Abstract
In wastewater treatment plants, nitrifying systems are usually operated with elevated levels of aeration to avoid nitrification failures. This approach contributes significantly to operational costs and the carbon footprint of nitrifying wastewater treatment processes. In this study, we tested the effect of aeration rate on nitrification by correlating ammonia oxidation rates with the structure of the ammonia-oxidizing bacterial (AOB) community and AOB abundance in four parallel continuous-flow reactors operated for 43 days. Two of the reactors were supplied with a constant airflow rate of 0.1 liter/min, while in the other two units the airflow rate was fixed at 4 liters/min. Complete nitrification was achieved in all configurations, though the dissolved oxygen (DO) concentration was only 0.5 ± 0.3 mg/liter in the low-aeration units. The data suggest that efficient performance in the low-DO units resulted from elevated AOB levels in the reactors and/or putative development of a mixotrophic AOB community. Denaturing gel electrophoresis and cloning of AOB 16S rRNA gene fragments followed by sequencing revealed that the AOB community in the low-DO systems was a subset of the community in the high-DO systems. However, in both configurations the dominant species belonged to the Nitrosomonas oligotropha lineage. Overall, the results demonstrated that complete nitrification can be achieved at low aeration in lab-scale reactors. If these findings could be extended to full-scale plants, it would be possible to minimize the operational costs and greenhouse gas emissions without risk of nitrification failure.
INTRODUCTION
The water industry is energy intensive. In the United Kingdom, the water industry uses 9 TWh, releasing 5 million tonnes (carbon dioxide equivalent) of greenhouse gases (GHGs) to the atmosphere per year (61). Driven by increasing operational power costs and the need to abate anthropogenic GHG release, the water industry is being forced to develop new low-energy and cost-effective technologies. In activated sludge systems, particular regard should be given to nitrification, the two-step biological oxidation of ammonia to nitrate via nitrite. This process prevents excessive hazardous discharges of nitrogen into receiving waters. Conventional nitrification is carried out by the ammonia-oxidizing bacteria (AOB) and the nitrite-oxidizing bacteria (NOB). The AOB, responsible for the first and often limiting step of nitrification (39), are generally considered to have a strictly chemolithoautotrophic aerobic metabolism, to grow slowly, and to be poor competitors for oxygen (22, 60). Consequently, nitrifying systems are usually operated at a solid retention time (SRT) longer than 5 days and dissolved oxygen (DO) concentration above 2 mg/liter to satisfy both carbon and nitrogen removal requirements and to overcome diffusional resistance in the flocs (59).
However, there is evidence that complete nitrification can also occur at low DO levels (<0.5 mg/liter) (35) or with intermittent aeration (30). Also, a recent theoretical study modeling the risk of failure in nitrification based on the overall oxygen transfer coefficient (KLa) suggested that the risk of failure increases at intermediate aeration rates, while high (KLa > 35 day−1) and relatively low (4 day−1 < KLa < 10 day−1) aeration rates promote process stability (33). These studies are promising, for though it may take many years to renew the physical assets of the water industry to meet the challenge of a low-energy and low-carbon footprint, in principle, the less efficient biological assets could be replaced in a few weeks. Studies investigating the type of AOB in both activated sludge flocs and biofilms, where low oxygen was available, have been performed already using culture-independent techniques (12, 25, 30, 34, 35), but the results are sometimes conflicting. For example, Park and Noguera (34, 35) reported that AOB communities in activated sludge were dominated by members of the Nitrosomonas europaea lineage and the Nitrosomonas oligotropha lineage under low-DO conditions (<0.24 mg/liter), while members of the N. oligotropha lineage were dominant at higher DO concentrations. These results partially contradict comparable research on biofilms, where N. oligotropha-like and N. europaea-like groups were both found in high-DO zones (12).
Each AOB group is assumed to be characterized by specific growth kinetic parameters (and thus different yields and affinities for oxygen and ammonia), which are used to define the aeration requirements (40). The true yield of the AOB (YAOB) depends on the reactions involved in cell synthesis and energy production. All nitrifiers are known to be chemolithotrophic bacteria, using ammonia as electron donor and oxygen as electron acceptor, and consequentially have low growth yields (0.34 kg volatile suspended solids [VSS]/kg NH4+-N) (42). However, recent studies reported chemoorganoheterotrophic growth of N. europaea and Nitrosomonas eutropha in the presence of pyruvate, lactate, acetate, serine, succinate, and fructose under anoxic conditions (48). Also, some Nitrosomonas species are able to replace molecular oxygen with nitrogen dioxide or dinitrogen tetroxide as an electron acceptor (49, 50). In the presence of oxygen, N. europaea has been reported to grow chemolithoheterotrophically, deriving energy from ammonia and using fructose as carbon source (17). The growth rate of N. europaea under such conditions was lower than the growth rate on CO2, but a high growth yield was observed (17). Enhanced growth yield and rate have also been reported for Nitrobacter hamburgensis (NOB) when d-lactate and NO2− were supplied as the sole carbon source and as energy source, respectively (52). Similarly, Nitrospira appeared to simultaneously assimilate inorganic and organic carbon (e.g., pyruvate), but it is still unclear whether Nitrospira spp. use organic substrates only as carbon sources or also for the production of energy. However, the growth rate of Nitrospira marina under mixotrophic conditions was higher than that under chemolithotrophic conditions (62).
The yield is related also to the cell-specific ammonia oxidation rate (CSAOR), which is a measure of the AOB community activity and defines the work rate of AOB in nitrification. It has been suggested that CSAORs between 4 and 10 fmol cell−1 h−1 represents a good balance between risk of nitrification failure and meeting the needs of the AOB (37). This is consistent with the results reported by Coskuner et al. (5) in which a plant close to failure had a high CSAOR and a low number of nitrifiers. In these studies, parameters such as AOB number and activity were emphasized rather than AOB diversity and composition in controlling and predicting nitrification failures. Nevertheless, it is generally assumed that a more diverse AOB community will be able to cope better with variations in operating conditions, maintaining process stability (8), especially given the fragile coupling of AOB and NOB in bioreactors (13).
This study focuses on the effect of aeration rate on nitrification and aims to provide a deeper insight into the structure of the AOB community selected under high- and low-DO conditions. Furthermore, we are interested in how AOB activity and oxygen demand can be maximized so that nitrifying bioreactors might be operated at shorter solid retention times, which has further cost advantages. Finally, we hypothesize that AOB abundance, activity, diversity, and community composition are influenced by oxygen availability, which is key to understanding process resilience in future applications.
MATERIALS AND METHODS
Bench-scale reactors.
