family and a fructose lysine kinase. Finally, the ZG1 genome encodes prevent host death/death on curing (phd/doc) toxin-antitoxin system. The 2 genes overlap by 10 bp and are thought to be in an operon. Toxin-antitoxin systems are widespread in bacteria, have been thought to be encoded on plasmids, and function to maintain only the plasmid-carrying progeny (also known as plasmid addiction systems) (23). However, the TA loci have also been found to be encoded on the chromosome of almost all prokaryotic genomes. They most probably function in helping cells cope with nutritional stress (23).
Identification of OP11-affiliated fragments from published metagenomic data sets.
The 16S rRNA gene sequence of ZG1 was used to query IMG/M system metagenomes, the environmental GenBank data set as a whole, and the environmental data set for the global ocean survey data set. The percent similarity to the OP11 sequence for the first 100 hits ranged from 71.7% to 77.3%. These hits were classified using the Greengenes database and their NCBI accession numbers. Unfortunately, none of these 16S rRNA gene sequences belonged to candidate division OP11.
Therefore, we used 22 of the core set of proteins from the ZG1 genome. Six proteins (ribosomal protein S6, thymidylate synthase, deoxycytidylate deaminase, homoserine kinase, DNA-dependent RNA polymerase subunit B′, and dephospho-CoA kinase) had no matches in the IMG/M system or the env_samples database with an E-value cutoff of less than or equal to the blastp system E value against the nr database. For the remaining 16 proteins, a total of 682 metagenomic sequences (ranging from 1 to 163 genes for individual proteins) met such criteria. Subsequent phylogenetic analysis (see Materials and Methods) indicated that only 12 metagenomic sequences could confidently be identified as belonging to members of candidate division OP11 (see Fig. S1 in the supplemental material). These fragments were retrieved from five different ecosystems: pristine groundwater from Oak Ridge, TN (29) (IMG system taxon object ID number 2007427000), global oceanic survey (51), and three different Yellowstone hot springs (IMG system taxon object ID numbers 2016842008, 2014031006, and 2013954000). Six of those were associated with thymidylate kinase (IMG system scaffold ID numbers orpgwFw301_FCAY24009_y1, YNP7_C2268, and YNP20_C4462, and NCBI accession numbers AACY023345562, AACY020068054, and AACY021966861), 2 with ribosomal protein L31 (IMG system scaffold ID number YNP5_C2757 and NCBI accession number AACY022893676), 2 with seryl-tRNA synthetase (IMG system scaffold ID number YNP20_C9994 and NCBI accession number AACY023310294), 1 with leucyl-tRNA synthetase (NCBI accession number AACY021531547), and 1 with nucleoside diphosphate kinase (NCBI accession number AACY023786466). These OP11 metagenomic fragments are shown in Table 2. Apart from the above-described proteins, the metagenomic contigs encoded some proteins that were part of the ZG1 partial genome (e.g., peptide chain release factor A, penicillin-binding protein family 1A, XTP pyrophosphatase), and in some cases, the gene order conservation was similar between ZG1 and the metagenomic fragment identified, e.g., peptide chain release factor A and ribosomal protein L31. Genes that were identified in these metagenomic fragments but that were not encountered in the partial ZG1 genome include deoxy UTP pyrophosphatase, ribosomal proteins L10 and L12, DNA gyrase subunit B, ferredoxin:NADP oxidoreductase, endonuclease III, DNA repair protein RadC, and 2 hypothetical proteins. The low number of metagenomic contigs identified and the absence of metabolically informative genes in these metagenomic fragments hindered our ability to identify any novel additional functions with this approach. Nevertheless, such an approach remains useful with the accumulation of more metagenomic sequences and for future single-cell genomic surveys.
Table 2.
