Abstract
The mevalonate pathway is utilized for the biosynthesis of isoprenoids in many bacterial, eukaryotic, and archaeal organisms. Based on previous reports of its feedback inhibition, mevalonate kinase (MVK) may play an important regulatory role in the biosynthesis of mevalonate pathway-derived compounds. Here we report the purification, kinetic characterization, and inhibition analysis of the MVK from the archaeon Methanosarcina mazei. The inhibition of the M. mazei MVK by the following metabolites derived from the mevalonate pathway was explored: dimethylallyl diphosphate (DMAPP), geranyl pyrophosphate (GPP), farnesyl pyrophosphate (FPP), isopentenyl monophosphate (IP), and diphosphomevalonate. M. mazei MVK was not inhibited by DMAPP, GPP, FPP, diphosphomevalonate, or IP, a proposed intermediate in an alternative isoprenoid pathway present in archaea. Our findings suggest that the M. mazei MVK represents a distinct class of mevalonate kinases that can be differentiated from previously characterized MVKs based on its inhibition profile.
INTRODUCTION
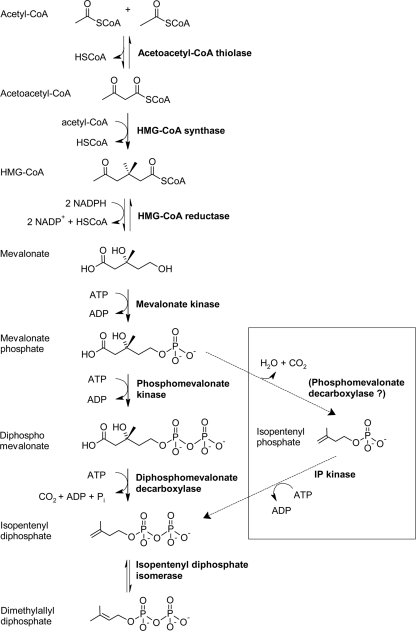
Isoprenoids are a large and diverse class of compounds containing more than 40,000 naturally occurring terpenes and terpenoids (33). They encompass many classes of bioactive molecules, including carotenoids, steroid hormones, phytols, redox carriers, secondary metabolites, and pheromones, that make them commercially attractive for the production of compounds varying from pharmaceuticals to biofuels (5, 15, 21, 23, 24). Currently, a number of groups are working on increasing the production of terpenoid compounds for a variety of medicinal, agricultural, sustainable biofuel, and biomaterial applications (21, 23, 24, 33). All isoprenoids are biosynthesized from the five carbon precursors, isopentenyl diphosphate (IPP), and its isomer, dimethylallyl diphosphate (DMAPP). Two pathways for the biosynthesis of these central metabolites have been described, the mevalonate pathway (28) and the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway (25). The mevalonate pathway typically is found in animals, plants, and in many Gram-positive bacteria, including Streptococcus pneumoniae (17, 31, 32). Some enzymes of the mevalonate pathway also have been identified in archaea; however, the complete pathway has not been elucidated (27). The mevalonate pathway catalyzes the conversion of three molecules of acetyl coenzyme A (CoA) to IPP and DMAPP. Briefly, two molecules of acetyl-CoA undergo a Claisen condensation to form acetoacetyl-CoA, which is catalyzed by acetoacetyl-CoA thiolase. 3-Hydroxy-3-methylglutaryl-CoA (HMG-CoA) synthase then catalyzes an aldol reaction between acetoacetyl-CoA and a third molecule of acetyl-CoA. The conversion of HMG-CoA to mevalonate subsequently is catalyzed by HMG-CoA reductase. Mevalonate kinase (MVK) and phosphomevalonate kinase (PMK) catalyze the phosphorylation of the primary alcohol of mevalonate and the phosphate of phosphomevalonate, respectively, to form diphosphomevalonate. The penultimate reaction in the pathway is the phosphorylative decarboxylation of diphosphomevalonate catalyzed by the diphosphomevalonate decarboxylase to yield IPP (10, 32). IPP isomerase (IDI) catalyzes the conversion of IPP to DMAPP (Fig. 1).
Fig. 1.
Mevalonate pathway. The proposed modified pathway for the production of isoprenoids in archaea organisms is illustrated in the box (17).
A distinguishing characteristic of archaeal organisms is that isoprenoids make up the major component of their membrane lipids. In contrast, the lipids of eukaryotic and bacterial organisms are composed primarily of fatty acids (6, 17, 20, 27). Studies of isoprenoid biosynthesis in archaea have demonstrated that both acetate and mevalonate are precursors for IPP formation, indicating that the mevalonate pathway is involved in their biosynthesis (11, 17). Putative homologues of all mevalonate pathway genes, excluding the diphosphomevalonate decarboxylase, have been identified in archaea by genomic analysis (3, 7, 17, 19). In addition, putative isopentenyl monophosphate kinases have been identified and characterized from archaea, suggesting the possible utilization of a modified mevalonate pathway for the production of isoprenoids in archaea (8, 17) (Fig. 1).
