Abstract
Although Vibrio cholerae is an important human pathogen, little is known about its populations in regions where the organism is endemic but where cholera disease is rare. A total of 31 independent isolates confirmed as V. cholerae were collected from water, sediment, and oysters in 2008 and 2009 from the Great Bay Estuary (GBE) in New Hampshire, a location where the organism has never been detected. Environmental analyses suggested that abundance correlates most strongly with rainfall events, as determined from data averaged over several days prior to collection. Phenotyping, genotyping, and multilocus sequence analysis (MLSA) revealed a highly diverse endemic population, with clones recurring in both years. Certain isolates were closely related to toxigenic O1 strains, yet no virulence genes were detected. Multiple statistical tests revealed evidence of recombination among strains that contributed to allelic diversity equally as mutation. This relatively isolated population discovered on the northern limit of detection for V. cholerae can serve as a model of natural population dynamics that augments predictive models for disease emergence.
INTRODUCTION
Vibrio cholerae is a ubiquitous waterborne bacterium found commonly in estuarine environments (37). Although V. cholerae is comprised of over 200 serotypes, only serotypes O1 and O139 are currently responsible for epidemic and pandemic cholera outbreaks (1, 9, 14, 15, 37, 49, 57, 60). Closely related clones of types O1 and O139 are found rarely in the environment and only in warm waters (16, 54). Whereas these pandemic serotypes and their associated populations have been the subjects of intense study, investigations of the remaining ecotypes are less common (29, 31–33, 36, 53). Yet other serotypes could be reservoirs of new pathogenic lineages that emerge by horizontal transfer and recombination. Recombination has been observed in several V. cholerae collections (31, 61) and is thought to have driven the emergence of new infective variants (16).
Only 40 domestically acquired toxigenic V. cholerae cases have been reported in the United States since 1995 (66), but epidemics are ongoing in warm, subtropical climates, the most recent occurring in Haiti, which had been cholera-free for decades (6). Variation in disease incidence is caused in part by transmission between patients by feces-contaminated drinking water and by endemic populations of toxigenic cholera organisms in warm subtropical environments (11, 12, 37). Endemic cholera has not posed a public health threat in temperate regions in modern times (38, 60). Toxigenic O1 and O139 serotypes of V. cholerae are not regularly isolated from temperate waters in the United States, but some environmental non-O1/non-O139 populations, a few of which are associated with disease, have been described (11, 31, 43, 58). Even so, the pathogenic potential and ecology of most endemic populations are not well understood.
Here we describe the genotypic and phenotypic characteristics of a newly identified and highly diverse population of non-O1 V. cholerae isolated from water, sediment, and live oysters from the Great Bay Estuary (GBE) in New Hampshire, a location where until now V. cholerae had not been isolated and where endemic disease has never been reported. Major rainfall events were the primary predictor for abundance, which correlated more weakly with increased temperature and decreased salinity. Multilocus sequence analysis (MLSA) identified members of clonal complexes from different samples and in consecutive years. These genotypes also displayed evidence of frequent recombination. This report will facilitate further monitoring for invading strains and the study of population dynamics in response to changing environmental conditions.
MATERIALS AND METHODS
Isolation, serotyping, and genotyping of V. cholerae.
Water and oyster (Crassostrea virginica) samples were collected approximately every 2 weeks during May to December 2008 and 2009 from two sites in the GBE. One oyster bed is located near Nannie Island and is within an area classified as approved for shellfish harvesting, and the other oyster bed is located in the Oyster River within an area classified as prohibited because of its proximity to the Durham, NH, wastewater treatment facility effluent discharge pipe. Oysters were sampled using oyster tongs at similar locations within the same general area of the oyster beds. Sediments were collected with a van Veen grab sampler. The top 2 cm of sediment was scooped into sterile plastic bags. Water samples were collected on site by filling and then capping sterile 1-liter plastic bottles at ∼30 cm below the water surface. Sampling was designed to screen a variety of conditions for V. cholerae detection, and all samples were single replicates for each date, site, and environmental matrix. All oyster, sediment, and water samples were immediately stored in coolers containing ice packs and brought back to the laboratory for analysis within 1 to 3 h.