Four continuous-flow lab-scale reactors were operated in parallel for 43 days. The reactors consisted of glass cylinders with a funnel bottom in which a porous glass grid was placed in the center. The systems were inoculated (day 1) with 3 liters of return activated sludge (RAS) from a municipal wastewater plant in Spennymoor (County Durham, England; 54°42′0″N, 1°35′24″W), and the liquor was constantly mixed by a stirrer (∼130 rpm). Two of the reactors (RH3 and RH4) were supplied with a constant airflow rate of 4 liters per minute, while in the other two units (RL1 and RL2) the airflow rate was initially fixed at 0.2 liter/min but was decreased to 0.1 liter/min after 4 days of operation. Synthetic wastewater, containing 66 mg/liter (NH4)2SO4, 320 mg/liter peptone, 190 mg/liter meat extract, 30 mg/liter yeast extract, 30 mg/liter urea, 28 mg/liter K2HPO4, 2 mg/liter CaCl2 · 2H2O, 2 mg/liter of MgSO4 · 7H2O, 1 ml/liter of trace element solution (0.75 g/liter FeCl3 · 6H2O, 0.075 g/liter H3BO3, 0.015 g/liter CuSO4 · 5H2O, 0.09 g/liter KI, 0.06 g/liter MnCl2 · 4H2O, 0.03 g/liter NaMoO4 · 2H2O, 0.06 g/liter ZnSO4 · 7H2O, 0.075 g/liter CoCl2 · 6H2O, 0.5 g/liter EDTA, and 1 ml/liter concentrated hydrochloric acid), and 0.8 mg/liter NaHCO3 (21), was continuously pumped into the reactors at a fixed solid and hydraulic retention time (SRT and HRT, respectively) of 3 days. The ammonia and chemical oxygen demand (COD) concentrations in the influent were 15.4 ± 2.6 mg/liter and 717 ± 107 mg/liter, respectively. Samples (200 ml) from each reactor and the feed synthetic wastewater were collected every 3 days for chemical, physical, and microbial community analyses.
Physical and chemical analyses.
The dissolved oxygen (DO) concentration, temperature, and pH were constantly monitored with specific probes (Broadley Technologies Ltd., United Kingdom). Nitrification performance was assessed by analyzing NH4+-N, NO2−-N, and NO3−-N levels in the bulk solution over time. NO2− and NO3− were measured using ion chromatography (Dionex DW-100 ion chromatograph; Dionex Corp., Sunnyvale, CA) The ion chromatograph was fitted with an IonPac AS14A analytical column and a 25-μl injection loop. It was operated at a flow rate of 1 ml/min with 8.0 mM Na2CO3-1.0 mM NaHCO3 as eluent. Ammonia concentration (NH4+-N), total suspended solids (TSS), and volatile suspended solids (VSS) were determined according to standard methods (4a). COD from nonfiltered samples (i.e., including the biomass) was measured using the COD cell test (Merck KGaA, Germany) according to the manufacturer's instructions.
DNA extraction and quantitative PCR (qPCR).
DNA was extracted from the mixed liquor (250 μl) collected from the four reactors at days 1, 10, 19, 28, and 43 and stored at −20°C until further analyses. After mechanical cell lysis using lysing matrix E (MP Biomedicals, Solon, OH) in a Ribolyser (Hybaid Ltd., United Kingdom), DNA extraction was performed using the FastDNA Spin kit for soil (MP Biomedicals, Solon, OH) in accordance with the manufacturer's instructions.
The abundance of AOB in the mixed liquor was evaluated by primers CTO 189fA/B (5′-GGAGRAAAGCAGGGGATCG-3′), CTO189fC (5′-GGAGGAAAGTAGGGGATCG-3′) (23), and RT1r (5′-CGTCCTCTCAGACCARCTACTG-3′) (16). Primers were supplied by Sigma-Genosys (The Woodlands, TX). The qPCRs were performed in a Bio-Rad iCycler equipped with an iCycler iQ fluorescence detector and associated software (Bio-Rad; version 2.3). PCRs used a 15-μl reaction mixture, which contained 2 μl of DNA template, 1 μl of primer mixture (7.5 pmol each), and 12 μl of PCR Precision Mastermix PCR reagent (Primer Design Ltd., United Kingdom). For fluorescence detection, SYBR green I (10,000×; Sigma, United Kingdom) was firstly diluted 1/100 in sterile and autoclaved molecular water and such solution was added to the PCR mixture as 1% (vol/vol) of the total volume of the reaction mix. The thermal cycling was carried out as previously reported by Hermansson and Lindgren (16). The 16S rRNA gene copy numbers were converted to equivalent cell numbers, assuming that one rRNA operon existed per AOB cell (20).
Calculation of theoretical AOB numbers, AOB fraction, yield, and cell-specific ammonia oxidation rate.
The theoretical AOB numbers were calculated as previously described by Rittmann et al. (42) using the following equation:
where θ is the solid retention time (3 days), b is the endogenous respiration rate (0.15 day−1), fd is the fraction of the active biomass that is biodegradable (0.8), and YAOB is the AOB yield (0.34 kg VSS/kg NH4+-N) (43). XAOB is expressed as mg/liter, and the biomass results were converted in cell numbers considering the diameter and total density of an AOB cell to be equal to 1 μm and 0.636 g/cm3, respectively (5). Similarly, the AOB cell numbers (cells/ml) obtained by qPCR were converted in biomass (XAOB) to infer the AOB percentage and yield (YAOB). The AOB fraction in the systems was calculated by dividing XAOB (mg VSSAOB/liter) by the total volatile suspended solids, X (mg VSS/liter). To evaluate YAOB (mg VSSAOB/mg NH4+-N), the equation was solved.
The cell-specific ammonia oxidation rates were inferred according to the method of Daims et al. (6).
Nested PCR and denaturant gradient gel electrophoresis (DGGE).
A nested PCR approach, followed by DGGE, was performed to monitor AOB population dynamics. In the first PCR, a 465-bp fragment of the 16S rRNA gene of AOB was amplified using CTO189F (5′-GAGRAAAGYAGGGGATCG-3′) and CTO654R (5′-CTAGCYTTGTAGTTTCAAACGC-3′) (23). The PCR products from this reaction were used as a template for a second PCR with primer 3 (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGGCCTACGGGAGGCAGCAG-3′) and primer 2 (5′-ATTACCGCGGCTGCTGG-3′) (31). All the PCRs were carried out in 25-μl reaction mixtures containing 23.5 μl of PCR buffer (MegaMix-Blue; Microzone Ltd., United Kingdom), 0.5 μl of each primer (10 pmol), and 0.5 μl of DNA template. The PCR conditions for the CTO primer set and primer 3 and primer 2 set were previously described (23, 31), and all the PCRs were performed using a Px2 thermal cycler (Thermo Hybaid, Hybaid Ltd., United Kingdom).