Eukaryotic, bacterial, and archaeal organisms must ensure the sufficient production of a variety of isoprenoid compounds that are essential for the proper growth, signaling, transport, and life cycle controls as well as the prevention of the overaccumulation of potentially toxic products, such as cholesterol (15, 27). Organisms manage these tasks through the intricate regulation of isoprenoid-producing pathways (15). MVK was demonstrated to be an important regulatory point in the mevalonate pathway in both bacteria (1, 2, 31) and eukaryotes (4, 9, 13, 16, 18). Previously the small-molecule regulation of MVKs could be divided into two classes. The first class is inhibited by metabolites downstream of the diphosphomevalonate decarboxylase reaction (IPP, DMAPP, GPP, FPP, and longer chain isoprenoids) (9, 16, 18, 31). The regulation of a eukaryotic MVK isolated from pig liver was first reported by Dorsey and Porter in 1968 (9). Their detailed kinetic analysis revealed the significant feedback regulation of this enzyme by GPP and FPP and, to a lesser degree, by DMAPP, IPP, and PPi (9). Human MVK subsequently was characterized and found to be inhibited by FPP, GPP, IPP, DMAPP, and geranylgeranyl pyrophosphate (18, 22). The characterization of four plant MVKs and S. cerevisiae MVK by Gray and Kekwick in 1972 revealed that they all are inhibited by GPP, FPP, geranylgeranyl pyrophosphate, and phytyl pyrophosphate (16). In addition, two MVKs from Gram-positive cocci, Staphylococcus aureus and Enterococcus faecalis, were found to be competitively inhibited by FPP with respect to ATP, with a Ki of 45 μM (31).
The second class of MVKs is inhibited by diphosphomevalonate but not by metabolites downstream of the diphosphomevalonate decarboxylase. Interestingly, DMAPP, IPP, GPP, and FPP were not feedback inhibitors of the Gram-positive bacterium S. pneumoniae MVK at concentrations of up to 12 μM; however, diphosphomevalonate inhibited S. pneumoniae MVK at nanomolar concentrations (2).
We report the overexpression, purification, kinetic analysis, and inhibition studies of the mvk gene product from the archaeon Methanosarcina mazei. The S. cerevisiae and S. pneumoniae MVKs have been recharacterized in this study and serve as positive controls for the two known classes of feedback-regulated MVKs. Our findings demonstrate that, unlike MVKs from S. cerevisiae and S. pneumoniae, M. mazei MVK is not inhibited by known feedback inhibitors of MVKs. A phylogenetic tree of 29 MVK representatives from Archaea, Eukarya, and Bacteria indicates a clear evolutionary separation of the mvk gene between these domains and leads to the hypothesis that these distinct branches utilize alternative regulation mechanisms (Fig. 2).
Fig. 2.
Phylogenetic tree for MVKs from the mevalonate pathway of Eukarya, Archaea, and Bacteria.
Accordingly, we conclude that there are at least three classes of MVKs that can be differentiated based on their inhibition profiles.
MATERIALS AND METHODS
Expression vectors, cell lines, and competent cells were purchased from Invitrogen (Carlsbad, CA). Carbenicillin, kanamycin, and chloramphenicol were obtained from Novagen (Gibbstown, NJ), IBI Scientific (Peosta, IA), and Calbiochem (Gibbstown, NJ), respectively. Isopropyl thiogalactoside (IPTG), geranyl pyrophosphate (GPP), farnesyl pyrophosphate (FPP), isopentenyl monophosphate (IP), ATP, phosphoenolpyruvate (PEP), NADH, MgCl2, NaCl, Tris, HEPES, dithiothreitol (DTT), DNase I, and lysozyme were purchased from Sigma (St. Louis, MO). Dimethylallyl diphosphate (DMAPP) was obtained from Cayman Chemicals (Ann Arbor, MI). Lactate dehydrogenase (LDH) was purchased from Calbiochem. Pyruvate kinase (PK) was purchased from MP Biomedicals LLC (Solon, OH). Mevalonate solution was prepared from mevalonic acid, which was purified by Stereo Chemicals, Inc. (Newark, DE). All columns used in purification were obtained from GE Healthcare (Piscataway, NJ). Purity was assessed by gel electrophoresis using precast gels and reagents purchased from Invitrogen. Kinetic studies were conducted using a SpectraMax 190 plate reader from Molecular Devices (Sunnyvale, CA). Kinetic data were analyzed using Kaleidagraph 4.0 (Synergy Software).
Preparation of MVK expression strains.
A synthetic gene encoding the M. mazei MVK was designed based on NP_633786 from Methanosarcina mazei Go1; synthesis and codon optimization were performed by DNA 2.0. This gene was amplified by PCR using the following primer set: forward, 5′-CACCATGGTATCCTGTTCTGCG-3′, and reverse, 5′-TTAATCTACTTTCAGACCTTGC-3′. The PCR cycles were 94°C for 2 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 75 s; 72°C for 7 min; and 4°C overnight. The 0.9-kb PCR product was cloned into the pET200D vector per the manufacturer's instructions. Transformants were selected on LA/Kan50 plates (Teknova) at 37°C. Plasmid was isolated, sequence verified, and transformed into Escherichia coli BL21(λDE3) pLysS cells per the manufacturer's protocol.
