Abstract
Marine group I Crenarchaeota (MGI) represents a ubiquitous and numerically predominant microbial population in marine environments. An understanding of the spatial dynamics of MGI and its controlling mechanisms is essential for an understanding of the role of MGI in energy and element cycling in the ocean. In the present study, we investigated the diversity and abundance of MGI in the East China Sea (ECS) by analysis of crenarchaeal 16S rRNA gene, the ammonia monooxygenase gene amoA, and the biotin carboxylase gene accA. Quantitative PCR analyses revealed that these genes were higher in abundance in the mesopelagic than in the euphotic zone. In addition, the crenarchaeal amoA gene was positively correlated with the copy number of the MGI 16S rRNA gene, suggesting that most of the MGI in the ECS are nitrifiers. Furthermore, the ratios of crenarchaeal accA to amoA or to MGI 16S rRNA genes increased from the euphotic to the mesopelagic zone, suggesting that the role of MGI in carbon cycling may change from the epipelagic to the mesopelagic zones. Denaturing gradient gel electrophoretic profiling of the 16S rRNA genes revealed depth partitioning in MGI community structures. Clone libraries of the crenarchaeal amoA and accA genes showed both “shallow” and “deep” groups, and their relative abundances varied in the water column. Ecotype simulation analysis revealed that MGI in the upper ocean could diverge into special ecotypes associated with depth to adapt to the light gradient across the water column. Overall, our results showed niche partitioning of the MGI population and suggested a shift in their ecological functions between the euphotic and mesopelagic zones of the ECS.
INTRODUCTION
Microbes are the most abundant and diverse life forms in oceans and play a critical role in energy flux and element cycling (14, 28). Determining the structure of marine microbial communities and their spatial and temporal variation is an essential step in an understanding of the role of microbes in the functioning of ecosystems (55). The application of molecular technology has led to great advances in microbial oceanography (14). On the horizontal dimension, marine microorganisms have been shown to have biogeographic distribution patterns like those of macroorganisms (39). Depth is considered a predominant factor in structuring of microbial communities across the oceanic water column, since the major physical and chemical gradients (e.g., light, temperature, pressure, nutrients, and organic matter) are apparent on the vertical scale (12). Numerous studies show the depth-stratified niche adaptation of marine bacteria, either at the population level (7) or at the community level (12).
Marine group I Crenarchaeota (MGI), one of the major phylogenetic groups of pelagic archaea, were first discovered by using 16S rRNA gene-based molecular methods (11, 15). A number of studies showed that MGI forms one of the most abundant populations of marine microorganisms: it is estimated that there are 1.3 × 1028 to 2.7 × 1028 MGI cells, representing up to 40% of the total prokaryotic biomass in the dark ocean (31, 49). Phylogenetic and genomic studies revealed that MGI together with mesophilic Crenarchaeota in soil environments likely form a third archaeal phylum, the Thaumarchaeota (6, 51). Recent observations of the crenarchaeal amoA gene, encoding ammonia monooxygenase subunit A, in marine (56) and soil (54) metagenomics and the isolation of the marine crenarchaeon “Candidatus Nitrosopumilus maritimus” (32) imply that most mesophilic Crenarchaeota have the ability to oxidize ammonia to nitrite, which was previously thought to be achieved only by ammonia-oxidizing bacteria (AOB) (34). Meanwhile, a novel mechanism of autotrophic carbon fixation, the 3-hydroxypropionate/4-hydroxybutyrate pathway, was identified in the genomes of “Candidatus Nitrosopumilus maritimus” and “Candidatus Cenarchaeum symbiosum” (20, 57). The acc gene, encoding acetyl coenzyme A (acetyl-CoA) carboxylase, one of the key enzymes in that pathway, and the amoA gene are used as phylogenetic markers to mirror the ecological function of MGI (5, 13, 25, 59, 60).
Previous investigations showed that the MGI population seems to be rather similar on the horizontal scale in the world's oceans (25, 35, 40), except where different water masses meet (1, 16, 30). Vertically, however, two distinct phylogenetic groups of MGI (MGI-α and MGI-γ), based on analyses of 16S rRNA genes, predominated in shallow and deep waters, respectively (3, 40). Additional evidence for depth-stratified phylogeny within MGI has also been observed by using the crenarchaeal 16S rRNA gene internal transcribed spacer region (18) and the amoA (5, 21, 48) and accA (25, 60) genes. The concordant depth-stratified phylogeny suggests that MGI populations have evolved into different ecotypes to adapt to specific habitats (niches) according to different environmental factors or resources (60).
Some studies suggested that the depth-related phylogeny of MGI may be due to photoinhibition-resistant adaptations (8, 42). Assuming this to be true, we hypothesized that (i) MGI might have a restricted distribution pattern (niche partitioning) across the water column due to the influence of light or other continuous environmental gradients and (ii) evolutionary divergence among MGI ecotypes can be observed since niche partitioning might contribute to restricted gene flow during their evolution (44a). To test these hypotheses, we investigated the vertical distribution of the abundance and diversity of MGI in the euphotic and upper mesopelagic zones at two stations along the Kuroshio Current using multiphasic molecular methods, including denaturing gradient gel electrophoresis (DGGE), clone libraries, quantitative PCR (qPCR), and ecotype simulation analysis. Our results showed that MGI in the upper ocean diverged into special ecotypes associated with depth in order to adapt to the light gradient in the water column.
MATERIALS AND METHODS
Sampling stations and environmental conditions.
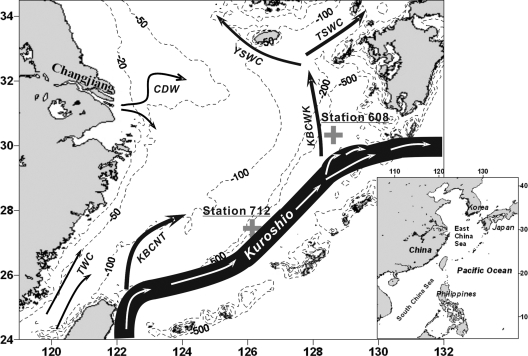
The East China Sea (ECS), located in the Northwest Pacific Ocean, is the largest continental shelf sea in the temperate zone. The Kuroshio Current going along the ECS shelf edge originates from the West Pacific warm pool and is characterized by high temperature and high salinity (29) (Fig. 1). Two sites along the shelf slope of the ECS on the west side of the Kuroshio Current were selected for the present study: station 712 (27.44°N, 126.14°E) and station 608 (30.33°N, 128.62°E) (Fig. 1). Station 712 is located by the main stream of the Kuroshio Current, while station 608 is situated near a branch of the Kuroshio Current. With similar water depths, both beyond 500 m, but different environmental conditions (please see below for detailed results), these two stations provided an opportunity for comparative studies of microbial dynamics along both geographic and vertical environmental dimensions.