Nested-PCR-amplified fragments were separated using a D-Code DGGE system (Bio-Rad Laboratories Ltd., United Kingdom). Each sample was loaded onto a 10% (wt/vol) polyacrylamide gel using a denaturing gradient ranging from 35% to 55% (100% of denaturant comprises 7 M urea and 40% [vol/vol] deionized formamide). The gel was prepared with 10% (wt/vol) acrylamide stock solution (acrylamide-N,N-methylenebisacrylamide; 37:1) in 1× TAE buffer (Tris-acetate-EDTA; 40 mM Tris-acetate, 1 mM EDTA, pH 8.0) with an appropriate amount of 100% denaturant solution. The electrophoresis was run in 1× TAE buffer at 60°C for about 4.5 h at constant voltage (200 V). The gels were stained in a solution of SYBR green I (Sigma, United Kingdom) diluted 1/10,000 in 1× TAE buffer for 30 min and examined under UV illumination (Bio-Rad Fluor-S MultiImager; Bio-Rad, United Kingdom). The gels were processed using Bionumerics 4.0 (Applied Maths BVBA, Saint-Martens-Latem, Belgium); pairwise similarities between DGGE profiles were calculated with Raup and Crick coefficients for cluster analyses (single linkage clustering), which were conducted using PAST (15). The Raup and Crick similarity index (SRC) is defined by the probability that the expected similarity (randomized data) would be greater than or equal to the observed similarity. Similarity values between 0.05 and 0.95 are indicative of random occurrence of the same organism (DGGE band) in two samples, whereas SRC above 0.95 and below 0.05 indicates significant similarity and significant dissimilarity, respectively. The algorithm assumes that the taxa are equally likely to be selected. The strongest bands detected on the DGGE gels were excised and sequenced. The sequences were then compared to the GenBank database using BLASTn (2) at the National Center for Biotechnology Information (NCBI).
Clone library preparation.
To evaluate and compare the AOB community structure selected in high- and low-DO reactors after 43 days of operation, gene clone libraries were constructed from AOB partial 16S rRNA genes amplified from samples collected from each reactor at the end of the experiment. PCR products amplified with the CTO primer set were cloned with a TOPO TA cloning kit (Invitrogen, Ltd., United Kingdom) according to the manufacturer's instructions. Following PCR amplification, the correct size of the fragment was checked by electrophoresis in a 1.5% agarose gel run in 1× TAE buffer stained with ethidium bromide. The bands were excised and purified with a Qiaquick PCR gel extraction kit (Qiagen Ltd., United Kingdom). The PCR products and the vector provided by the kit were ligated and used to transform OneShot competent cells according to the manufacturer's instructions (Invitrogen, Ltd., United Kingdom). Clones (∼40) were randomly picked and transferred to a 30-μl PCR mixture containing the primers T3 (5′-ATTAACCCTCACTAAAGGGA-3′) and T7 (5′-TAATACGACTCACTATAGGG-3′). PCR products of the correct size were prepared for sequencing using ExoSAP-IT (GE Healthcare, United Kingdom), and the sequences of the partial 16S rRNA gene fragment (ca. 465 bp) were determined by Genevision, United Kingdom.
The sequences obtained were aligned using ClustalX v1.83 (56), and vector and primer sequences were removed. The triggered sequences with >97% identity were then grouped by FastGroupII (http://biome.sdsu.edu/fastgroup/fg_tools.htm) into operational taxonomic units (OTUs), and the representative sequences of each group were analyzed using BLASTn. The representative OTUs were also checked for putative chimeras with the online software Bellerophon (18) using a window size of 200 bp. Chimeric sequences were identified and removed. Comparisons between cloned sequences of the representative OTUs and DGGE-derived sequences were performed using the sequence identity matrix function implemented in BioEdit freeware (14). MEGA version 4 (54) was used to conduct phylogenetic analysis. The evolutionary history was inferred using the minimum evolution (ME) method (44) using the OTUs found in this study and selected AOB and Ferribacterium sequences for which 16S rRNA gene sequences longer than 1,200 bp are available in the ribosomal database project, RDP (26). A bootstrap consensus tree was determined from 1,000 replicates (9). Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The evolutionary distances were computed using the maximum composite likelihood method (55) and are in units of the number of base substitutions per site. The ME tree was searched using the Close-Neighbor-Interchange (CNI) (32) algorithm at a search level of 1. The neighbor-joining algorithm (46) was used to generate the initial tree. All positions containing alignment gaps and missing data were eliminated in pairwise sequence comparisons.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under accession numbers JF327348, JF327350, JF327351, and JF327354 to JF327363.
RESULTS
Reactor performance.
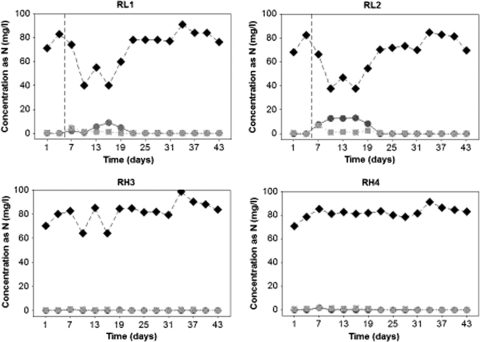
There was no apparent difference in ammonia removal over time between the four reactors (P value > 0.05, nested analysis of variance [ANOVA]) (Fig. 1). The DO concentrations in the systems were assessed (Fig. 2), and the reactors supplied with air at a flow rate of 0.2 liter/min had a lower DO concentration than did the reactors supplied at a high rate (P value = 0.015, nested ANOVA); the DO concentrations of duplicate reactors were, however, comparable (P value = 0.185, nested ANOVA). When the airflow was decreased from 0.2 liter/min to 0.1 liter/min in RL1 and RL2, the DO concentrations significantly decreased in the low-aeration reactors (P value < 0.05, ANOVA), enhancing the difference in DO concentration between the two treatments (P value < 0.05, nested ANOVA). Although oxygen supply rates were the same in the two low- and two high-DO reactor pairs, respectively, actual DO levels differed among the four reactors (P value < 0.05, nested ANOVA), even if the scale of differences in DO concentrations between the low- and high-DO reactors was an order of magnitude greater than that between reactor pairs with the same oxygen supply rates. The average temperature in the systems was 23.7 ± 0.5°C, while the pH ranged between 7 and 7.4 and 8 and 8.10 in RH3-RH4 and in RL1-RL2, respectively. The mixed liquor VSS averaged 297 ± 140 mg/liter and 243 ± 69 mg/liter in RL1-RL2 and RH3-RH4, respectively. The large variance in the low-DO reactors was due to an increase in VSS in RL1, where the VSS concentration averaged 198 ± 38 mg/liter between day 1 and 28 and 539 ± 41 mg/liter between day 31 and 43. Nevertheless, the COD removal efficiency was indistinguishable among configurations (P value > 0.05, nested ANOVA), averaging 77.5% ± 0.2%. The COD removal in the systems was lower than expected by mass balance calculations (82.7% ± 16.3%; P value = 0.012, ANOVA) (43), accounting for the determined VSS concentrations and the SRT, probably because the COD was measured in nonfiltered bulk samples.