The mvk gene from S. cerevisiae, containing the NdeI restriction site, was amplified by PCR from yeast chromosomal DNA using the following primer set: forward, 5′-CAGCAGCAGCATATGTCATTACCGTTCTTAACTTC-3′, and reverse, 5′-CAGCAGCAGCATATGGCCTATCGCAAATTAGCTTATG-3′. The PCR cycles were 95°C for 2 min; 29 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 21 s; 72°C for 3 min; and 4°C overnight. The 1.4-kb products were purified using a QIAquick gel extraction kit (Qiagen), treated with shrimp alkaline phosphatase and NdeI, ligated overnight to the pET-16b (Invitrogen) vector harboring a hexahistidine tag, and transformed into chemically competent TOP10 cells per the manufacturer's protocol. Plasmids from transformants were purified via a QIAprep spin Miniprep kit (Qiagen), and the insert was sequenced using T7 primers (forward, 5′-TAATACGACTCACTATAGGG-3′; reverse, 5′-GCTAGTTATTGCTCAGCGG-3′). Verified clones were transformed into E. coli BL21(λDE3) pLysS cells per the manufacturer's protocol.
The S. pneumoniae DNA region coding for MVK was amplified by PCR from ATCC strain BAA-255D-5 using gene-specific primers (forward, 5′-CACCATGACAAAAAAAGTTGGTGTCGGTCAGGCAC-3′; reverse, 5′-CTGTCACAGGCTCTCTATCCATGTCTGAAC-3′). The PCR cycles were 95°C for 4 min; 5 cycles of 95°C for 20 s, 52°C for 20 s, and 72°C for 30 s; 25 cycles of 95°C for 20 s, 55°C for 20 s, and 72°C for 30 s; 72°C for 10 min; and 4°C overnight. The 0.9-kb fragment was TOPO cloned into the pET200D-TOPO expression vector and transformed into chemically competent E. coli TOP10 cells according to the manufacturer's recommended protocol. Colonies were screened by PCR using the primers T7 forward (5′-TAATACGACTCACTATAGGG-3′) and T7 reverse (5′-CTGTCACAGGCTCTCTATCCATGTCTGAAC-3′). Positive plasmids were purified via a QIAprep spin Miniprep kit (Qiagen) and transformed into chemically competent E. coli BL21 Star (λDE3) cells for expression analysis.
Expression and purification of recombinant MVKs from M. mazei, S. cerevisiae, and S. pneumoniae.
Cells containing the M. mazei MVK expression plasmid were grown in Terrific broth (26) supplemented with 50 mg/liter kanamycin and 30 mg/liter chloramphenicol and were induced overnight with the addition of 0.5 mM IPTG. Cells containing the MVK expression plasmids were grown in Luria-Bertani broth (26) supplemented with 50 mg/liter carbenicillin and 30 mg/liter chloramphenicol for the expression of S. cerevisiae MVK or 50 mg/liter kanamycin for the expression of S. pneumoniae MVK, and they were induced overnight with the addition of 0.2 mM IPTG at an optical density at 600 nm (OD600) of ∼0.4 to 0.6. All cells were harvested by centrifugation at 10,000 × g for 10 min and resuspended in 0.05 M sodium phosphate, 0.3 M sodium chloride, 0.02 M imidazole (pH 8.0) buffer containing lysozyme and DNase I. Resuspended cells were lysed by repeated passes through a French pressure cell at 20,000 lb/in2. Cell lysates were clarified by ultracentrifugation at 229,000 × g for 1 h. The supernatants were loaded onto a HiTrap IMAC HP column charged with nickel sulfate and equilibrated with 0.05 M sodium phosphate, 0.3 M sodium chloride, 0.02 M imidazole (pH 8.0). Enzymes were isolated using a linear gradient from 0.02 to 0.5 M imidazole. Fractions containing MVK were identified using SDS-PAGE (Invitrogen), pooled, and desalted into 0.05 M HEPES, 0.05 M sodium chloride (pH 7.4) with 1 mM DTT using a Hi Prep 26/10 desalting column. MVK from S. cerevisiae was further purified on an anion-exchange HiTrap Q HP column. The column was washed with 0.05 M Tris, 0.05 M sodium chloride (pH 7.6) with 1 mM DTT and eluted with a 0.05 to 1.0 M sodium chloride gradient. Fractions containing MVK were desalted into 0.05 M HEPES, 0.05 M sodium chloride (pH 7.4) containing 1 mM DTT. The purity of all three enzymes was greater than 95% as judged by SDS-PAGE and Coomassie staining. The proteins were optically quantitated at 280 nm using the following conversion factors: 0.343 OD/mg/ml for M. mazei MVK, 0.597 OD/mg/ml for S. cerevisiae MVK, and 0.516 OD/mg/ml for S. pneumoniae MVK. These values were obtained using the ExPASy ProtParam tool (14).
Expression and purification of recombinant PMK enzyme from S. cerevisiae.