Fig. 1.
Map of the ECS showing the approximate bottom topography and hydrographic structure and the location of the sampling stations. CDW, Changjiang diluted water; TWC, Taiwan Warm Current; KBCNT, Kuroshio Branch Current north of Taiwan; KBCWK, Kuroshio Branch Current west of Kyushu; TSWC, Tsushima Strait Warm Current; YSWC, Yellow Sea Warm Current; Kuroshio, Kuroshio Current. The map was created by use of Planiglobe, with information on currents taken from references 7a and 29.
Water samples were collected on board the RV Dongfanghong #2 between 5 and 12 November 2007. A SeaBird SBE 9/11 Plus conductivity-temperature-depth (CTD) system fitted with a rosette sampler was used to measure temperature and salinity and to collect water samples. The potential density (σt) was calculated based on the equation for the state of seawater as proposed previously (41). The mixed-layer depths (MLDs) were estimated by using the σt values. The euphotic zone depth of these two stations was derived from the monthly mean Aqua-MODIS euphotic depth [Lee] data set (34a) in the corresponding months (http://oceancolor.gsfc.nasa.gov/cgi/l3). Water samples were collected at 10 intervals in the euphotic and upper mesopelagic zones (0 to 400 m) for each station (Table 1). Subsamples (2 to 3 liters) were prefiltered through a 20-μm mesh (Millipore, Billerica, MA) and subsequently filtered onto 0.2-μm-pore-size polycarbonate filters (Millipore) at a pressure of <0.03 MPa. The filters were immediately frozen and stored at −80°C until further analysis.
Table 1.
Properties of the water bodies of two stations located in the ECS
| Depth (m) | Station 712 |
Station 608 |
||||
|---|---|---|---|---|---|---|
| Temp (°C) | Salinity (‰) | σt (kg m−3) | Temp (°C) | Salinity (‰) | σt (kg m−3) | |
| 0 | 25.80 | 34.58 | 22.782 | 24.36 | 34.38 | 23.065 |
| 10 | 25.46 | 34.59 | 22.889 | 24.37 | 34.39 | 23.070 |
| 30 | 25.45 | 34.58 | 22.893 | 24.37 | 34.30 | 23.002 |
| 50 | 25.27 | 34.56 | 22.928 | 24.31 | 34.40 | 23.098a |
| 75 | 25.26 | 34.55 | 22.932a | 22.22 | 34.63 | 23.882 |
| 100 | 23.65 | 34.39 | 23.291 | 20.03 | 34.62 | 24.469 |
| 150 | 18.46 | 34.64 | 24.892 | 17.38 | 34.62 | 25.140 |
| 200 | 15.99 | 34.62 | 25.463 | 13.19 | 34.50 | 25.969 |
| 300 | 11.96 | 34.44 | 26.170 | 10.00 | 34.36 | 26.455 |
| 400 | 10.42 | 34.36 | 26.391 | 7.96 | 34.33 | 26.762 |
Depth of the mixed layer.
Environmental DNA extraction.
Microbial community DNA was extracted by using the UltraClean Soil DNA kit (MoBio, San Diego, CA) as described elsewhere previously (26). DNA integrity and size were checked in a 0.8% agarose gel stained with SYBR green I (Molecular Probes, Eugene, OR), and the concentrations were quantified in duplicate by using a FlexStation 3 instrument (Molecular Devices, Sunnyvale, CA) with a Quant-iT dsDNA HS assay kit (Molecular Probes). A standard curve was generated by using known amounts of lambda DNA (Molecular Probes).
qPCR analysis.
qPCR was performed on an ABI Prism 7500 system (Applied Biosystems, Foster City, CA) with the primers listed in Table S1 in the supplemental material. Plasmids carrying the respective 16S rRNA or functional gene fragments (archaeal and MGI 16S rRNA genes, crenarchaeal amoA and accA genes, and β-AOB amoA genes) as an insert were constructed (25). The concentrations of plasmid DNAs were determined by using a Quant-iT dsDNA HS assay kit (Molecular Probes). Tenfold serial dilutions of a known number of plasmids were subjected to qPCR assays in triplicate to generate an external standard curve.
The abundances of the 16S rRNA genes of the archaea and MGI and the functional genes of the Crenarchaeota (amoA and accA) and β-AOB (amoA) in all samples were measured in triplicate for each sample. For the quantification of 16S rRNA genes, a 20-μl reaction mixture consisting of 10 μl of SYBR GreenER-qPCR SuperMix Universal (Molecular Probes), 50 nM ROX dye, and 5 μg bovine serum albumin (BSA) plus 0.4 μM primers and 1 μl of template (1 to 10 ng) was used. For the quantification of the functional genes, the following reaction mixture was used: 10 μl of SYBR Premix Ex Taq (TaKaRa, Dalian, China), 50 nM ROX dye, 5 μg BSA, 0.4 μM primers, and 1 μl template DNA of 1 to 10 ng in a final volume of 20 μl. The specificity of qPCRs was confirmed by using melting curve analysis and agarose gel electrophoresis after amplification. The thermocycling parameters and efficiency of the qPCRs are described in Table S2 in the supplemental material.
T-RFLP analysis of bacterial 16S rRNA genes.
Bacterial community structures were analyzed by using terminal restriction fragment length polymorphism (T-RFLP) analysis for PCR amplification with primer pair 27F/926R, with primer 27F labeled by 6-carboxyfluorescein phosphoramidite at the 5′ end. The PCR conditions and chemistry were described elsewhere previously (62). Briefly, 1 to 10 ng of the extracted DNA was added as a template in a 50-μl PCR mixture. Purified PCR products were digested with RsaI (TaKaRa) at 37°C for 12 h. Digested products were recovered in a final volume of 20 μl of Mill-Q water using ethanol precipitation. Purified products (10 μl) were mixed with 0.5 μl of the internal ET ROX-900 size standard (Amersham Bioscience) and then detected by using a MegaBACE genetic analyzer (Amersham) operated in genotyping mode (62). The T-RFLP data were exported by using MegaBACE Genetic Profiler software and were processed with T-REX software for filtering out noise, aligning terminal restriction fragments (T-RFs), and constructing a data matrix (9). The obtained matrix was further imported into PAST v1.92 (23) to perform the cluster analysis with both Sorensen and Bray-Curtis algorithms. T-RFs of <50 bp or contributing <0.5% to the total fluorescence signal were excluded from the analysis.