Identification of OP11-affiliated fragments from published metagenomic data sets
| Contig | Environment | Size (bp) | ORF(s)a |
|---|---|---|---|
| 1 | Oak Ridge pristine groundwater FRC FW301 | 898 | dUTP diphosphatase (EC 3.6.1.23), thymidylate kinase |
| 2 | Microbial community from a Yellowstone hot springs (Bath Lake Vista Annex) | 2,411 | LSU ribosomal protein L31P, LSU ribosomal protein L10, LSU ribosomal protein L12P, DNA gyrase subunit B (EC 5.99.1.3) |
| 3 | Microbial community from a Yellowstone hot springs (Chocolate Pots) | 1,070 | Thymidylate kinase (EC 2.7.4.9), hypothetical protein |
| 4 | Microbial community from Yellowstone hot springs (Bath Lake Vista Annex, Purple Sulfur Mats) | 1,397 | Hypothetical protein, thymidylate kinase |
| 5 | Microbial community from Yellowstone hot springs (Bath Lake Vista Annex, Purple Sulfur Mats) | 4,002 | XTP pyrophosphatase, seryl-tRNA synthetase (EC 6.1.1.11), penicillin-binding protein 1A family |
| 6 | Global ocean survey | 864 | Protein chain release factor A, LSU ribosomal protein L31 |
| 7 | Global ocean survey | 1,658 | Seryl-tRNA synthetase |
| 8 | Global ocean survey | 883 | Leucyl-tRNA synthetase |
| 9 | Global ocean survey | 1,286 | Aspartyl tRNA synthetase, nucleoside diphosphate kinase |
| 10 | Global ocean survey | 1,348 | Thymidylate kinase |
| 11 | Global ocean survey | 1,826 | Endonuclease III, Ferredoxin:NADP oxidoreductase, thymidylate kinase |
| 12 | Global ocean survey | 817 | Thymidylate kinase, DNA repair protein RadC |
Genes originally identified in ZG1 and used for fragment recruiting from metagenomic data sets are in boldface. LSU, large subunit.
DISCUSSION
In this study, using a laser-assisted microfluidic device, a single cell belonging to candidate division OP11 was isolated, and its genomic DNA was partially amplified and sequenced, yielding a 270-kb partial genome. In-depth analysis revealed a heterotrophic, fermentative lifestyle with the capacity for cellulose, starch, and potentially lignin metabolism. In addition, evidence for a cell wall of Gram-negative bacteria, multiple secretion systems, antibiotic and bactericidal peptidase production, antibiotic resistance, and stress response mechanisms was identified. Within Zodletone Spring source sediments from which the OP11 cell has been sorted, phototrophic and chemolithoautotrophic CO2 fixation coupled to sulfide and sulfur oxidation results in an extremely eutrophic habitat with an extremely biomass-rich prokaryotic community. Therefore, the reported heterotrophic, polymer-degrading capabilities of OP11 suggest a possible role for members of this candidate phylum, or at least the OP11 cell from which ZG1 assembly is obtained, in carbon cycling. Specifically, OP11 could be involved in the breakdown of polymers derived from microbial biomass and subsequent degradation of the fermentable products obtained to fermentation end products (e.g., fatty acids and CO2) that could be utilized by other metabolic guilds within the community (e.g., sulfate and sulfur-reducing and photoautotrophic and chemolithautotrophic bacteria), respectively.
Members of OP11 have been encountered in a wide variety of habitats. In general, ecosystems where OP11 has been identified are eutrophic, with high prokaryotic biomass and high phylogenetic diversity (e.g., hydrocarbon-impacted habitats and hydrothermal vents [16, 27, 63]). In addition, OP11 members were also identified in various soils (20), ecosystems characterized by constant fluctuations in environmental conditions and high competition for nutritional resources by soil prokaryotes and soil fungi. Bacterial survival in such ecosystems requires constant adaptation to environmental fluctuations and possession of multiple survival weapons to enhance competitiveness. The ZG1 partial genome appears to have a moderately high ratio of transcription-associated genes (3.55%), indicating a high capability to respond to environmental fluctuations (11). In addition, ZG1 genomes have multiple antibiotic production and resistance mechanisms, which enhance its survivability in highly diverse and competitive ecosystems. The possession and retention of a large array of regulatory and other nonhousekeeping genes are in stark contrast to the characteristics of streamlined genomes of marine picoplankton, e.g., Pelagibacter ubique (25) or genomes of prokaryotic parasites (e.g., Mycoplasma spp.), where the lack of environmental fluctuations and relaxation of positive selection for genes used in biosynthesis or regulation favor genome reduction as an energy-saving strategy.