The S. cerevisiae DNA region coding for the PMK protein was amplified by PCR using gene specific primers (forward, 5′-CACCTCAGAGTTGAGAGCCTTCAGTGC-3′; reverse, 5′-GAATTCTGCATGCAGCTACCTTAAG-3′), TOPO cloned into the pET200D-TOPO expression vector (Invitrogen), and transformed into chemically competent E. coli TOP10 cells according to the manufacturer's recommended protocol. Colonies were screened by PCR using T7 forward (5′-TAATACGACTCACTATAGGG-3′) and a gene-specific reverse (5′-GAATTCTGCATGCAGCTACCTTAAG-3′) primer. Positive plasmids were purified via a QIAprep spin Miniprep kit (Qiagen) and transformed into chemically competent E. coli BL21(λDE3) cells for expression analysis. Cells containing the PMK expression plasmid were grown in Terrific broth supplemented with 50 mg/liter kanamycin. The culture was induced with 0.2 mM IPTG at an OD600 of 0.9 and harvested by centrifugation after 6 h at 30°C. The purification of PMK involved nickel affinity and anion-exchange chromatography and followed the same protocol as that described above for S. cerevisiae MVK. The purity was greater than 95% as judged by SDS-PAGE and Coomassie staining. The protein was optically quantitated at 280 nm using a conversion factor of 1.099 OD/mg/ml.
Native molecular mass determination.
The native molecular masses of the MVKs and PMK were determined by size-exclusion chromatography using a Superdex 200 10/300 GL column. The column was equilibrated using the following seven molecular standards with masses ranging from 6.5 to 669 kDa: aprotinin (6.5 kDa), RNase A (13.7 kDa), carbonic anhydrase (29 kDa), ovalbumin (43 kDa), conalbumin (75 kDa), aldolase (158 kDa), and thyroglobulin (669 kDa). The column void volume was calculated using the elution volume of blue dextran 2000. Column equilibration and sample runs were performed in 50 mM HEPES, 150 mM NaCl (pH 7.4) buffer containing 1 mM DTT at room temperature. The masses of M. mazei, S. cerevisiae, and S. pneumoniae MVKs, as well as that of S. cerevisiae PMK, were calculated using the linear fit to the plot of log masses versus the elution volume obtained for the molecular standards.
Enzyme activity and inhibition by DMAPP, GPP, and FPP.
The catalytic activities of the MVKs were measured using a modified spectrophotometric assay that couples ADP formation to pyruvate synthesis and reduction to lactate (13). The initial rate of disappearance of NADH serves as a measure of the phosphorylation of mevalonate by MVK. The assays were performed in triplicate in a 96-well plate (Costar catalog number 9017) format at 30°C. Each 100-μl reaction mixture contained 0.4 mM PEP, 0.05 mM DTT, 0.32 mM NADH, 10 mM MgCl2, 2 U of LDH, and 2 U of PK in 50 mM Tris, 50 mM NaCl (pH 7.6).
The Michaelis constants for MVK (designated Km-Mev) from M. mazei and S. pneumoniae were determined at a saturating concentration of ATP (5 mM) and with mevalonate concentrations ranging from 0.005 to 5 mM. The reaction was initiated with the addition of 80 nM (0.25 μg) purified M. mazei or 60 nM (0.21 μg) S. pneumoniae MVK. The Km-ATP for these MVKs was determined similarly, using saturating concentrations of mevalonate (1.25 mM) and ATP concentrations ranging from 0.005 to 5 mM. Km values for S. cerevisiae MVK were determined using the same procedure with the following exceptions: substrate concentrations ranged from 0.039 to 5 mM, and the reaction was initiated by adding 10 nM (50.1 ng) of purified S. cerevisiae MVK. Absorbance changes associated with the amount of NADH oxidized to NAD+ were monitored continuously at 340 nm and plotted against time to determine the rate of the MVK-coupled reactions. Protein inhibition studies were performed in quadruplicate by adding terpenyl diphosphates (DMAPP, GPP, FPP, and diphosphomevalonate) at various concentrations to the reaction mix. The inhibition studies of M. mazei MVK also included studies with isopentenyl monophosphate.
Inhibition of MVKs by diphosphomevalonate.
The inhibition of three MVKs by diphosphomevalonate was investigated using a spectrophotometric pyruvate kinase- and lactate dehydrogenase-coupled assay, as previously described (2). This approach couples two reactions of the mevalonate pathway, the initial phosphorylation of mevalonate by MVK and the subsequent conversion of phosphomevalonate to diphosphomevalonate by PMK. Reaction mixtures contained 100 mM Tris-HCl, 100 mM NaCl, 1 mM DTT, 10 mM MgCl2, 5 mM ATP, 2.5 mM NADH, 4 mM PEP, 10 U of LDH, 10 U of PK, and 1 mM mevalonate. Initially, MVK was added to the reaction mixture and the depletion of NADH was monitored at 386 nm. After all of the mevalonate was converted to phosphomevalonate, S. cerevisiae PMK was added to the mixture to catalyze the reaction from phosphomevalonate to diphosphomevalonate. To test for the feedback inhibition of the MVK by diphosphomevalonate, both PMK and MVK were added simultaneously to the reaction mixture. The inhibition of M. mazei MVK by diphosphomevalonate was evaluated using 1.7 μM MVK and 2 μM PMK. Inhibition studies of S. cerevisiae and S. pneumoniae MVKs by diphosphomevalonate utilized 0.1 μM MVK and 1 μM PMK as well as 1 μM MVK and 2 μM PMK, respectively.