DGGE analysis of crenarchaeal 16S rRNA genes.
Crenarchaeal 16S rRNA gene fragments were amplified by employing a nested PCR strategy as described previously (44). Briefly, crenarchaeal 16S rRNA gene fragments were amplified by using primer 21F (11) and modified primer 1492R (44) in the first round and primers 771F (45) and GI_956R (42) in the second round, with primer GI_956R containing a 40-bp GC clamp. DGGE was performed by using a Bio-Rad DCode universal mutation detection system (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. The PCR products of the crenarchaeal 16S rRNA gene were applied onto 8% (wt/vol) gels in 1× Tris-acetate-EDTA (TAE) buffer with a denaturing gradient of 30 to 55% denaturant (100% denaturing solution containing 40% formamide and 7 M urea). Electrophoresis was performed at a constant temperature of 60°C and at 75 V for 16 h. The DGGE images, stained with SYBR green I (Molecular Probes), were captured and analyzed by using GeneSnap and GeneTools software (SynGene-Synoptics, Cambridge, United Kingdom).
Crenarchaeal amoA and accA gene clone library analyses.
Clone libraries were constructed for the crenarchaeal amoA genes targeting ammonia monooxygenase subunit A and for the crenarchaeal accA genes targeting acetyl-CoA carboxylase subunit A for the six depth zones at each station. The amoA gene fragments were amplified with Arch-amoAF/Arch-amoAR (13), and the accA gene fragments were amplified with Cren529F/Cren981R (59), except that the nested PCR strategy was employed when few positive PCR products were obtained from the euphotic samples. Subsequently, purified PCR products were ligated into the pMD18-T vector (TaKaRa) and then transformed into competent Escherichia coli DH5α cells (TaKaRa). Positive clones were screened by using PCR reamplification with vector primers M-13F and M-13R and selected for sequencing by using an ABI 3730 XL sequencer (Applied Biosystems).
Phylogenetic analysis and ecotype simulation.
The crenarchaeal amoA and accA gene sequences, along with their closest relatives retrieved from GenBank, were imported into ARB (37). The sequences were first translated and aligned by using Clustal W in ARB, and the nucleotides were then realigned according to their protein alignments. Ambiguously and incorrectly aligned positions were corrected manually by using the ARB-edit tool. The sequence base frequency filters were used to exclude ambiguous positions and columns containing gaps. The Bayesian tree was generated by using the MrBayes v. 3.1.2 program (47) and the following parameters: the general time reversal model of evolution with gamma-invariable-distributed rate, the number of chains set to 6, and the temperature set to 0.1. Five Markov chains in parallel were run with 5,000,000 generations and sampled every 100 generations (the first 7,500 to 10,000 “burn-in” trees were excluded from the consensus tree). Tree topologies were also evaluated with the neighbor-joining and maximum parsimony methods by using PAUP*4.0 (53).
Ecotype simulation analyses were performed by using AdaptML software (27). Since Bayesian trees constructed with archaeal amoA or accA gene sequences contained several multifurcation nodes, maximum likelihood trees constructed by using RAxML v7.0.4 (52) were used as inputs for AdaptML analyses with default parameters.
Statistical analysis.
Operational taxonomic units (OTUs) for clone library analyses were defined by using the furthest-neighbor algorithm in DOTUR (50) and a cutoff of ≤5% as described in previous studies (4, 13, 25). Rarefaction, the nonparametric richness estimator Chao1, the Shannon diversity index, and Simpson's index were also calculated by using DOTUR. The coverage of each clone library was calculated as coverage (C) = 1 − (n/N) × 100, where n is the number of unique clones detected in the sample and N is the total number of clones analyzed (19).
Community classification of the crenarchaeal assemblages for both the amoA and accA genes was determined by using phylogeny-based weighted UniFrac environmental clustering (36) or ecotype-abundance-based clustering analyses. Analyses of similarity (ANOSIM) were performed to verify the significance of microbial community structures from different groups or stations. The significance of the correlations between two distance matrices or between microbial community structures and environmental variables was tested with the Mantel test. Analyses were performed with the PAST v1.92 program (23). Since a normality of distribution of the individual data sets was not always met, nonparametric statistical analyses were performed by using SPSS v13.0 (SPSS, Inc., Chicago, IL).
Nucleotide sequence accession numbers.
Sequences reported in this study have been deposited in the GenBank database under accession numbers GU181423 to GU181799 (crenarchaeal amoA genes), GU195200 to GU195605 (crenarchaeal accA genes), and GU195606 to GU195629 (bacterial accA genes).
RESULTS
The main hydrographic characteristics of the two stations investigated in this study are summarized in Table 1. The temperature was significantly higher but the σt was significantly lower at station 712 than at station 608, while the salinity values were not significantly different between the two stations. As indicated by the σt, the MLD was slightly greater at station 712 (∼75 m) than at station 608 (∼50 m) (Table 1). At both stations, the temperature showed a strong decrease with increasing depth from the MLD, whereas salinity had constant values (Table 1). As seen from the MODIS euphotic zone depth data for November 2007, the euphotic depth at station 608 was around 84 m, whereas the euphotic depth at station 712 was slightly greater, at ∼106 m.
Quantification of 16S rRNA, amoA, and accA genes of pelagic Crenarchaeota.
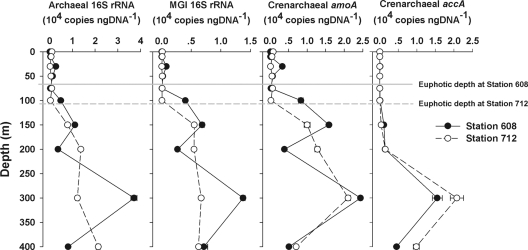
Archaeal 16S rRNA gene abundances ranged from 1.41 × 102 to 3.72 × 104 copies ng DNA−1 within the euphotic zone at the two stations (Fig. 2). Depth profiles of MGI 16S rRNA gene abundance ranging from 3.3 × 101 to 1.38 × 104 copies ng DNA−1 were similar to those of archaeal 16S rRNA genes (Fig. 2). The MGI 16S rRNA gene abundance was significantly correlated with the archaeal 16S rRNA gene abundance (R2 = 0.86; P < 0.001). Furthermore, qPCR analysis showed that MGI dominated in the archaeal community below the euphotic zone, with an averaged relative abundance of 57.2% of the total.