Fig. 1.
Concentrations of ammonia (circles), nitrite (squares), and nitrate (diamonds) in reactors with low aeration (RL1 and RL2) and high aeration (RH3 and RH4). The vertical dashed line represents the time when the airflow rate was decreased from 0.2 liter/min to 0.1 liter/min in RL1 and RL2.
Fig. 2.
Average DO concentrations measured in the systems during the two phases of the experiment.
AOB abundance assessed by qPCR and CSAOR.
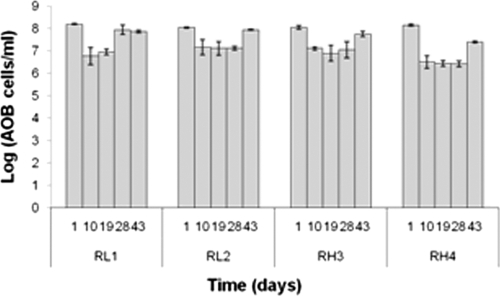
The abundance of AOB species fluctuated over time (Fig. 3); however, there was no significant difference (nested ANOVA) between reactors supplied with high and low aeration (P value = 0.6) and between duplicate reactors (P value = 0.23). Overall, the numbers of AOB ranged between 2.8 × 106 and 1.6 × 108 cells per ml, slightly higher than the results predicted by the Rittmann model (1.2 × 106 and 1.4 × 107 cells per ml) (42, 43). The AOB averaged 9% ± 1.4% of the total biomass in the original sludge, but the AOB fraction decreased over the experiment to 5.2% ± 2.1% (P value = 0.027, nested ANOVA). In general, the percentage of nitrifying bacteria was not affected by the aeration rate (P values = 0.2, nested ANOVA). Standard process models (42) predicted a lower percentage of AOB, ranging between 0.2% and 2.4%. This suggests that the parameters used in the model of Rittmann et al. are not appropriate for the AOB found in the reactors studied here and/or operating conditions used.
Fig. 3.
AOB abundance in the systems over time assessed by qPCR. Bars show the means of duplicate samples, while error bars represent standard deviations.
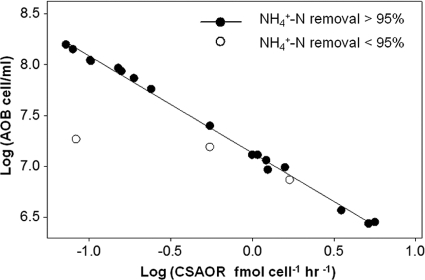
Cell-specific ammonia oxidation rates ranged from 0.07 to 5.6 fmol cell−1 h−1, and a linear relationship between log CSAORs and log AOB abundance was observed when the ammonia removal was higher than 95% (y = 7.136 to 0.9537x, Fig. 4). Three outliers were observed when the ammonia removal efficiency was lower, and these points correspond to the times when the airflow rate was decreased from 0.2 liter/min to 0.1 liter/min in RL1-RL2 (Fig. 4). In general, the CSAORs were not significantly different under conditions of high and low aeration nor between duplicate reactors (all P values > 0.05, nested ANOVA).
Fig. 4.
Correlation between cell-specific ammonia oxidation rate (CSAOR) and AOB abundance in the systems when the NH4+-N removal is higher and when it is lower than 95%.
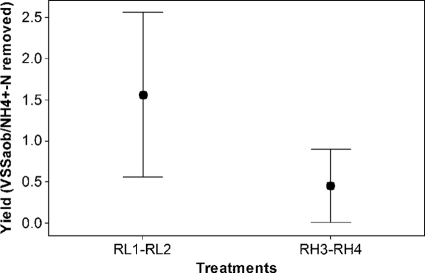
Calculations of the growth yield under the two aeration levels revealed higher yield values (1.56 ± 1.2 kg VSS/kg NH4+-N) in the RL1 and RL2 reactors (operated at 0.1 liter/min) than in highly aerated reactors (Fig. 5) (P value = 0.021, Kruskal-Wallis test). Furthermore, the AOB growth yield in the high-DO reactors (0.45 ± 0.53 kg VSS/kg NH4+-N) was comparable to the yield of 0.34 reported by Rittmann et al. (42). No differences in growth yield were detected between duplicate reactors (P value = 0.985, nested ANOVA).
Fig. 5.
Average yield values observed in the reactors with low aeration (RL1 and RL2) and high aeration (RH3 and RH4) (P value = 0.021, Kruskal-Wallis test).
Analysis of AOB community structure by DGGE.
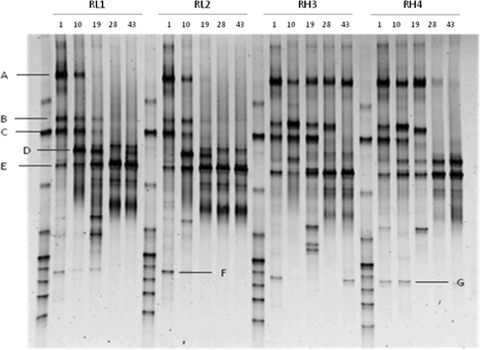
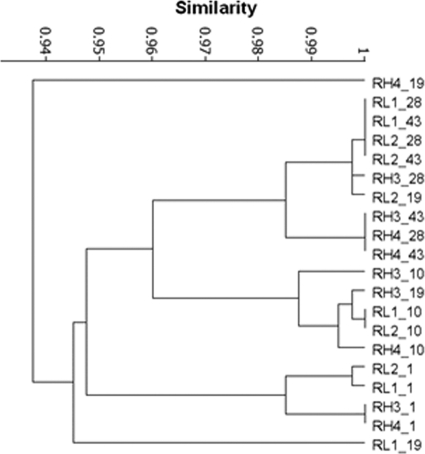
Changes in AOB community structure were detected by DGGE over time in all reactors (Fig. 6); the strong bands from the inoculum became fainter or disappeared, and new bands became dominant. Cluster analyses based on Raup and Crick similarity indices (SRC) showed a parallel change over time of the putative AOB communities in duplicate reactors (Fig. 7). Initially, the samples from the reactors treated with different aeration rates grouped together according to the sampling day. By day 43, all the reactors were clustered more strongly according to the aeration mode than to the sampling day. The average number of bands detected at day 1 (13 ± 0.81) was not significantly different from that observed at the end of the experiment (9 ± 1.8) (P value = 0.127, nested ANOVA), while the samples from the low-DO systems revealed a lower number of bands than did the reactors supplied with high aeration (P value = 0.028, nested ANOVA).