Shellfish were cleaned of debris and shucked using standard aseptic procedures (30). Tissue was transferred to sterile beakers, weighed, and then diluted 1:1, 1:2, 1:3, or 1:4, depending on the sample size, with alkaline peptone water (APW [pH 8.6]; 1% NaCl) prior to homogenization for 90 s in a Waring blender. Sediment samples were homogenized by hand, and 1.0 g (wet weight) was transferred to 9.0 ml of APW for further dilution. Water samples were shaken for 20 s. Following homogenization, volumes of 10, 1.0, 0.1, and/or 0.001 ml of oyster homogenates, sediment, and water samples were inoculated into a three-tube multiple tube fermentation (most probable number [MPN]) analysis series with APW selective enrichment for 16 h at 37°C. Turbid MPN tubes were streaked onto thiosulfate-citrate-bile salts-sucrose (TCBS) (46) and colistin-polymyxin B-cellobiose (CPC+) (67) agar plates. Following incubation, a single representative isolate of each colony morphology type recognized putatively as Vibrio spp. from each environmental sample (for 2008) or from each MPN dilution tube (2009) was restreaked for isolation onto T-SOY agar for additional analysis. This measure was taken to eliminate the collection of clones that were a product of enrichment, but such a collection design could also underrepresent diversity. Individuals were genotyped and identified as V. cholerae by using multiplex PCR for the conserved virulence-associated regulatory genes toxR and tcpI, as well as the virulence-associated genes used for strain typing, ompU and hlyA (56). Amplicons were analyzed by gel electrophoresis and compared to positive- and negative-control strains reported by Panicker et al. (56). Any isolates from which these genes were amplified were confirmed as V. cholerae by 16S V2-3 amplification and sequencing (40). Additional accessory virulence genes whose products contribute to disease, including zot, ace, tcpA, and the major toxin-encoding gene ctxA, were subsequently amplified by following published protocols (8, 22, 62).
Isolates were serotyped using V. cholerae polyantiserum (BD Difco, Sparks, MD). Briefly, isolates were grown on LB agar plates, and single colonies were transferred by a sterile loop onto 1 drop (approximately 40 μl) of antiserum on a glass slide. Glass slides were rotated and observed for agglutination over 5 min and compared to a serotype O1 positive control, V. cholerae C6707 (48), and a negative nonpandemic control strain, V. cholerae DAL-2919.
Analysis of abundance with environmental conditions.
Salinity, water temperature, and dissolved oxygen were recorded by two YSI 6 series multiparameter datasondes (Yellow Springs Inc., Yellow Springs, OH) located near the Oyster River and Nannie Island oyster beds and managed by the Great Bay system-wide monitoring program (SWMP; http://www.greatbay.org). A YSI 85 instrument was used to measure water temperature, salinity, and dissolved oxygen on site at each sample event; these data were compared with the sonde data to ensure consistent recording. Daily rainfall totals were also recorded and accessed via the University of New Hampshire Weather Station website (http://www.weather.unh.edu). Multivariate stepwise regression analysis was used to determine environmental condition correlations with the overall maximum MPN, maximum MPN in oysters, and maximum MPN in water for each site at each collection date, using JMP version 8.01.1. Effects of environmental factors on V. cholerae abundance are reported as the squared partial correlation coefficient, as determined using a Pcorr script. Pearson correlations were determined for each single environmental condition and each MPN value with the SPSS PASW statistic software version 18.0.
Phenotypic analysis.
All phenotypic assays were performed using standard published protocols. Biochemical tests were performed as described previously (7). Briefly, lysine and ornithine decarboxylase assays were performed using the Moeller decarboxylase base medium (Difco) with the addition of amino acids at 1% (wt/vol). The methyl red reaction was performed using MR-VP medium (Difco), and the mixture was incubated at 37°C for 48 h. The Vogues-Proskauer assay was performed on inoculated MR-VP medium at 37°C for 48 h. Growth in 0, 6, and 8% (wt/vol) NaCl in nutrient broth was assessed by turbidity after overnight incubation at 37°C. Biofilm production was measured for cultures grown for 24 h in heart infusion (HI) broth followed by crystal violet staining (44). The quantitative hemolysin assay (44) was performed by incubating defibrinated sheep blood (Northeast Laboratory, Waterville, ME) with overnight cultures of cells conditioned for 24 h in HI. Blood cell lysis was assessed by absorbance of cell-free supernatants at 415 nm using a Tecan Infinite M200 plate reader (Tecan, Durham, NC). Antimicrobial susceptibility was determined in three replicate experiments by measuring the diameters of the zones of inhibition produced on a lawn of cells from each of two paper discs saturated with 10 μg gentamicin, 30 μg kanamycin, 30 μg chloramphenicol, 30 μg naladixic acid, or 30 μg tetramycin (Tet). Polymyxin B plates had a concentration of 50 U/ml. Citrate and sialic acid media were prepared by supplementing M9 minimal medium with 1 mg/ml of citrate or 1 mg/ml of sialic acid (N-acetylneuraminic acid; Sigma), respectively. Motility plates contained 0.3% agar. The detection of vibrio pathogenicity island 2 (VPI-2), vibrio seventh pathogenicity island 1 (VSP-I), and VSP-II and their products was performed as described previously (2, 53).