This study yielded interesting metabolic insights (e.g., cellulose- and starch-degrading capability, resistance to β-lactam and aminoglycoside antibiotics, and potential capability to metabolize galactose) that could help in designing enrichment strategies for some members of OP11 in Zodletone Spring source sediments. However, the capability of a single genome assembly, much like a single bacterial isolate from a high-order bacterial lineage (e.g., phylum or class), is not an adequate representation of the panmetabolic capability of an entire lineage. Therefore, we strongly caution against regarding these findings as a general indication of the overall metabolic capabilities of all members of OP11. This is partially due to the small size of the ZG1 genome assembly but, more importantly, to the ubiquity and expected high level of genomic diversity within various members and lineages within candidate division OP11. As such, multiple diverse capabilities in different OP11 lineages or even within the same lineage are entirely possible. Indeed, a previous study enriching for anaerobes from an uranium-contaminated environment in field research center (FRC) sediments (Oak Ridge, TN) enriched for members of OP11 (GenBank accession numbers EF508021 and EF507960) during syntrophic growth on ethanol (46).
The sequencing of nucleic acids derived from a phenotypically identified single cell in a chemically fixed sample constitutes a powerful new tool available to microbiologists. This tool is capable of providing great insights both in cases where complete or nearly complete genome sequences can be recovered (6, 61) and when partial genomes are recovered, as in the study described in this report. The analysis of a partial single-cell genome is reminiscent of the analysis of environmental fosmid sequences (8, 24) or fragments assembled from metagenomic data (57). One difference is that single-cell data sets can span much greater genomic distances than fosmids or conventional metagenomic assemblies. The key to confident interpretation of single-cell data is an assurance that only one cell is, in fact, being analyzed and that the sample is not contaminated, as the potential for contamination of material amplified from a single cell is extraordinarily high.
Metagenomic mining using a stringent genome recruitment approach was utilized to identify hitherto unclassified metagenomic fragments belonging to OP11. Our goal was to demonstrate the utility of these OP11 gene markers in improving phylogenetic binning in metagenomics and to determine whether additional characteristics of OP11 in other environments could be gleaned from such efforts. Unfortunately, only an additional 18 kb of total fragments was identified from three different metagenomic data sets. Multiple reasons could possibly explain the limited success in such effort: (i) the fact that members of OP11 are almost always encountered in low numbers in highly diverse ecosystems (16, 20, 54, 63), which renders obtaining large (if any) OP11 fragments unlikely, except in extremely ambitious metagenomic surveys (e.g., GOS); (ii) the stringent genome recruitment criteria applied to positively identify OP11 metagenomic fragments coupled to the potentially high level of intralineage diversity within members of OP11; and (iii) the rapid increase in dependence on short-read technology (pyrosequencing/Illumina) in recent metagenomic studies and the subsequent difficulty and uncertainty associated with assembly under such conditions due to potential chimeras (39). It is telling that all the metagenomic OP11 fragments obtained were from Sanger sequencing-based metagenomic studies. Nonetheless, there is undoubtedly a tremendous potential for synergy between single-cell genomic data and metagenomic data (6, 30). Our limited success with recruitment of database sequences highlights the novelty of the sequence that we report here, the extreme diversity of environmental microbial communities, and the importance of pairing analyses of single-cell data sets and metagenomic data sets derived from the same sample. Additional OP11 single-cell data sets, long-read sequencing technologies (17) with sufficient throughput to deeply sample complex communities, and improvements in assembly and binning strategies could each lead to much better identification of OP11 fragments from current and future data sets.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation Microbial Observatories Program (grant EF0801858) to M.S.E., an NIH Director's Pioneer Award, and NIH grant 5R01HG004863-02 to S.R.Q.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Abulencia C., et al. 2006. Environmental whole genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72:3291–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Apweiler R., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bateman A., et al. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beloqui A., et al. 2006. Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen. J. Biol. Chem. 281:22933–22942 [DOI] [PubMed] [Google Scholar]