Fig. 6.
AOB 16S rRNA gene community fingerprints detected at days 1, 10, 19, 28, and 43 by DGGE. Bands A, B, C, D, E, F, and G were excised and sequenced for phylogenetic analyses.
Fig. 7.
Cluster analysis of the AOB DGGE profiles based on the Raup and Crick coefficient.
The strongest bands detected on the DGGE gel were excised and sequenced to characterize predominant members of the AOB community. Five bands out of seven were related to Nitrosomonas spp., while two were associated with non-AOB species (Table 1). Bands A, B, and C were dominant in the inoculum, but only A was found in the DGGE pattern of RH3 and RH4 after 43 days of operation; all the others dropped below the detection limit. In contrast, bands E and D were faintly present in the inoculum, but band E became the strongest band at the end of the experiment in all systems.
Table 1.
Closest matches (BLASTn) of the seven AOB bands excised from the DGGE gela
| Band | QC (%) | MI (%) | Accession no. | Description |
|---|---|---|---|---|
| A | 100 | 97 | FM200933 | Uncultured Nitrosomonas sp., clone MBR-30_LF_AS36 |
| B | 100 | 100 | AF527015 | Uncultured Nitrosomonas sp. strain 26Ft |
| C | 99 | 98 | AF527015 | Uncultured Nitrosomonas sp. strain 26Ft |
| D | 100 | 100 | EU285310 | Uncultured bacterium clone AOB7 |
| E | 100 | 99 | EU285313 | Uncultured bacterium clone AOB10 |
| F | 100 | 98 | AJ890204 | Uncultured Ferribacterium sp. |
| G | 100 | 96 | EU381112 | Uncultured Ferribacterium sp. |
QC, query coverage; MI, maximum identity.
Composition of the AOB community.
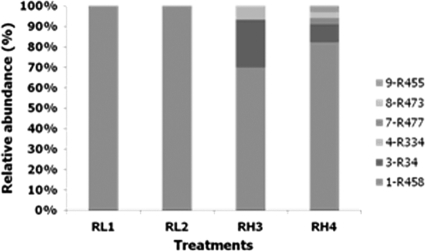
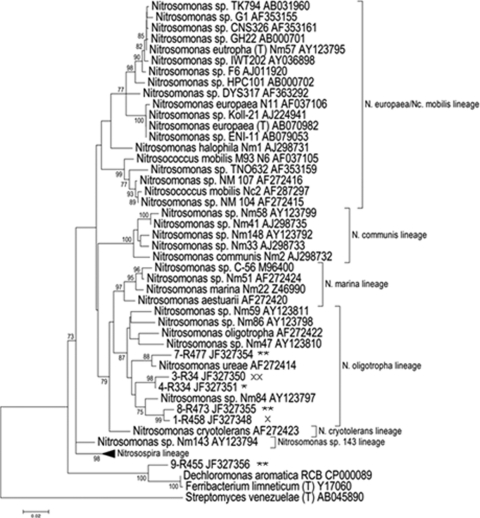
Of the 133 partial sequences obtained from the clone libraries, only one was identified as non-AOB. The remaining clones had 98 to 99% sequence identity to Nitrosomonas 16S rRNA sequences present in GenBank. The 16S rRNA sequences were sorted into six groups, and the distribution of the representative OTUs in the reactors is summarized in Fig. 8. All sequences retrieved from the reactors supplied with a small amount of oxygen were grouped in one OTU, while the reactors with high aeration showed more diverse communities: three OTUs and five OTUs were found in RH3 and RH4, respectively. The representative sequence from each OTU was analyzed using BLASTn to identify the most closely related sequence in the GenBank database (Table 2), and phylogenetic analysis was conducted (Fig. 9). The OTU retrieved from the low- and high-DO reactors (1-R458) was affiliated with uncultured Nitrosomonas clones related to the N. oligotropha lineage. In the high-DO reactors, one OTU (7-R477) retrieved only in RH4 was closely related to Nitrosomonas ureae, while the other representative OTUs (3-R34, 4-R334, and 8-R473) were similar to uncultured AOB clones related to Nitrosomonas species. OTU 9-R455, found only in RH4, was instead related to Ferribacterium spp. Since the number of OTUs can be considered a crude indicator of the species richness, it was probable that the low-aeration reactors had a lower number of AOB taxa than did the high-aeration reactors.
Fig. 8.
Distribution of the retrieved clones in the reactors. The clones were sorted in six OTUs, represented by the bars in different shades of gray.
Table 2.
Closest matches (BLASTn) of the representative OTUs retrieved from the clone librariesa
| OTU | QC (%) | MI (%) | Accession no. | Description | ID, % (DGGE band[s]) |
|---|---|---|---|---|---|
| 1-R458 | 95 | 99 | EU285313 | Uncultured bacterium clone AOB10 | 99 (E) |
| 3-R34 | 94 | 99 | FM200933 | Uncultured Nitrosomonas sp., clone MBR-30_LF_AS36 | 99 (A) |
| 4-R334 | 92 | 100 | FM201063 | Uncultured Nitrosomonas sp., clone MBR-8_LF_BF28 | 98 (A) |
| 7-R477 | 95 | 98 | AF272414 | Nitrosomonas ureae | 95 (A, B, D, F) |
| 8-R473 | 95 | 98 | EU285313 | Uncultured bacterium clone AOB10 | 99 (E) |
| 9-R455 | 93 | 98 | AJ238205 | Uncultured Ferribacterium sp. | 99 (F) |
QC, query coverage; MI, maximum identity; ID, identity. The DGGE band is the closest DGGE-derived sequence.
Fig. 9.
Phylogenetic tree of the AOB 16S rRNA gene sequences of the representative OTUs and their phylogenetic relatives constructed by MEGA4. The tree is based on the minimum evolution method, and the bootstrap consensus tree is inferred from 1,000 replicates. Clones marked × were retrieved in all reactors, clones marked ×× were found in both high-DO reactors, and clones marked * and ** were retrieved only in RH3 and RH4, respectively.
Most of the sequences retrieved from the AOB clone library and the DGGE gel were closely related to the same species. The few discrepancies between the cloned and the DGGE-derived sequences could be due to the length of the latter (198 bp), which are too short for conclusive phylogenetic analyses. Also, the DGGE sequences cover a highly variable region of the 16S rRNA gene, whereas the cloned fragments encompass both conserved and variable regions. Nevertheless, the most abundant clones, 1-R458 and 3-R34, had more than 99% identity with the DGGE band sequences A and E (Table 2). In addition, the low identities found between clones 4-R334 and 7-R477 and the pool of DGGE sequences were due to the low frequency of these clones, presumably associated with faint bands not excised from the DGGE gels.