The collected phenotypic data were used to build a trait matrix, from which a phenogram was produced. Continuous phenotypic data (biofilm, hemolysin, and antimicrobial susceptibility to five antibiotics) were converted into categorical data (pairwise Sidak t tests; P < 0.05) at two or three levels each. The methyl red levels were categorized by either a positive or negative result. The unordered multistate discrete characters parsimony (pars) algorithm method in the PHYLIP package was utilized to build the phenogram (17).
Phylogenetic characterization.
MLSA was performed on DNA purified by a cetyltrimethylammonium bromide-NaCl precipitation followed by phenol-chloroform extraction (3) on five loci, gapA, gyrB, pyrH, recA, and topA, using published primers and PCR conditions (65). PCR was performed with a MasterTaq PCR kit (Eppendorf, Hauppauge, NY), with 2.0 mM MgCl2 and a 0.2 μM concentration of each primer. PCR products were analyzed on a 1.2% agarose gel, and the remainder of the reaction mixtures that yielded a single band of the proper size were treated by ExoSAP (USB, Fremont, CA) and sequenced at the DNA sequencing core facility at the Hubbard Center for Genome Studies at the University of New Hampshire (Durham, NH) using Applied Biosystems BigDye terminator cycle sequencing kits (v3.1) and analyzed with an ABI3130 DNA analyzer.
Sequence analysis and phylogeny analysis were completed with MEGA version 4.04.0 unless otherwise noted (63). Contigs were assembled from the consensus of both forward and reverse sequences of each locus for each strain. The assembled sequences of each locus were then aligned for all strains and the sequences further trimmed to include only overlapping DNA. The five loci were then concatenated, and the sequences were aligned using ClustalW. Phylogenetic trees were constructed by the neighbor-joining algorithm with 1,000 bootstrap replacements and with the Jukes-Cantor substitution model. Sequences from reference toxigenic V. cholerae strains, including four O1 El Tor biotypes (M66-2, N16961, 2740-80, and MJ-1236) and one O1 classical biotype (0395), as well as Vibrio mimicus VM603 and Vibrio parahaemolyticus RIMD 2210633, were accessed from NCBI GenBank and included in the phylogenetic analyses. Sequences for additional reference V. cholerae strains MO10 (O139; Bangladesh), V51 (O37; United States), 1587 (O12; Peru), AM-19226 (O39; Bangladesh), MZO-2 (O14; Bangladesh), and MZO-3 (O37; Bangladesh) were retrieved from the Broad Institute (http://www.broadinstitute.org/) and included in phylogeny analyses.
Additional phylogenies were constructed, and the extent of recombination was analyzed by SplitsTree version 4 utilizing the Phi test module (24). A nonredundant allele database was created using NRDB Align (26), and a subsequent allelic profile was determined for the total collection. Using the allelic profile, recombination was analyzed by using the LIAN 3.5 linkage analysis program (21). The PHYLIP phyML algorithm was used to build additional habitat trees (17) and for the adaptML script (23), in which isolates were labeled by location and year collected. ClonalFrame 1.1 was used to determine the relative influence of variation of recombination compared to mutation (r/m) (13).
RESULTS
Rainfall as a predictor of abundance of environmental V. cholerae in the Great Bay Estuary.
In an ongoing study of resident Vibrio species in New Hampshire (44), we unexpectedly isolated V. cholerae from the GBE. From July 2008 to September 2009, 31 bacterial isolates from oysters, sediment, and overlying water from two oyster beds located 6 miles apart in the GBE were identified as V. cholerae by multiplex PCR (56) (Table 1) and confirmed as such by 16S rRNA genes sequencing. Isolates were identified more frequently from water or oyster samples (87% of isolates) than from sediment. Isolation of V. cholerae from the GBE is rare in light of the total number of isolates screened by multiplex PCR, representing only 2.5% of all putative Vibrio isolates.