Fig. 2.
Depth profiles of abundances of archaeal and MGI 16S rRNA genes and crenarchaeal amoA and accA genes measured by using qPCR at stations 608 and 712 in the ECS. Bars denote 1 standard error of triplicate qPCR determinations and are not visible when they are less than the width of the data point.
The crenarchaeal amoA gene abundance increased from near the detection limit in the surface waters to a maximum of 2.44 × 104 copies ng DNA−1 in the upper mesopelagic zones of both stations (Fig. 2). A linear regression analysis indicated that the crenarchaeal amoA and MGI 16S rRNA genes were significantly correlated with each other (R2 = 0.83; P < 0.001). The abundance of the amoA gene of β-AOB was below the detection limits in all samples.
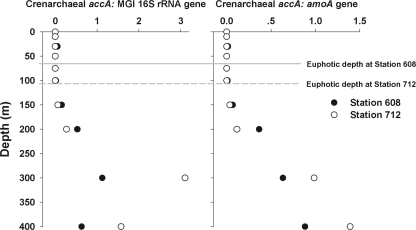
The abundance of the crenarchaeal accA gene was almost always below the detection limit within the euphotic zones but then increased with depth to maximal values at a depth of 300 m at both stations (Fig. 2). The abundance ratios of the crenarchaeal accA gene to the MGI 16S rRNA gene or to the crenarchaeal amoA gene increased with depth, and the peak ratios occurred in the upper mesopelagic zone (Fig. 3).
Fig. 3.
Ratios of crenarchaeal accA to MGI 16S rRNA genes or crenarchaeal amoA genes.
Community structure of bacterial and crenarchaeal 16S rRNA genes.
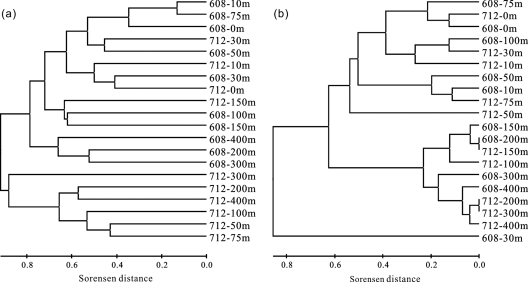
The T-RFLP pattern of the bacterial communities revealed a total of 134 T-RFs at the 16S rRNA gene level. The number of T-RFs showed no depth-related pattern at both stations (data not shown). In contrast, ANOSIM showed distinct site- or water mass-specific bacterial communities between the two stations (P < 0.05). Furthermore, the cluster analysis demonstrated that the bacterial communities at station 608 were stratified in the water column, but most samples from station 712 clustered together and did not show a depth-related pattern (Fig. 4a). Mantel tests indicated that the bacterial communities at station 608 were strongly influenced by depth, temperature, and σt (r > 0.5; P < 0.001), while this influence at station 712 was not obviously seen (P > 0.1).
Fig. 4.
Clustering of the T-RFLP profiles of bacterial 16S rRNA genes (a) and clustering of the DGGE profiles of crenarchaeal 16S rRNA genes (b) in the ECS. Clustering analyses were performed based on the Sorensen algorithm, and the scale bar indicates the Sorensen distance.
Crenarchaeal community structures at both stations were characterized on the basis of the 16S rRNA gene by using DGGE. The number of DGGE bands detected ranged from 1 to 14 per sample (see Fig. S1 in the supplemental material). There were relatively lower band numbers in the euphotic zone (≤100 m) than in greater depths at both stations (P < 0.05 by Mann-Whitney test) (Fig. S1), resulting in depth-related variations. Cluster analysis demonstrated a general depth-stratified pattern for the crenarchaeal communities at both stations (Fig. 4b): a deep clade (≥150 m) and a euphotic zone cluster (≤100 m). Moreover, the deep clade could be further divided into two subclades (Fig. 4b): a deep epipelagic subclade (150 m and 200 m) and a mesopelagic subclade (300 m and 400 m), except for the depth of 712 to 200 m. Mantel tests indicated that the crenarchaeal communities in both stations were closely related to water depth, temperature, and σt (r > 0.4; P < 0.001). In contrast, homogeneous crenarchaeal communities were observed at both stations (P > 0.5 by ANOSIM).
Diversity and phylogeny of crenarchaeal amoA and accA genes.
To assess the evolutionary divergence and restricted distribution of MGI ecotypes along the water column, samples from six depths from each station were chosen, based on the above-described DGGE profiles, to construct clone libraries of the crenarchaeal amoA and accA genes. In total, 377 amoA and 406 accA gene sequences were obtained (Table 2). These sequences contained 31 unique amoA OTUs and 53 unique accA OTUs based on a 5% cutoff value at the DNA level. The numbers of OTUs per sample varied between 2 and 15 for amoA and between 3 and 17 for accA (Table 2). The values of library coverage ranged from 83.7 to 100% for amoA and 82 to 97.4% for accA. The diversity of amoA varied at both stations: clone libraries below the euphotic zones (≥150 m) had a higher level of diversity than did those recovered from the shallower depths (≤100 m), as indicated by the Shannon, reciprocal Simpson's, and Chao1 diversity indices (P < 0.01 by Mann-Whitney test) (Table 2). In contrast, there were no significant differences in accA diversity between the euphotic zone samples and those from greater depths (P > 0.1 by Mann-Whitney test) (Table 2). Overall, the numbers of OTUs and the Shannon and Chao1 richnesses of accA were generally higher than those of amoA, with three exceptions: at stations at depths of 608 to 400 m, 712 to 150 m, and 712 to 400 m.
Table 2.