Phylogenetic analysis of MVK.
Sequences of MVK from a range of different organisms were retrieved and aligned using ClustalW multiple sequence alignment (30). A rooted phylogenetic tree (phenogram) was derived using the program DrawGram (12).
RESULTS
Characterization of MVKs.
Three MVKs, from M. mazei, S. cerevisiae, and S. pneumoniae, as well as PMK from S. cerevisiae, were expressed in E. coli, extracted, and purified using affinity chromatography to >95% apparent homogeneity. The apparent masses of the MVKs and PMK were determined by gel filtration to be 78 kDa for M. mazei MVK, 97 kDa for S. cerevisiae MVK, 72 kDa for S. pneumoniae MVK, and 47 kDa for PMK. The calculated molecular masses using the amino acid sequence are 35.5, 51, 39, and 53 kDa for M. mazei, S. cerevisiae, and S. pneumoniae MVKs and S. cerevisiae PMK, respectively. This suggests that the MVKs tested form dimers and that S. cerevisiae PMK is a monomer in solution.
The rates of mevalonate phosphorylation by the archaeal, eukaryotic, and bacterial MVKs were monitored (2). The apparent Km (Km app) values were evaluated for each enzyme with respect to ATP (Km app-ATP) and mevalonate (Km app-Mev) using the Michaelis-Menten equation (Table 1). Of the three enzymes assayed, M. mazei MVK had the slowest turnover (kcat) at 30°C of 4.3 s−1. S. cerevisiae MVK had a kcat nearly four times faster than that of M. mazei MVK. However, M. mazei MVK had the lowest apparent Km of 68 μM; S. cerevisiae and S. pneumoniae MVKs had apparent Km values of 131 and 236 μM, respectively.
Table 1.
Kinetic characterization of MVKs
| MVK origin | Kinetic constant |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kmapp-Meva (μM) | Kmapp-ATPa (μM) | kcata (s−1) | Ki(DPM)b | Ki-Mev (DMAPP)b (μM) | Ki-ATP (DMAPP)b (μM) | Ki-Mev (GPP)b (μM) | Ki-ATP (GPP)b (μM) | Ki-Mev (FPP)b (μM) | Ki-ATP (FPP)b (μM) | Ki-(IP)b | |
| S. cerevisiae | 131 ± 8 | 650 ± 72 | 38 ± 5 | NDc | 389 ± 25e | 34 ± 17d | 1.8 ± 0.4e | 0.25 ± 0.09d | 1.9 ± 0.6e | 0.13 ± 0.08d | NAg |
| S. pneumoniae | 236 ± 14 | 372 ± 9 | 11 ± 4 | Inhibitedf | >5,000 | >5,000 | >100 | >100 | >100 | >100 | NAg |
| M. mazei | 68 ± 4 | 464 ± 12 | 4.3 ± 0.2 | ND | >5,000 | >5,000 | >100 | >100 | >100 | >100 | NDc |
Km app and kcat values were determined by fitting the Michaelis-Menten equation to the data with Kaleidagraph (Synergy Software). Error values represent one standard deviation from three replicates.
Ki values were determined by fitting the Lineweaver-Burk equation to the data. Error values represent one standard deviation from four replicates.
Not determined (ND) means no inhibition detected.
Competitive inhibition.
Uncompetitive inhibition.
S. pneumoniae MVK is inhibited by diphosphomevalonate (DPM), but this was not quantifiable in this assay.
NA, not applicable.
M. mazei MVK is not inhibited by DMAPP, GPP, FPP, or IP.
The potential inhibition of the M. mazei MVK by the downstream products (DMAPP, GPP, and FPP) of the mevalonate pathway was evaluated. The catalytic activity of M. mazei MVK was not inhibited by 5 mM DMAPP, 100 μM GPP, or 100 μM FPP. The archaeal mevalonate pathway has been postulated to contain an IP kinase that catalyzes the formation of IPP, therefore we examined the inhibition of M. mazei MVK by isopentenyl monophosphate (IP) (17). Our experiments demonstrated that M. mazei MVK is not inhibited by concentrations of IP of up to 100 μM.
S. cerevisiae MVK is inhibited by DMAPP, GPP, and FPP, products of the mevalonate pathway.
The MVK from yeast was reported to be inhibited by GPP, FPP, geranylgeranyl pyrophosphate, and phytyl pyrophosphate (16, 29) and serves as a positive control for a class of MVKs that are inhibited by intermediates downstream of diphosphomevalonate decarboxylase. In this study, the inhibition of S. cerevisiae MVK was probed with the isoprenoid precursors DMAPP, GPP, and FPP. Our results demonstrate that DMAPP, GPP, and FPP are competitive inhibitors of S. cerevisiae MVK with respect to ATP and uncompetitive inhibitors with respect to mevalonate. The inhibition constants (Kis) of DMAPP, GPP, and FPP for the S. cerevisiae MVK with respect to ATP were 34 ± 17 (DMAPP), 0.25 ± 0.09 (GPP), and 0.13 ± 0.08 μM (FPP). The Kis of DMAPP, GPP, and FPP for S. cerevisiae MVK with respect to mevalonate were 389 ± 25 (DMAPP), 1.8 ± 0.4 (GPP), and 1.9 ± 0.6 μM (FPP). The inhibition constants are summarized in Table 1.