DISCUSSION
This study suggests that effective nitrification can be achieved in reactors with low DO and provides preliminary insights into the basis for this capacity. Oxygen supply rate did not seem to limit the performance of the systems, as low effluent ammonia levels were consistently observed. The results obtained, together with previous reports that showed complete nitrification at low DO concentrations (<0.5 mg/liter) in chemostats (35), lab-scale reactors treating swine sewage (30), and biofilms (12), suggest that the aeration requirements in nitrifying systems can be substantially lowered as long as there is no associated increase in the release of the GHGs (especially N2O) (53). Although we did not measure N2O here, its release is typically associated with the accumulation of ammonia and nitrite and/or shock loadings (1, 10, 19, 41, 64), none of which occurred in our system. We are optimistic that stable and efficient nitrifying systems kept at constant low DO concentrations will have acceptable emissions of nitrous oxide (1). We therefore believe that our findings hold out the possibility of reducing energy and operational costs, as well as greenhouse gas emissions, without increasing the risk of nitrification failures.
Nevertheless, nitrification in the low-DO reactors studied here seems to be different from textbook accounts of autotrophic nitrification. It is possible that the ammonia was removed by mixotrophic AOB communities, able to use organic carbon sources and inorganic electron donors (43). Although carbon assimilation by the nitrifying biomass was not directly assessed, calculated growth yields in the low-DO reactors were much higher than those reported in the literature for chemolithoautotrophic bacteria (39, 43). A likely explanation for this discrepancy is that the AOB were utilizing organic compounds as a carbon source in the low-DO systems, decreasing the need for an energetically expensive reductive process to convert CO2 to cellular carbon inherent in autotrophic metabolism. In contrast, mixotrophic metabolism reduces the energy requirement for cell synthesis as organic carbon is used, thereby increasing the cell yield (17, 43, 52, 62). Chemolithoorganotrophic growth of AOB has been demonstrated under anoxic conditions where molecular oxygen is replaced with nitrogen dioxide or nitrogen tetroxide as electron acceptor and pyruvate or lactate is replaced with CO2 as carbon source (24, 48). Also, the combination of aerobic and anaerobic ammonia oxidation has been reported to play an important role in the anoxic and oxic zones of plug flow reactors (28) and in a phosphate-removing biofilm (12). Therefore, the selection of an AOB community capable of chemolithoorganotrophic growth under low-oxygen conditions reduces the energy requirements, and thus CO2 emissions, associated with aeration. This is an interesting hypothesis suggested by the data reported, and direct measurement of organic carbon assimilation by AOB in reactors under different levels of aeration will be required to provide more direct evidence to test the hypothesis.
The reactors were capable of reliable operation at short SRTs and irrespective of the DO level, as COD and ammonia removals were consistent in all systems. The COD removal efficiency was low in comparison with what is required in wastewater treatment plants (WWTPs), probably because the short SRT reduced the VSS in the systems. This, in turn, could have also decreased the total volumetric nitrification rate but increased the ratio of active nitrifying bacteria in the total biomass as suggested in previous batch kinetic and modeling tests (51). Nonetheless, the relatively high percentage of AOB (more than 5%) and the elevated abundances (up to >108 cells/ml) present in the systems were likely responsible for the high and reliable nitrifying performance (37). Furthermore, the overall low cell-specific oxidation rates suggest that the systems were not close to failure and were being supplied with more oxygen than needed (5, 37). However, when the oxygen supply was decreased from 0.2 liter/min to 0.1 liter/min in the low-DO reactors, a transient reduction in AOB metabolic activity was observed (Fig. 4, outlier points), suggesting that the ammonia oxidizer community was inhibited and/or that it needed to acclimatize to the new conditions. It seems that the AOB community was not working too hard and hence the nitrification process was prone to failure as the cell numbers transiently appeared to be too low to support effective nitrification. Presumably, some of the AOB populations were lost, while the rest of the nitrifying biomass made the necessary physiological adjustments before increasing in number. Variations in kinetic parameters of the ammonia oxidation in lab-scale sequencing batch reactors operated under oxygen-limiting conditions at SRTs ranging between 3 and 24 days have been already suggested by Pollice et al. (38). They also observed comparable ammonia oxidation rates irrespective of the aeration level.
The community developed in the low-DO reactors was a subset of the high-DO community. The DGGE banding patterns from the AOB communities clustered according to the aeration conditions at the end of the operating time. The low DO concentration apparently drove parallel variations in the structure of the AOB community in agreement with a study monitoring AOB dynamics in chemostats operated at low DO (35). However, in our study this process resulted in loss of species rather than changes in species composition. The microbial communities in the high-DO reactors were also not static. The SRT could have been one of a number of factors that underlie such change. Probably, the SRT in our systems, much shorter than the SRT of the WWTP of the original inoculum, might have influenced the structure of the AOB community (45).
Surprisingly, none of the sequences retrieved from RL1 and RL2 was phylogenetically close to N. europaea or N. eutropha, the only AOB species reported to be able to grow chemolithoorganotrophically so far (48). In contrast, most of the clones and the strongest band belonged to organisms from the N. oligotropha lineage. Members of this lineage have been detected in various environments (22), including less oxygenated layers of biofilms (12), chemostat reactors operated with high and low DO (34), and the upstream region of the Schelde estuary with low DO and salinity levels (7), suggesting flexibility in the physiology of this AOB lineage. The adaptation of Nitrosomonas oligotropha-like AOB to low-DO conditions has been reported to be due to an increase of the expression of ammonia monooxygenase enzymes and the high affinity for ammonia rather than a high affinity for oxygen per se (34). Furthermore, members of the N. oligotropha lineage are known to produce urease (22), and this may have also favored the growth of such bacteria in our reactors, which were fed with synthetic wastewater containing urea. This suggests that the presence of urea might induce the AOB community to grow chemolithoheterotrophically under oxygen-depleted conditions.
The low number of different AOB taxa found in the low-DO reactors did not affect nitrification performance. Those who contend that high-species-diversity ecosystems are more resistant to sudden environmental variations, i.e., operating conditions (8, 58), would predict an elevated risk of failure in the low-DO systems with reduced diversity. Nevertheless, stable wastewater treatment plants with extensive aeration also harbored only one or two dominant AOB species (37), which implies that high diversity is not essential. In fact, resource ratio theory (57) predicts less diversity in a community when the ratio of different resources is high, i.e., low oxygen and high ammonia. It seems, therefore, that engineers must make a choice between operational costs and functional redundancy. However, it has been proposed that stability is correlated with functional redundancy rather than the population diversity per se (3), and our findings appear to support this suggestion. It could be speculated that ammonia-oxidizing archaea (AOA), which are functionally redundant but phylogenetically separate from the AOB, might have enhanced the performance and stability of our systems, as they have been already found in bioreactors (36, 63, 65) and freshwater and seawater column/sediment (4, 11, 29, 47) with low DO concentrations. However, AOA were not detected in our systems by qPCR (results not shown); presumably, the relatively high ammonia loading did not promote their growth in our systems (27). This hypothesis can be confirmed only when the mechanisms selecting for AOA, and their real role, in activated sludge are better understood.