Table 1.
Summary of V. cholerae isolates collected from the Great Bay Estuary
| Isolate no. | Isolation date | Sitea | Source | MPNb | Presence of genec |
|||
|---|---|---|---|---|---|---|---|---|
| toxR | ompU | hlyA | tcpI | |||||
| 428 | 7/30/2008 | OR | Oyster | 1.5 × 102 | + | + | + | − |
| 442 | 7/30/2008 | NI | Oyster | 1.5 × 101 | + | − | − | − |
| 466 | 8/5/2008 | NI | Water | 1.5 × 101 | + | + | + | + |
| 474 | 8/5/2008 | OR | Water | 2.4 × 101 | + | − | − | − |
| 476 | 8/5/2008 | OR | Water | 1.5 × 101 | + | + | + | + |
| 504 | 8/12/2008 | OR | Water | 2.8 | + | + | − | − |
| 509 | 8/12/2008 | OR | Oyster | 2.9 × 101 | + | − | − | − |
| 658 | 9/19/2008 | NI | Water | 3.6 × 10−1 | + | − | − | − |
| 661 | 9/19/2008 | NI | Water | 3.6 × 10−1 | + | − | − | − |
| 684 | 9/24/2008 | OR | Water | 3.6 × 10−1 | + | − | − | − |
| 704 | 10/8/2008 | NI | Oyster | 9.1 × 10−1 | + | + | + | − |
| 705 | 10/8/2008 | NI | Oyster | 3.6 × 10−1 | + | − | − | − |
| 901 | 6/29/2009 | OR | Water | 1.1 | + | + | + | − |
| 907 | 6/29/2009 | OR | Oyster | 1.5 × 101 | + | − | − | − |
| 917 | 6/29/2009 | NI | Water | 2.1 | + | + | + | − |
| 925 | 6/29/2009 | OR | Water | 1.5 | + | − | − | − |
| 937 | 6/29/2009 | NI | Water | 1.5 | + | − | − | − |
| 1063 | 7/27/2009 | OR | Oyster | 3.6 | + | + | + | − |
| 1068 | 7/27/2009 | NI | Oyster | 3.6 | + | − | − | − |
| 1069 | 7/27/2009 | NI | Sediment | 2.3 × 101 | + | + | + | − |
| 1070 | 7/27/2009 | NI | Sediment | 2.3 × 101 | + | − | − | − |
| 1075 | 7/27/2009 | NI | Oyster | 7.3 | + | − | − | − |
| 1105 | 7/27/2009 | NI | Oyster | 1.5 × 102 | + | + | + | − |
| 1106 | 7/27/2009 | OR | Water | 1.5 × 102 | + | − | − | − |
| 1114 | 7/27/2009 | OR | Water | 2.0 | + | − | + | − |
| 1116 | 7/27/2009 | OR | Water | 1.1 | + | − | − | − |
| 1118 | 7/27/2009 | NI | Water | 9.1 × 10−1 | + | − | − | − |
| 1173 | 8/11/2009 | OR | Sediment | 2.0 × 102 | + | − | − | − |
| 1194 | 8/11/2009 | OR | Water | 1.2 × 101 | + | + | + | − |
| 1200 | 8/11/2009 | OR | Water | 1.5 × 102 | + | − | − | − |
| 1261 | 9/1/2009 | NI | Sediment | 9.1 | + | − | − | − |
NI, Nannie Island; OR, Oyster River.
Reported as MPN/liter for water samples and MPN/g for sediment and oyster samples.
The genes zot, ace, tcpA, and ctxA were absent from all isolates.