Similarly to M. mazei MVK, the S. pneumoniae MVK was previously demonstrated to be uninhibited by DMAPP, GPP, and FPP at concentrations of up to 12 μM (2). Significantly greater concentrations of these metabolites may be encountered during the metabolic engineering of terpenoid pathways; therefore, we assayed the inhibition of S. pneumoniae MVK using 5 mM DMAPP, 100 μM GPP, and 100 μM FPP and confirmed that the S. pneumoniae MVK is not inhibited at these concentrations.
Inhibition of MVK by diphosphomevalonate.
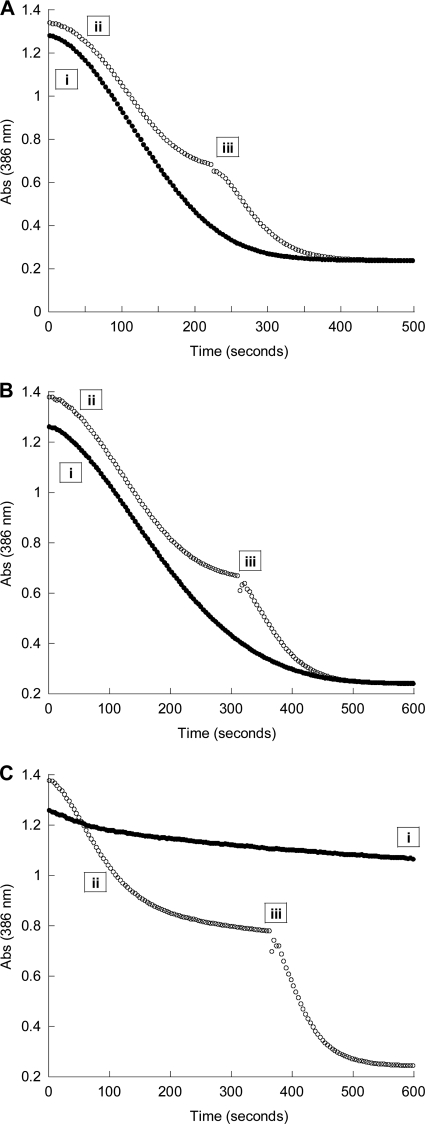
The effect of diphosphomevalonate on the rate of MVK reactions was analyzed using a pyruvate kinase and lactate dehydrogenase coupling system, as previously described (2). Briefly, the addition of mevalonate to the reaction mixture containing MVK resulted in the quantitative conversion of the substrate to phosphomevalonate. The subsequent addition of PMK resulted in the conversion of phosphomevalonate to diphosphomevalonate. To demonstrate feedback inhibition, both MVK and PMK were added at the initiation of the assay. The inhibition of MVK was indicated if the rates of mevalonate conversion to phosphomevalonate and diphosphomevalonate were significantly decreased compared to those from the assays performed by the sequential addition of MVK and PMK. In our studies, when S. cerevisiae PMK and M. mazei MVK were present at the initiation of the reaction, the mevalonate was completely converted into diphosphomevalonate (Fig. 3B). The same result was obtained when S. cerevisiae MVK was assayed with PMK (Fig. 3A), demonstrating that neither M. mazei nor S. cerevisiae MVK is inhibited by diphosphomevalonate. However, when S. pneumoniae MVK and PMK were present at the initiation of the reaction, the velocity of mevalonate conversion significantly decreased (Fig. 3C), verifying the inhibition of S. pneumoniae MVK by diphosphomevalonate (2).
Fig. 3.
Conversion of mevalonate to phosphomevalonate catalyzed by S. cerevisiae (A), M. mazei (B), and S. pneumoniae (C) MVKs was monitored in the presence and absence of S. cerevisiae PMK. The rate of conversion of mevalonate to phosphomevalonate and subsequently to diphosphomevalonate was detected indirectly by the oxidation of NADH at 386 nm. Reactions that were initiated by the simultaneous addition of MVK and PMK are indicated by line i on each graph. Reactions that were initiated with MVK in the absence of PMK are indicated by line ii in each graph. Reactions were allowed to proceed until mevalonate was completely converted to phosphomevalonate. PMK then was added to the reaction mixture at line iii to complete conversion to diphosphomevalonate. Reaction mixtures contained the following components: 100 mM Tris-HCl, 100 mM NaCl, 1 mM DTT, 10 mM MgCl2, 5 mM ATP, 2.5 mM NADH, 4 mM PEP, 10 U of LDH, 10 U of PK, and 1 mM mevalonate. To this mixture was added 0.1 μM S. cerevisiae MVK and 1 μM S. cerevisiae PMK (A), 1.7 μM M. mazei MVK and 2 μM S. cerevisiae PMK (B), and 1 μM S. pneumoniae MVK and 2 μM S. cerevisiae PMK (C).