Diversity indices of crenarchaeal amoA and accA clone libraries recovered from the stations investigated in the ECS
| Sample depth range (m) | Crenarchaeal amoA |
Crenarchaeal accA |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of clones sequenced | No. of OTUsb | C (%) | H′ | 1/D | Chao1 index value | No. of clones sequenceda | No. of OTUsb | C (%) | H′ | 1/D | Chao1 index value | |
| 608–10c | 29 | 2 | 100 | 0.69 | 2.06 | 2 | − | |||||
| 608–75c | 30 | 3 | 100 | 0.83 | 2.09 | 3 | 36 | 8 | 91.7 | 1.37 | 2.6 | 9 |
| 608–100 | 29 | 4 | 93.1 | 0.79 | 1.81 | 5 | 31 | 12 | 87.1 | 2.15 | 7.05 | 13 |
| 608–150 | 30 | 6 | 100 | 1.62 | 4.95 | 6 | 31 | 11 | 87.1 | 2.14 | 8.62 | 13 |
| 608–200 | 37 | 12 | 89.2 | 2.27 | 10.53 | 14 | 52 | 17 | 82.7 | 2.3 | 7.41 | 26 |
| 608–400 | 30 | 6 | 96.7 | 1.62 | 5.24 | 6 | 40 | 4 | 97.5 | 0.57 | 1.38 | 4 |
| 712–10c | 27 | 3 | 96.3 | 0.79 | 2.09 | 3 | 28 | 7 | 89.3 | 1.4 | 3.11 | 8 |
| 712–75c | 31 | 3 | 100 | 0.94 | 2.5 | 3 | 38 | 9 | 86.8 | 1.46 | 2.85 | 19 |
| 712–100 | 31 | 3 | 96.8 | 0.8 | 2.17 | 3 | 30 | 6 | 93.3 | 1.4 | 3.6 | 7 |
| 712–150 | 30 | 8 | 93.3 | 1.7 | 4.5 | 8 | 32 | 8 | 90.6 | 1.56 | 3.55 | 9 |
| 712–200 | 43 | 15 | 83.7 | 2.43 | 11.9 | 22 | 50 | 16 | 82 | 2.14 | 5.81 | 26 |
| 712–400 | 30 | 8 | 90 | 1.67 | 4.37 | 10 | 38 | 3 | 97.4 | 0.55 | 1.46 | 3 |
A minus sign indicates the unsuccessful application of crenarchaeal accA genes.
OTUs were defined as 5% divergence at the DNA level.
A nested PCR strategy was employed for crenarchaeal accA gene amplification for these samples.
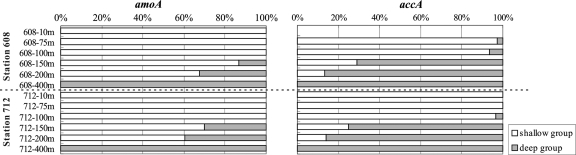
Phylogenetic analyses demonstrated that all sequences of both genes were affiliated with the two primary marine clusters described previously, the “shallow group” and the “deep group” (1, 25, 60) (see Fig. S2 in the supplemental material), indicating that there was a high level of phylogenetic congruence between the crenarchaeal amoA and accA genes. The shallow group for both genes contained sequences derived exclusively from epipelagic water (≤200 m) (Fig. S2), but their relative abundances decreased with increasing depth (Fig. 5). Accordingly, the deep group for both genes appeared near the base of the euphotic zone (150 m for amoA genes and 75 m for accA genes), and the relative abundance of this group increased with depth (Fig. 5).
Fig. 5.
Depth distribution of phylogenetic community structures of crenarchaeal amoA and accA genes recovered from the ECS. The relative abundance of each phylotype named in Fig. S2 in the supplemental material was calculated and is represented in a column diagram.
Ecotype simulation and community classification of crenarchaeal amoA and accA.
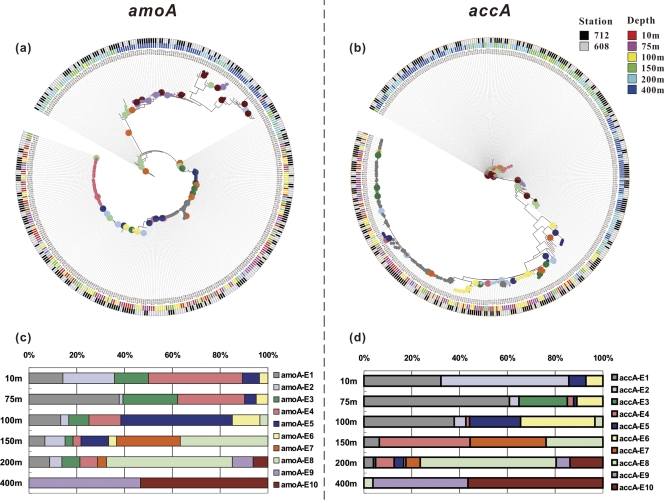
The UniFrac significance and P-test significance analyses indicated that there was a nonrandom clustering of the community structures of crenarchaeal amoA and accA across the sampling depths at both stations (P < 0.001). We therefore demarcated ecotypes using the AdaptML model, with the sampling depths and sites as habitat source inputs. AdaptML identified 10 amoA ecotypes and 10 accA ecotypes, both of which were clearly associated with sampling depth but not with sampling site (Fig. 6a and b). Noticeably, some amoA and accA ecotypes (8 out of 10 ecotypes) had similar depth-related variations (Fig. 6c and d). For example, the relative abundances of amoA-E1/accA-E1 and amoA-E2/accA-E2 generally decreased with increasing depth, while the abundances of amoA-E5/accA-E5 and amoA-E6/accA-E6 increased from the surface to the bottom of the euphotic zone (∼100 m). On the other hand, amoA-E7/accA-E7 and amoA-E8/accA-E8 appeared near the bottom of the euphotic zone and showed an opposite trend. Both amoA-E9/accA-E9 and amoA-E10/accA-E10 thrived in the mesopelagic zone. These results supported the phylogenetic congruence between the amoA and accA genes, implying that most pelagic Crenarchaeota adapt to the changing environments along the vertical dimension of the water column.
Fig. 6.
Ecotype simulation of crenarchaeal amoA (a) and accA (b) gene sequences recovered from the ECS and shift in relative abundances of different predicted ecotypes for amoA (c) and accA (d) across the water column of the ECS. Sampling stations and sampling depths are coded by colored squares. Predicted ecotypes are coded by colored circles at the phylogenetic nodes or the colored squares shown in panels c and d. Two more-detailed images for panels a and b are shown in Fig. S3 in the supplemental material.