ACKNOWLEDGMENTS
This project was supported by the EU Marie Curie Excellence Programme (MEXT-CT-2006-023469). I.D.O. was supported also by CNCSIS-UEFISCSU (project no. PN II-RU 29/09.08.2010) and European Reintegration Grant FLOMAS (FP7/2007-2013 no. 256440).
We thank C. W. Knapp for technical advice and scientific discussions.
Footnotes
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Ahn J. H., et al. 2010. N2O emissions from activated sludge processes, 2008-2009: results of a national monitoring survey in the United States. Environ. Sci. Technol. 44:4505–4511 [DOI] [PubMed] [Google Scholar]
- 2. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 3. Briones A., Raskin L. 2003. Diversity and dynamics of microbial communities in engineered environments and their implications for process stability. Curr. Opin. Biotechnol. 14:270–276 [DOI] [PubMed] [Google Scholar]
- 4. Caffrey J. M., Bano N., Kalanetra K., Hollibaugh J. T. 2007. Ammonia oxidation and ammonia-oxidizing bacteria and archaea from estuaries with differing histories of hypoxia. ISME J. 1:660–662 [DOI] [PubMed] [Google Scholar]
- 4a. Clesceri L. S., Greenberg A. E., Eaton A. D. (ed.). 1998. Standard methods for the examination of water and wastewater, 20th ed American Public Health Association, Washington, DC [Google Scholar]
- 5. Coskuner G., et al. 2005. Agreement between theory and measurement in quantification of ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 71:6325–6334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Daims H., Ramsing N. B., Schleifer K.-H., Wagner M. 2001. Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescence in situ hybridization. Appl. Environ. Microbiol. 67:5810–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. de Bie M. J. M., et al. 2001. Shifts in the dominant populations of ammonia-oxidizing β subclass Proteobacteria along the eutrophic Schelde estuary. Aquat. Microb. Ecol. 23:225–236 [Google Scholar]
- 8. Egli K., et al. 2003. Community analysis of ammonia and nitrite oxidizers during start-up of nitritation reactors. Appl. Environ. Microbiol. 69:3213–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 10. Foley J., de Haas D., Yuan Z., Lant P. 2010. Nitrous oxide generation in full-scale biological nutrient removal wastewater treatment plants. Water Res. 44:831–844 [DOI] [PubMed] [Google Scholar]
- 11. Francis C. A., Roberts K. J., Beman J. M., Santoro A. E., Oakley B. B. 2005. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U. S. A. 102:14683–14688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gieseke A., Purkhold U., Wagner M., Amann R., Schramm A. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Graham D. W., et al. 2007. Experimental demonstration of chaotic instability in biological nitrification. ISME J. 1:385–393 [DOI] [PubMed] [Google Scholar]
- 14. Hall T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95–98 [Google Scholar]
- 15. Hammer Ø., Harper D. A. T., Ryan P. D. 2001. PAST: paleontological statistics software package for education and data analysis. Palaeontol. Electronica 4:1–9 [Google Scholar]
- 16. Hermansson A., Lindgren P.-E. 2001. Quantification of ammonia-oxidizing bacteria in arable soil by real-time PCR. Appl. Environ. Microbiol. 67:972–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hommes N. G., Sayavedra-Soto L. A., Arp D. J. 2003. Chemolithoorganotrophic growth of Nitrosomonas europaea on fructose. J. Bacteriol. 185:6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huber T., Faulkner G., Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 19. Kampschreur M. J., Temmink H., Kleerebezem R., Jetten M. S. M., van Loosdrecht M. C. M. 2009. Nitrous oxide emission during wastewater treatment. Water Res. 43:4093–4103 [DOI] [PubMed] [Google Scholar]
- 20. Klappenbach J. A., Saxman P. R., Cole J. R., Schmidt T. M. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knapp C. W., Graham D. W. 2007. Nitrite-oxidizing bacteria guild ecology associated with nitrification failure in a continuous-flow reactor. FEMS Microbiol. Ecol. 62:195–201 [DOI] [PubMed] [Google Scholar]
- 22. Koops H. P., Purkhold U., Pommerening-Roser A., Timmermann G., Wagner M. 2003. The lithoautotrophic ammonia-oxidizing bacteria, p. 778–811 In Dworkin M., et al. (ed.), The prokaryotes, 3rd ed., vol. 5 Springer-Verlag, New York, NY [Google Scholar]
- 23. Kowalchuk G. A., et al. 1997. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee N., et al. 1999. Combination of fluorescent in situ hybridization and microautoradiography—a new tool for structure-function analyses in microbial ecology. Appl. Environ. Microbiol. 65:1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lydmark P., et al. 2007. Effects of environmental conditions on the nitrifying population dynamics in a pilot wastewater treatment plant. Environ. Microbiol. 9:2220–2233 [DOI] [PubMed] [Google Scholar]
- 26. Maidak B. L., et al. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martens-Habbena W., Berube P. M., Urakawa H., de la Torre J. R., Stahl D. A. 2009. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461:976–979 [DOI] [PubMed] [Google Scholar]
- 28. Milner M. G., Curtis T. P., Davenport R. J. 2008. Presence and activity of ammonia-oxidising bacteria detected amongst the overall bacterial diversity along a physico-chemical gradient of a nitrifying wastewater treatment plant. Water Res. 42:2863–2872 [DOI] [PubMed] [Google Scholar]
- 29. Molina V., Belmar L., Ulloa O. 2010. High diversity of ammonia-oxidizing archaea in permanent and seasonal oxygen-deficient waters of the eastern South Pacific. Environ. Microbiol. 12:2450–2465 [DOI] [PubMed] [Google Scholar]
- 30. Mota C., Head M. A., Ridenoure J. A., Cheng J. J., de los Reyes F. L. 2005. Effects of aeration cycles on nitrifying bacterial populations and nitrogen removal in intermittently aerated reactors. Appl. Environ. Microbiol. 71:8565–8572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Muyzer G., de Waal E. C., Uitterlinden A. G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nei M., Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY [Google Scholar]