DISCUSSION
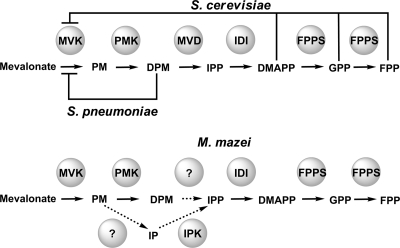
These studies demonstrate that at least three classes of MVKs can be distinguished based on their inhibition profiles (Fig. 4). Unlike previously reported MVKs, the MVK of the archaeon M. mazei was not inhibited by DMAPP, GPP, FPP, diphosphomevalonate, or IP, a proposed intermediate of the mevalonate pathway in archaea (17). The MVK of S. cerevisiae also was not inhibited by diphosphomevalonate accumulation in our studies, but it was inhibited by DMAPP, GPP, and FPP, similarly to the human enzyme. Furthermore, the inhibition of S. pneumoniae MVK was probed using 100 μM GPP and FPP as well as 5 mM DMAPP, but no significant inhibition of enzyme activity was observed.
Fig. 4.
Diagram of the regulation of MVKs from S. pneumoniae, S. cerevisiae, and M. mazei by the intermediates of the mevalonate pathway. Shown is a schematic of the mevalonate pathway in S. cerevisiae, S. pneumoniae, and M. mazei with enzymes MVK, PMK, diphosphomevalonate decarboxylase (MVD; unidentified in archaea), IDI, and farnesyl diphosphate synthase (FPPS), as well as their corresponding intermediates, phosphomevalonate (PM), diphosphomevalonate (DPM), IPP, DMAPP, GPP, and FPP. Inhibition studies were performed with DMAPP, GPP, FPP, DPM, and IP. S. pneumoniae MVK is inhibited by DPM, whereas S. cerevisiae MVK is inhibited by DMAPP, GPP, and FPP. M. mazei MVK is not inhibited by DMAPP, GPP, FPP, or DPM. The inhibition of M. mazei MVK also was tested with IP, the proposed intermediate of an alternative archaeal mevalonate pathway involving a putative phosphomevalonate decarboxylase and isopentenyl monophosphate kinase (IPK). IP did not inhibit M. mazei MVK in our studies.
We hypothesize that MVKs have evolved with different regulation mechanisms to accommodate their specific utilization of isoprenoids (17, 20, 27). A phylogenetic tree of the MVKs from Bacteria, Archaea, and Eukarya was constructed to assess the similarity between MVKs from the three domains of life (Fig. 2). Interestingly, the 29 MVKs that were surveyed clearly separated into three classes, suggesting the vertical transfer of the mvk gene. It should be noted that the MVK of the thermostable archaeon Methanocaldococcus jannaschii has been studied by Huang et al. and was found to be inhibited by micromolar concentrations of GPP, FPP, and IPP metabolites (19). However, the MVKs from M. mazei and M. jannaschii are distantly related, with 32% amino acid sequence identity, and occupy different branches of the archaeal dendrogram (Fig. 2). The specific activity of the M. jannaschii MVK at an optimum temperature of 70 to 75°C was reported to be 387 μmol/min/mg. Approximately 25% of the maximal activity was observed at 30°C, the temperature at which our studies were conducted (19). The specific activity of M. mazei MVK we report is more than 20 times less than the specific activity of M. jannaschii MVK at 30°C. The regulation of these enzymes seems widespread and, therefore, likely is important for maintaining properly functioning cells. Further studies are necessary to determine if MVKs that are not inhibited by metabolites are regulated at the transcriptional, translational, or posttranslational level, or if the low catalytic efficiency of this enzyme is important for regulation.