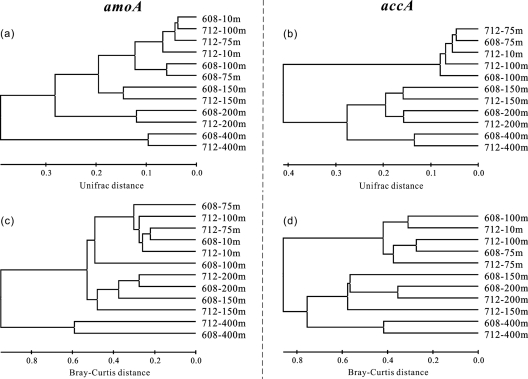
Both phylogeny- and ecotype-based cluster analyses demonstrated almost identical depth-related patterns for the amoA and accA genes (Fig. 7). Generally, the community structures of both genes could be divided into three major clusters (Fig. 7): a euphotic zone cluster (≤100 m), a deep epipelagic cluster (150 m and 200 m), and a mesopelagic cluster (400 m). Mantel tests verified that there were significant correlations among these clusters (r > 0.5; P < 0.001).
Fig. 7.
Clustering of the crenarchaeal amoA and accA clone libraries based on the weighted UniFrac algorithm (a and b) or based on the Bray-Curtis algorithm of ecotype abundance (c and d). The scale bar indicates the UniFrac distance (a and b) or the Bray-Curtis distance (c and d).
DISCUSSION
Spatial distribution of MGI in the ECS.
The spatial dynamics of pelagic microbial populations is a central concern in microbial oceanography (14). Several recent studies indicate that ocean water masses play an important role in shaping the community structures and distribution patterns of plegia bacteria (2, 17, 22, 63) and archaea (1, 16, 30). The results of the present study did verify the recognition with bacterial data from the two investigation stations with heterogeneity in hydrographic characteristics. However, our archaeal data, including those from crenarchaeal 16S rRNA gene-based DGGE analysis (Fig. 4b and see Fig. S1 in the supplemental material) and amoA and accA gene-based clone library analyses (Fig. 7a and b), showed highly similar crenarchaeal communities at both investigation stations. The case of archaea is unexpected but consistent with some previous studies showing that communities of pelagic archaea from similar water depths appear to be similar in structure regardless of geographic location (25, 35, 61). The observed distinct bacterial communities and similar archaeal communities between the two hydrological sites suggest different controlling mechanisms for bacteria and archaea in spatial dynamics, which is of great interest for future studies.
Niche partitioning of MGI in the upper ocean of the ECS.
As shown by the DGGE profiles of 16S rRNA genes showing that depth-stratified MGI populations inhabited both stations, archaeal communities are less variable in geographic dimension but more variable along the water depth. The functional gene-based clone library analyses revealed that the crenarchaeal amoA and accA genes diverged into two lineages (shallow and deep groups) associated with different water layers (epipelagia versus mesopelagia) (Fig. 5 and see Fig. S2 in the supplemental material). These results were similar to those observed previously for other ocean regions (5, 40, 42, 60). In the central California Current, members of the “deep group” are strongly associated with deepwater transportation but are less active than their shallow-water-adapted counterparts, providing evidence that these two independent MGI groups indeed represent shallow- and deep-water-adapted “ecotypes” (48). In the present work, we applied AdaptML modeling and found 10 ecotypes for the amoA and accA genes, while only two general lineages were demarcated based on common phylogenetic analyses (Fig. 6). Definition of ecotypes with higher resolution would provide new insights into the evolutionary mechanisms of MGI speciation. More interestingly, although the accA gene abundance was extremely low in the epipelagic zone, 8 out of 10 ecotypes of both genes had similar depth-related distribution patterns (Fig. 6 and 7c and d). This finding suggested that MGI functional groups containing amoA and accA underwent a similar ecological adaptation and evolutionary history. This conclusion is further supported by the almost consistent clustering network observation based on phylogenetic branch length or ecotype abundance (Fig. 7).
Although the mechanisms of the niche partitioning of MGI with depth remain unknown, light and oxygen have been proposed to play a significant role in structuring of communities of ammonia-oxidizing crenarchaea in the ocean (42, 43). In the present study, the ecotypes of MGI in the oxygenated upper ocean of the ECS had a strongly restricted distribution with depths where the light intensity was attenuated across the water column, suggesting that light might have been a potential key factor resulting in this depth-related phylogenetic partitioning of MGI. However, it was impossible to exclude the effects of other environmental gradients existing in the water column, such as dissolved organic matter (DOM) and pressure. Further studies are needed to establish a certain relationship between environmental variables and crenarchaeal ecotypes.
Ecological roles of MGI in the ocean.
The qPCR assays indicated an amoA-to-16S rRNA gene ratio close to 2 at both stations, which is within the reference ranges reported previously for other regions of the Pacific Ocean (5, 8, 25, 48). Numerous studies have demonstrated that crenarchaeal amoA genes are more abundant than those of AOB in various open oceans (5, 10, 48, 58). In agreement with the results of those studies, our data suggest that ammonia-oxidizing archaea outcompete AOB in the open region of the study sites, which might be explained by the conspicuous adaptability of MGI to low concentrations of ammonia (38).
It was suggested previously that meso- and bathypelagic Crenarchaeota may lack the amoA gene, given the low ratios of amoA to 16S rRNA genes, and therefore are heterotrophic (1, 10). This conclusion remains debatable due to the less comprehensive primers used in those studies (8, 33, 60). In this study, besides the amoA genes, we quantified the abundance of accA genes, one of the key genes in the 3-hydroxypropionate/4-hydroxybutyrate pathway of mesophilic Crenarchaeota. Our results indicated that crenarchaeal accA genes were almost absent in the euphotic zone but were more abundant below the euphotic zone. The scarcity of crenarchaeal accA genes in the euphotic zone raised the possibility that epipelagic Crenarchaeota may rely on chemolithoheterotrophy and play a minor role in dissolved inorganic carbon fixation. However, a ratio of 1 amoA gene copy to 1 accA gene copy was found previously in the metagenomic data set recovered from surface water of the Sargasso Sea (21). One possible explanation for this discrepancy is that some accA genes of epipelagic Crenarchaeota might have been missed in our qPCR assays, since the primers commonly used were designed based on only a few available sequences (25). In contrast, in the upper mesopelagic zone, the ratios of crenarchaeal accA to MGI 16S rRNA genes or to crenarchaeal amoA genes were close to 1, which was in agreement with our previous investigation in the South China Sea (25). This finding suggested that ammonia oxidization may be an important energy source for autotrophic CO2 fixation by MGI in deep waters (1, 24). Such chemoautotrophy can reduce the respiratory consumption of DOM and further provide fresh DOM for other microbial carbon demands. An earlier study revealed that CO2 fixation by marine Crenarchaeota can meet the substantial carbon demand of the deep-sea microbial food web (46). A portion of such fresh DOM could ultimately be transformed into recalcitrant DOM through the microbial carbon pump contributing ocean carbon sequestration (28). The increasing ratio of crenarchaeal accA to amoA or to MGI 16S rRNA genes from the euphotic zone to the mesopelagic zone suggests that MGI could play a more important role in the dark ocean. The recognition of such a function of archaea at taxonomic- and functional-group levels would shed light on the mechanisms of carbon cycling in the ocean. A comprehensive view of the archaeal community structure and its ecological functioning is to be acquired through multiple approaches, including metagenomics, proteomics, and metabolomics, in the future.