- 5. Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27:573–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blainey P. C., Mosier A. C., Potanina A., Francis C. A., Quake S. R. 2011. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS One 6:e16626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blainey P. C., Quake S. R. 2011. Digital MDA for enumeration of total nucleic acid contamination. Nucleic Acids Res. 39:e19–e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brochier-Armanet C., et al. 2011. Complete-fosmid and fosmid-end sequences reveal frequent horizontal gene transfers in marine uncultured planktonic archaea. ISME J. 5:1291–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cantarel B. L., et al. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cascales E., Christie P. J. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cases I., de Lorenzo V., Ouzounis C. A. 2003. Transcription regulation and environmental adaptation in bacteria. Trends Microbiol. 11:248–253 [DOI] [PubMed] [Google Scholar]
- 12. Caspi R., et al. 2010. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 38:D473–D479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Davis J. P., Youssef N. H., Elshahed M. S. 2009. Assessment of the diversity, abundance, and ecological distribution of candidate division SR1 reveals a high level of phylogenetic diversity but limited morphotypic diversity. Appl. Environ. Microbiol. 75:4139–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dean F. B., et al. 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. U. S. A. 99:5261–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeSantis T. Z., et al. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dojka M. A., Hugenholtz P., Haack S. K., Pace N. R. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869–3877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eid J., et al. 2009. Real-time DNA sequencing from single polymerase molecules. Science 323:133–138 [DOI] [PubMed] [Google Scholar]
- 18. Elshahed M. S., et al. 2003. Bacterial diversity and sulfur cycling in a mesophilic sulfide-rich spring. Appl. Environ. Microbiol. 69:5609–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Elshahed M. S., et al. 2007. Phylogenetic and metabolic diversity of Planctomycetes from anaerobic, sulfide- and sulfur-rich Zodletone Spring, Oklahoma. Appl. Environ. Microbiol. 73:4707–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Elshahed M. S., et al. 2008. Novelty and uniqueness patterns of rare members of the soil biosphere. Appl. Environ. Microbiol. 74:5422–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gardner P. P., et al. 2009. Rfam: updates to the RNA families database. Nucleic Acids Res. 37:D136–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gerdes K., Christensen S. K., Lobner-Olesen A. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371–382 [DOI] [PubMed] [Google Scholar]
- 24. Gilbert J. A., Muhling M., Joint I. 2008. A rare SAR11 fosmid clone confirming genetic variability in the ‘Candidatus Pelagibacter ubique’ genome. ISME J. 2:790–793 [DOI] [PubMed] [Google Scholar]
- 25. Giovannoni S. J., et al. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245 [DOI] [PubMed] [Google Scholar]
- 26. Gomis-Rüth F. X., de la Cruz F., Coll M. 2002. Structure and role of coupling proteins in conjugal DNA transfer. Res. Microbiol. 153:199–204 [DOI] [PubMed] [Google Scholar]
- 27. Harris J. K., Kelley S. T., Pace N. R. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Havarstein L. S., Diep D. B., Nes I. F. 1995. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol. Microbiol. 16:229–240 [DOI] [PubMed] [Google Scholar]
- 29. Hemme C. L., et al. 2010. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J. 4:660–672 [DOI] [PubMed] [Google Scholar]
- 30. Hess M., et al. 2011. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467 [DOI] [PubMed] [Google Scholar]
- 31. Hongoh Y., et al. 2008. Complete genome of the uncultured Termite Group 1 bacteria in a single host protist cell. Proc. Natl. Acad. Sci. U. S. A. 105:5555–5560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hugenholtz P., Goebel B. M., Pace N. R. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765–4774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hugenholtz P., Pitulle C., Hershberger K. L., Pace N. R. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jacoby G. A. 1994. Extrachromosomal resistance in Gram-negative organisms: the evolution of [beta]-lactamase. Trends Microbiol. 2:357–360 [DOI] [PubMed] [Google Scholar]