Footnotes
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Andreassi J. L., Bilder P. W., Vetting M. W., Roderick S. L., Leyh T. S. 2007. Crystal structure of the Streptococcus pneumoniae mevalonate kinase in complex with diphosphomevalonate. Protein Sci. 16:983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andreassi J. L., Dabovic K., Leyh T. S. 2004. Streptococcus pneumoniae isoprenoid biosynthesis is downregulated by diphosphomevalonate: an antimicrobial target. Biochemistry 43:16461–16466 [DOI] [PubMed] [Google Scholar]
- 3. Barkley S. J., Desai S. B., Poulter C. D. 2004. Type II isopentenyl diphosphate isomerase from Synechocystis sp. strain PCC 6803. J. Bacteriol. 186:8156–8158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beytia E., Dorsey J. K., Marr J., Cleland W. W., Porter J. W. 1970. Purification and mechanism of action of hog liver mevalonic kinase. J. Biol. Chem. 245:5450–5458 [PubMed] [Google Scholar]
- 5. Bohlmann J., Keeling C. I. 2008. Terpenoid biomaterials. Plant J. 54:656–669 [DOI] [PubMed] [Google Scholar]
- 6. Boucher Y., Kamekura M., Doolittle W. F. 2004. Origins and evolution of isoprenoid lipid biosynthesis in archaea. Mol. Microbiol. 52:515–527 [DOI] [PubMed] [Google Scholar]
- 7. Bult C. J., et al. 1996. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science 273:1058–1073 [DOI] [PubMed] [Google Scholar]
- 8. Chen M., Poulter C. D. 2010. Characterization of thermophilic archaeal isopentenyl phosphate kinases. Biochemistry 49:207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dorsey J. K., Porter J. W. 1968. The inhibition of mevalonic kinase by geranyl and farnesyl pyrophosphates. J. Biol. Chem. 243:4667–4670 [PubMed] [Google Scholar]
- 10. Doun S. S., Burgner I. I., Briggs J. W. S. D., Rodwell V. W. 2005. Enterococcus faecalis phosphomevalonate kinase. Protein Sci. 14:1134–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ekiel I., Smith I. C. P., Sprott G. D. 1983. Biosynthetic pathway in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J. Bacteriol. 156:316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Felsenstein J. 1989. PHYLIP–phylogeny inference package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 13. Fu Z., Voynova N. E., Herdendorf T. J., Miziorko H. M., Kim J. P. 2008. Biochemical and structural basis for feedback inhibition of mevalonate kinase and isoprenoid metabolism. Biochemistry 47:3715–3724 [DOI] [PubMed] [Google Scholar]
- 14. Gasteiger E., et al. 2005. Protein identification and analysis tools on the ExPASy server, p. 571–607 In Walker J. M. (ed.), The proteomics protocols handbook. Humana Press, Totowa, NJ [Google Scholar]
- 15. Goldstein J. L., Brown M. S. 1990. Regulation of the mevalonate pathway. Nature 343:425–430 [DOI] [PubMed] [Google Scholar]
- 16. Gray J. C., Kekwick R. G. O. 1972. The inhibition of plant mevalonate kinase preparations by prenyl pyrophosphates. Biochim. Biophys. Acta 279:290–296 [DOI] [PubMed] [Google Scholar]
- 17. Grochowski L. L., Xu H., White R. H. 2006. Methanocaldococcus jannaschii uses a modified mevalonate pathway for biosynthesis of isopentenyl diphosphate. J. Bacteriol. 188:3192–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hinson D. D., Chambliss K. L., Toth M. J., Tanaka R. D., Gibson K. M. 1997. Post-translational regulation of mevalonate kinase by intermediates of the cholesterol and nonsterol isoprene biosynthetic pathway. J. Lipid Res. 38:2216–2223 [PubMed] [Google Scholar]
- 19. Huang K., Scott A. I., Bennett G. N. 1999. Overexpression, purification, and characterization of the thermostable mevalonate kinase from Methanococcus jannaschii. Protein Expression Purif. 17:33–40 [DOI] [PubMed] [Google Scholar]
- 20. Koga Y., Morii H. 2007. Biosynthesis of ether-type polar lipids in archaea and evolutionary considerations. Microbiol. Mol. Biol. Rev. 71:97–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martin V. J. J., Pitera D. J., Withers S. T., Newman J. D., Keasling J. D. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat. Biotechnol. 21:796–802 [DOI] [PubMed] [Google Scholar]
- 22. Potter D., Wojnar J. M., Narasimhan C., Miziorko H. M. 1997. Identification and functional characterization of an active-site lysine in mevalonate kinase. J. Biol. Chem. 272:5741–5746 [DOI] [PubMed] [Google Scholar]
- 23. Roberts S. C. 2007. Production and engineering of terpenoids in plant cell culture. Nat. Chem. Biol. 3:387–395 [DOI] [PubMed] [Google Scholar]
- 24. Rohdich F., Bacher A., Eisenreich W. 2005. Isoprenoid biosynthetic pathways as anti-infective drug targets. Biochem. Soc. Trans. 33:785–791 [DOI] [PubMed] [Google Scholar]
- 25. Rohmer M., Knani M., Simonin P., Sutter B., Sahm H. 1993. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochemistry 295:517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 3 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 27. Smit A., Mushegian A. 2000. Biosynthesis of isoprenoids via mevalonate in Archaea: the lost pathway. Genome Res. 10:1468–1484 [DOI] [PubMed] [Google Scholar]
- 28. Spurgeon S. L., Porter J. W. 1981. Biosynthesis of isoprenoid compounds, vol. 1 John Wiley and Sons, New York, NY [Google Scholar]
- 29. Tchen T. T. 1958. Mevalonic kinase: purification and properties. J. Biol. Chem. 233:1100–1103 [PubMed] [Google Scholar]
- 30. Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Voynova N. E., Rios S. E., Miziorko H. M. 2004. Staphylococcus aureus mevalonate kinase: isolation and characterization of an enzyme of the isoprenoid biosynthetic pathway. J. Bacteriol. 186:61–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wilding E. I., et al. 2000. Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. J. Bacteriol. 182:4319–4327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Withers A. T., Keasling J. D. 2007. Biosynthesis and engineering of isoprenoid small molecules. Appl. Microbiol. Biotechnol. 73:980–990 [DOI] [PubMed] [Google Scholar]