Supplementary Material
ACKNOWLEDGMENTS
We thank the captain and crew of the RV Dongfanghong #2, L. K. Hao and H. Y. Cai for assistance during sampling, J. W. Tian for providing the temperature and salinity data, and Lawrence David and Albert Wang for assistance in using the Adapt ML software. We thank John Hodgkiss for his help in polishing the English.
This work was supported by the NSFC project (grants 91028001, 41076063, 40906059), the SOA project (grant 201105021), and the NCET (09-0683). Anyi Hu was partially supported by the MEL Young Scientist Visiting Fellowship (MELRS1026) from the State Key Laboratory of Marine Environmental Science at Xiamen University.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Agogué H., Brink M., Dinasquet J., Herndl G. J. 2008. Major gradients in putatively nitrifying and non-nitrifying Archaea in the deep North Atlantic. Nature 456:788–791 [DOI] [PubMed] [Google Scholar]
- 2. Agogué H., Lamy D., Neal P. R., Sogin M. L., Herndl G. J. 2011. Water mass specificity of bacterial communities in the North Atlantic revealed by massively parallel sequencing. Mol. Ecol. 20:258–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bano N., Ruffin S., Ransom B., Hollibaugh J. T. 2004. Phylogenetic composition of Arctic Ocean archaeal assemblages and comparison with Antarctic assemblages. Appl. Environ. Microbiol. 70:781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beman J. M., Francis C. A. 2006. Diversity of ammonia-oxidizing archaea and bacteria in the sediments of a hypernutrified subtropical estuary: Bahia del Tobari, Mexico. Appl. Environ. Microbiol. 72:7767–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beman J. M., Popp B. N., Francis C. A. 2008. Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J. 2:429–441 [DOI] [PubMed] [Google Scholar]
- 6. Brochier-Armanet C., Boussau B., Gribaldo S., Forterre P. 2008. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 6:245–252 [DOI] [PubMed] [Google Scholar]
- 7. Carlson C. A., et al. 2009. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 3:283–295 [DOI] [PubMed] [Google Scholar]
- 7a. Chen C., Zhu J., Beardsley R. C., Franks P. J. S. 2003. Physical-biological sources for dense algal blooms near the Changjiang River. Geophys. Res. Lett. 30:22–1–22-4 [Google Scholar]
- 8. Church M. J., Wai B., Karl D. M., DeLong E. F. 2010. Abundances of crenarchaeal amoA genes and transcripts in the Pacific Ocean. Environ. Microbiol. 12:679–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Culman S., Bukowski R., Gauch H., Cadillo-Quiroz H., Buckley D. 2009. T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinformatics 10:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Corte D., Yokokawa T., Varela M. M., Agogue H., Herndl G. J. 2009. Spatial distribution of Bacteria and Archaea and amoA gene copy numbers throughout the water column of the Eastern Mediterranean Sea. ISME J. 3:147–158 [DOI] [PubMed] [Google Scholar]
- 11. DeLong E. F. 1992. Archaea in coastal marine environment. Proc. Natl. Acad. Sci. U. S. A. 89:5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DeLong E. F., et al. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496–503 [DOI] [PubMed] [Google Scholar]
- 13. Francis C. A., Roberts K. J., Beman J. M., Santoro A. E., Oakley B. B. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fuhrman J. A. 2009. Microbial community structure and its functional implications. Nature 459:193–199 [DOI] [PubMed] [Google Scholar]
- 15. Fuhrman J. A., McCallum K., Davis A. A. 1992. Novel major archaebacterial group from marine plankton. Nature 356:148–149 [DOI] [PubMed] [Google Scholar]
- 16. Galand P. E., et al. 2009. Archaeal diversity and a gene for ammonia oxidation are coupled to oceanic circulation. Environ. Microbiol. 11:971–980 [DOI] [PubMed] [Google Scholar]
- 17. Galand P. E., Potvin M., Casamayor E. O., Lovejoy C. 2009. Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J. 4:564–576 [DOI] [PubMed] [Google Scholar]
- 18. Garcia-Martinez J., Rodriguez-Valera F. 2000. Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of group I. Mol. Ecol. 9:935–948 [DOI] [PubMed] [Google Scholar]
- 19. Good I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237–264 [Google Scholar]
- 20. Hallam S. J., et al. 2006. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. U. S. A. 103:18296–18301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hallam S. J., et al. 2006. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 4:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hamilton A. K., Lovejoy C., Galand P. E., Ingram R. G. 2008. Water masses and biogeography of picoeukaryote assemblages in a cold hydrographically complex system. Limnol. Oceanogr. 53:922–935 [Google Scholar]
- 23. Hammer Ø., Harper D. A. T., Ryan P. D. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4:9 [Google Scholar]
- 24. Hansman R. L., et al. 2009. The radiocarbon signature of microorganisms in the mesopelagic ocean. Proc. Natl. Acad. Sci. U. S. A. 106:6513–6518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu A., Jiao N., Zhang C. L. 2011. Community structure and function of planktonic Crenarchaeota: changes with depth in the South China Sea. Microb. Ecol. 61:549–563 [DOI] [PubMed] [Google Scholar]
- 26. Hu A., et al. 2010. Community structures of ammonia-oxidising archaea and bacteria in high-altitude lakes on the Tibetan Plateau. Freshw. Biol. 55:2375–2390 [Google Scholar]
- 27. Hunt D. E., et al. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085 [DOI] [PubMed] [Google Scholar]
- 28. Jiao N., et al. 2010. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8:593–599 [DOI] [PubMed] [Google Scholar]
- 29. Jiao N., Yang Y., Koshikawa H., Watanabe M. 2002. Influence of hydrographic conditions on picoplankton distribution in the East China Sea. Aquat. Microb. Ecol. 30:37–48 [Google Scholar]