- 35. Johnson M., et al. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36:W5–W9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kanehisa M., Goto S., Furumichi M., Tanabe M., Hirakawa M. 2010. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 38:D355–D360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kirk T. K. 1987. Enzymatic “combustion”: the microbial degradation of lignin. Annu. Rev. Microbiol. 41:465–505 [DOI] [PubMed] [Google Scholar]
- 38. Koshland D., Botstein D. 1980. Secretion of beta-lactamase requires the carboxy end of the protein. Cell 20:749–760 [DOI] [PubMed] [Google Scholar]
- 39. Kumin V., Copeland A., Lapidus A., Mavromatis K., Hugenholtz P. 2008. A bioinformatician's guide to metagenomics. Microbiol. Mol. Biol. Rev. 72:557–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Larkin M. A., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 41. Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marcy Y., et al. 2007. Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet. 3:e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marcy Y., et al. 2007. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. U. S. A. 104:11889–11894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Markowitz V. M., et al. 2010. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 38:D382–D390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Martin H. G., et al. 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 24:1263–1269 [DOI] [PubMed] [Google Scholar]
- 46. Michalsen M. M., et al. 2007. Changes in microbial community composition and geochemistry during uranium and technetium bioimmobilization. Appl. Environ. Microbiol. 73:5885–5896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nurizzo D., et al. 2003. The crystal structure of aminoglycoside-3′-phosphotransferase-IIa, an enzyme responsible for antibiotic resistance. J. Mol. Biol. 327:491–506 [DOI] [PubMed] [Google Scholar]
- 48. Pelletier E., et al. 2008. “Candidatus Cloacamonas acidaminovorans”: genome sequence reconstruction provides a first glimpse of a new bacterial division. J. Bacteriol. 190:2572–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rawlings N. D., Barrett A. J., Bateman A. 2010. MEROPS: the peptidase database. Nucleic Acids Res. 38:D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rodrigue S. B., et al. 2009. Whole genome amplification and de novo assembly of single bacterial cells. PLoS One 4:e6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rusch D. B., et al. 2007. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rutherford K., et al. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 53. Senko J. M., et al. 2004. Barite deposition resulting from phototrophic sulfide-oxidizing bacterial activity. Geochim. Cosmochim. Acta 68:773–780 [Google Scholar]
- 54. Teske A., et al. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Treusch A. H., et al. 2004. Characterization of large-insert DNA libraries from soil for environmental genomic studies of Archaea. Environ. Microbiol. 6:970–980 [DOI] [PubMed] [Google Scholar]
- 56. Valvano M. A., Messner P., Kosma P. 2002. Novel pathways for biosynthesis of nucleotide-activated glycero-manno-heptose precursors of bacterial glycoproteins and cell surface polysaccharides. Microbiology 148:1979–1989 [DOI] [PubMed] [Google Scholar]
- 57. Venter J. C., et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74 [DOI] [PubMed] [Google Scholar]
- 58. Warburton P. E., Giordano J., Cheung F., Gelfand Y., Benson G. 2004. Inverted repeat structure of the human genome: the X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 14:1861–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. White D. 2000. The physiology and biochemistry of prokaryotes, 2nd ed Oxford University Press, New York, NY [Google Scholar]
- 60. White R., Blainey P., Fan H. C., Quake S. 2009. Digital PCR provides sensitive and absolute calibration for high throughput sequencing. BMC Genomics 10:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Woyke T., et al. 2010. One bacterial cell, one complete genome. PLoS One 5:e10314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yooseph S., et al. 2010. Genomic and functional adaptation in surface ocean planktonic prokaryotes. Nature 468:60–66 [DOI] [PubMed] [Google Scholar]
- 63. Youssef N. H., Couger M. B., Elshahed M. S. 2010. Fine-scale bacterial beta diversity within a complex ecosystem (Zodletone Spring, OK, USA): the role of the rare biosphere. PLoS One 5:e12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zareian S., et al. 2010. Purification and characterization of a novel amylopullulanase that converts pullulan to glucose, maltose, and maltotriose and starch to glucose and maltose. Enzyme Microb. Technol. 46:57–63 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.