- 33. Ofiteru I. D., Curtis T. P. 2009. Modeling risk of failure in nitrification: simple model incorporating abundance and diversity. J. Environ. Eng. 135:660–665 [Google Scholar]
- 34. Park H. D., Noguera D. R. 2007. Characterization of two ammonia-oxidizing bacteria isolated from reactors operated with low dissolved oxygen concentrations. J. Appl. Microbiol. 102:1401–1417 [DOI] [PubMed] [Google Scholar]
- 35. Park H. D., Noguera D. R. 2004. Evaluating the effect of dissolved oxygen on ammonia-oxidizing bacterial communities in activated sludge. Water Res. 38:3275–3286 [DOI] [PubMed] [Google Scholar]
- 36. Park H. D., Wells G. F., Bae H., Criddle C. S., Francis C. A. 2006. Occurrence of ammonia-oxidizing archaea in wastewater treatment plant bioreactors. Appl. Environ. Microbiol. 72:5643–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pickering R. L. 2008. How hard is the biomass working? Ph.D. thesis Newcastle University, Newcastle upon Tyne, United Kingdom [Google Scholar]
- 38. Pollice A., Tandoi V., Lestingi C. 2002. Influence of aeration and sludge retention time on ammonium oxidation to nitrite and nitrate. Water Res. 36:2541–2546 [DOI] [PubMed] [Google Scholar]
- 39. Prosser J. I. 1989. Autotrophic nitrification in bacteria. Adv. Microb. Physiol. 30:125–181 [DOI] [PubMed] [Google Scholar]
- 40. Randall C. W., Barnard J. L., Stensel H. D. 1992. Design and retrofit of wastewater treatment plants for biological nutrient removal. Technomic Publishing Company, Inc., Lancaster, PA [Google Scholar]
- 41. Rassamee V., Sattayatewa C., Pagilla K., Chandran K. 2011. Effect of oxic and anoxic conditions on nitrous oxide emissions from nitrification and denitrification processes. Biotechnol. Bioeng. 108:2036–2045 [DOI] [PubMed] [Google Scholar]
- 42. Rittmann B. E., et al. 1999. Molecular and modeling analyses of the structure and function of nitrifying activated sludge. Water Sci. Technol. 39:51–59 [Google Scholar]
- 43. Rittmann B. E., McCarty P. L. 2001. Environmental biotechnology: principles and applications. McGraw-Hill Book Co., Singapore [Google Scholar]
- 44. Rzhetsky A., Nei M. 1992. A simple method for estimating and testing minimum-evolution trees. Mol. Biol. Evol. 9:945 [Google Scholar]
- 45. Saikaly P. E., Oerther D. B. 2004. Bacterial competition in activated sludge: theoretical analysis of varying solids retention times on diversity. Microb. Ecol. 48:274–284 [DOI] [PubMed] [Google Scholar]
- 46. Saitou N., Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425 [DOI] [PubMed] [Google Scholar]
- 47. Santoro A. E., Francis C. A., De Sieyes N. R., Boehm A. B. 2008. Shifts in the relative abundance of ammonia-oxidizing bacteria and archaea across physicochemical gradients in a subterranean estuary. Environ. Microbiol. 10:1068–1079 [DOI] [PubMed] [Google Scholar]
- 48. Schmidt I. 2009. Chemoorganoheterotrophic growth of Nitrosomonas europaea and Nitrosomonas eutropha. Curr. Microbiol. 59:130–138 [DOI] [PubMed] [Google Scholar]
- 49. Schmidt I., Bock E. 1998. Anaerobic ammonia oxidation by cell-free extracts of Nitrosomonas eutropha. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 73:271–278 [DOI] [PubMed] [Google Scholar]
- 50. Schmidt I., Bock E. 1997. Anaerobic ammonia oxidation with nitrogen dioxide by Nitrosomonas eutropha. Arch. Microbiol. 167:106–111 [PubMed] [Google Scholar]
- 51. Spérandio M., Espinosa M. C. 2008. Modelling an aerobic submerged membrane bioreactor with ASM models on a large range of sludge retention time. Desalination 231:82–90 [Google Scholar]
- 52. Starkenburg S. R., Arp D. J., Bottomley P. J. 2008. D-lactate metabolism and the obligate requirement for CO2 during growth on nitrite by the facultative lithoautotroph Nitrobacter hamburgensis. Microbiology 154:2473–2481 [DOI] [PubMed] [Google Scholar]
- 53. Tallec G. L., Garnier J., Billen G., Gousailles M. 2008. Nitrous oxide emissions from denitrifying activated sludge of urban wastewater treatment plants, under anoxia and low oxygenation. Bioresour. Technol. 99:2200–2209 [DOI] [PubMed] [Google Scholar]
- 54. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 55. Tamura K., Nei M., Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. U. S. A. 101:11030–11035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tilman D. 1982. Resource competition and community structure. Monogr. Popul. Biol. 17:1–296 [PubMed] [Google Scholar]
- 58. Tilman D., et al. 2001. Diversity and productivity in a long-term grassland experiment. Science 294:843–845 [DOI] [PubMed] [Google Scholar]
- 59. U.S. Environmental Protection Agency 1993. Process design manual: nitrogen control. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 60. van Niel E. W. J., Robertson L. A., Kuenen J. G. 1993. A mathematical description of the behaviour of mixed chemostat cultures of an autotrophic nitrifier and a heterotrophic nitrifier/aerobic denitrifier; a comparison with experimental data. FEMS Microbiol. Lett. 102:99–108 [Google Scholar]
- 61. Water UK. 22 December 2010, posting date Sustainability indicators 2009-10 (report). Water UK, London, United Kingdom: http://www.water.org.uk/home/policy/publications/archive/sustainability/2009-10-report/sustainability-2010-final.pdf [Google Scholar]
- 62. Watson S. W., Bock E., Valois F. W., Waterbury J. B., Schlosser U. 1986. Nitrospira marina gen. nov. sp. nov.—a chemolithotrophic nitrite-oxidizing bacterium. Arch. Microbiol. 144:1–7 [Google Scholar]
- 63. Ye L., Zhang T. 2011. Ammonia-oxidizing bacteria dominates over ammonia-oxidizing archaea in a saline nitrification reactor under low DO and high nitrogen loading. Biotechnol. Bioeng. [Epub ahead of print.] doi:10.1002/bit.23211 [DOI] [PubMed] [Google Scholar]
- 64. Yu R., Kampschreur M. J., van Loosdrecht M. C. M., Chandran K. 2010. Mechanisms and specific directionality of autotrophic nitrous oxide and nitric oxide generation during transient anoxia. Environ. Sci. Technol. 44:1313–1319 [DOI] [PubMed] [Google Scholar]
- 65. Zhang T., et al. 2009. Occurrence of ammonia-oxidizing archaea in activated sludges of a laboratory scale reactor and two wastewater treatment plants. J. Appl. Microbiol. 107:970–977 [DOI] [PubMed] [Google Scholar]