- 30. Kalanetra K. M., Bano N., Hollibaugh J. T. 2009. Ammonia-oxidizing Archaea in the Arctic Ocean and Antarctic coastal waters. Environ. Microbiol. 11:2434–2445 [DOI] [PubMed] [Google Scholar]
- 31. Karner M. B., DeLong E. F., Karl D. M. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507–510 [DOI] [PubMed] [Google Scholar]
- 32. Könneke M., et al. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543–546 [DOI] [PubMed] [Google Scholar]
- 33. Konstantinidis K. T., Braff J., Karl D. M., DeLong E. F. 2009. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific Subtropical Gyre. Appl. Environ. Microbiol. 75:5345–5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kowalchuk G. A., Stephen J. R. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485–529 [DOI] [PubMed] [Google Scholar]
- 34a. Lee Z. P., et al. 2007. Euphotic zone depth: its derivation and implication to ocean-color remote sensing. J. Geophys. Res. 112:C03009 [Google Scholar]
- 35. Liu B., et al. 2009. Community structure of Archaea in the water column above gas hydrates in the Gulf of Mexico. Geomicrobiol. J. 26:363–369 [Google Scholar]
- 36. Lozupone C., Hamady M., Knight R. 2006. UniFrac—an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ludwig W., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martens-Habbena W., Berube P. M., Urakawa H., de la Torre J. R., Stahl D. A. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979 [DOI] [PubMed] [Google Scholar]
- 39. Martiny J. B. H., et al. 2006. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 4:102–112 [DOI] [PubMed] [Google Scholar]
- 40. Massana R., DeLong E. F., Pedros-Alio C. 2000. A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl. Environ. Microbiol. 66:1777–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Millero F. J., Chen C., Bradshaw A., Schleicher K. 1980. A new high pressure equation of state for seawater. Deep Sea Res. 27A:255–264 [Google Scholar]
- 42. Mincer T. J., et al. 2007. Quantitative distribution of presumptive archaeal and bacterial nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 9:1162–1175 [DOI] [PubMed] [Google Scholar]
- 43. Molina V., Belmar L., Ulloa O. 2010. High diversity of ammonia-oxidizing archaea in permanent and seasonal oxygen-deficient waters of the eastern South Pacific. Environ. Microbiol. 12:2450–2465 doi:10.1111/j.1462-2920.2010.02218.x [DOI] [PubMed] [Google Scholar]
- 44. Nicol G. W., Leininger S., Schleper C., Prosser J. I. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966–2978 [DOI] [PubMed] [Google Scholar]
- 44a. Oakley B. B., Carbonero F., van der Gast C. J., Hawkins R. J., Purdy K. J. 2010. Evolutionary divergence and biogeography of sympatric niche-differentiated bacterial populations. ISME J. 4:488–497 [DOI] [PubMed] [Google Scholar]
- 45. Ochsenreiter T., Selezi D., Quaiser A., Bonch-Osmolovskaya L., Schleper C. 2003. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 5:787–797 [DOI] [PubMed] [Google Scholar]
- 46. Reinthaler T., van Aken H. M., Herndl G. J. 2010. Major contribution of autotrophy to microbial carbon cycling in the deep North Atlantic's interior. Deep Sea Res. II 57:1572–1580 [Google Scholar]
- 47. Ronquist F., Huelsenbeck J. P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574 [DOI] [PubMed] [Google Scholar]
- 48. Santoro A. E., Casciotti K. L., Francis C. A. 2010. Activity, abundance and diversity of nitrifying archaea and bacteria in the central California current. Environ. Microbiol. 12:1989–2006 [DOI] [PubMed] [Google Scholar]
- 49. Schattenhofer M., et al. 2009. Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ. Microbiol. 11:2078–2093 [DOI] [PubMed] [Google Scholar]
- 50. Schloss P. D., Handelsman J. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Spang A., et al. 2010. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 18:331–340 [DOI] [PubMed] [Google Scholar]
- 52. Stamatakis A., Ludwig T., Meier H. 2005. RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics 21:456–463 [DOI] [PubMed] [Google Scholar]
- 53. Swofford D. L. 2003. PAUP*, phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- 54. Treusch A. H., et al. 2005. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ. Microbiol. 7:1985–1995 [DOI] [PubMed] [Google Scholar]
- 55. Treusch A. H., et al. 2009. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 3:1148–1163 [DOI] [PubMed] [Google Scholar]
- 56. Venter J. C., et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74 [DOI] [PubMed] [Google Scholar]
- 57. Walker C. B., et al. 2010. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc. Natl. Acad. Sci. U. S. A. 107:8818–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wuchter C., et al. 2006. Archaeal nitrification in the ocean. Proc. Natl. Acad. Sci. U. S. A. 103:12317–12322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yakimov M. M., Conoa V. L., Denaroa R. 2009. A first insight into the occurrence and expression of functional amoA and accA genes of autotrophic and ammonia-oxidizing bathypelagic Crenarchaeota of Tyrrhenian Sea. Deep Sea Res. II 56:748–754 [Google Scholar]
- 60. Yakimov M. M., et al. 2011. Contribution of crenarchaeal autotrophic ammonia oxidizers to the dark primary production in Tyrrhenian deep waters (Central Mediterranean Sea). ISME J. 5:945–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zeng Y. H., Li H. Y., Jiao N. Z. 2007. Phylogenetic diversity of planktonic archaea in the estuarine region of East China Sea. Microbiol. Res. 162:26–36 [DOI] [PubMed] [Google Scholar]
- 62. Zhang R., Thiyagarajan V., Qian P. Y. 2008. Evaluation of terminal-restriction fragment length polymorphism analysis in contrasting marine environments. FEMS Microbiol. Ecol. 65:169–178 [DOI] [PubMed] [Google Scholar]
- 63. Zhang Y., Jiao N. 2007. Dynamics of aerobic anoxygenic phototrophic bacteria in the East China Sea. FEMS Microbiol. Ecol. 61:459–469 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.