Abundance of toxigenic V. cholerae typically correlates with warmer temperatures and reduced salinity associated with rainfall (12, 55). In some temperate regions where populations of V. cholerae are described, these two factors are significant predictors (34, 42). In the more extreme northern temperate location of the GBE, V. cholerae was detected seasonally and only between the months of June and October. Even so, isolates were recovered from relatively cold waters, as cold as 13.6°C in October 2008. Salinity, temperature, and dissolved oxygen from point of collection data each correlated significantly with abundance in pairwise tests (Table 2). We noticed that V. cholerae was typically detected several days after major rainfall events. To explore this dynamic, we performed a stepwise regression of the highest V. cholerae MPN concentration at each collection site and date with salinity, temperature, and rainfall averaged over several days prior to collection. We reasoned that an extended window prior to collection would more accurately portray conditions favorable for microbial growth than single data points at the time of collection. We evaluated (i) average salinity over 12 and 72 h prior to collection, (ii) average water temperature over 12 and 72 h prior to collection, and (iii) total prior rainfall over 72 and 96 h prior to collection. This analysis revealed a significant correlation between MPN and prior rainfall at both 72 h and 96 h (Table 2), explaining 32% and 13% of V. cholerae abundance, respectively. When the highest detected MPN concentrations were considered for samples from oyster or water samples separately, the only significant regressors were rainfall during 72 h prior to collection for water isolates (P = 0.002) and rainfall during 96 h prior to collection for oyster isolates (P = 0.001). Because the 12-h mean temperature and salinity were no longer significantly associated with MPN in this regression, recent rainfall became the best predictor of V. cholerae incidence in this study. That different environments, oysters or overlying water, accumulate V. cholerae at different rates could influence future predictive models of occurrence.
Table 2.
Abundance and environmental factor correlations
| Environmental factor and time period or other parameter | Statistical correlationa of maximum MPN for source |
|||||
|---|---|---|---|---|---|---|
| Total |
Oyster |
Water |
||||
| Pearson correlation | Stepwise regression | Pearson correlation | Stepwise regression | Pearson correlation | Stepwise regression | |
| Temp | ||||||
| 12 h | 0.355* | NC | 0.230 | NC | 0.271 | NC |
| 72 h | 0.389* | NC | 0.270 | NC | 0.266 | NC |
| Salinity | ||||||
| 12 h | −0.369* | NC | −0.355* | NC | −0.236 | NC |
| 72 h | −0.461* | 0.1768 | −0.375* | 0.378 | −0.367 | 0.450 |
| Rain | ||||||
| 72 h | 0.472** | 0.049* | 0.112 | 0.4753 | 0.623** | <0.001** |
| 96 h | 0.566** | 0.006* | 0.555** | 0.001** | 0.228 | 0.976 |
| Dissolved O2 | ||||||
| Percent | −0.333* | NC | −0.261 | NC | −0.223 | NC |
| Concn (mg/liter) | −0.355* | NC | −0.247 | NC | −0.255 | NC |
| Turbidity | 0.112 | NC | 0.134 | NC | 0.112 | NC |
The r statistic is given for Pearson correlations. For stepwise regression analyses, the P statistic is given, unless the environmental condition was excluded from the stepwise model, in which case no correlation (NC) is reported.
, P < 0.05;
, P < 0.001.
V. cholerae Great Bay Estuary isolates lack virulence markers and belong to a highly diverse and recombining population.
V. cholerae genotyping and determination of toxigenic potential rely upon a number of markers of virulence and strain type (20, 25, 49, 50, 68). The genes ompU and hlyA, which correlate with certain serotypes, were present in 12 isolates, occurring together in each case except for isolates 504 (ompU) and 1114 (hlyA) (Table 1). Only two isolates were positive for a regulator of toxin-coregulated pilus expression (tcpI). Virulence-associated genes zot, ace, tcpA, and ctxA (encoding cholera toxin subunit A) were absent from all of the GBE isolates. All of the isolates were confirmed as non-O1 serotypes.
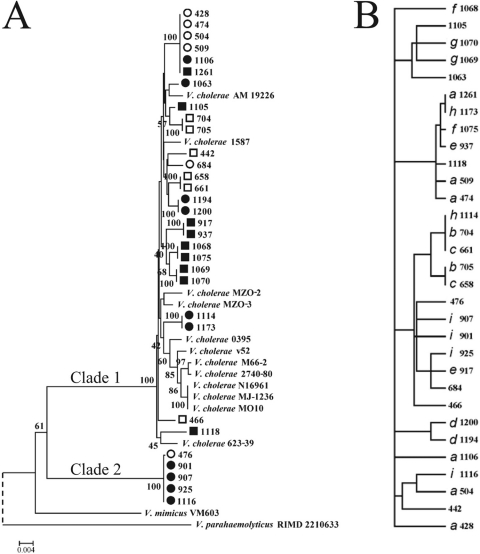
To examine the diversity and genetic relationships among these 31 GBE V. cholerae isolates, we performed MLSA with five loci: gapA, gyrB, pyrH, recA, and topA. Although an MLSA scheme was preexisting and used to generate a V. cholerae database (18), it was developed specifically for closely related O1 and O139 serotype strains and failed to amplify the orthologous alleles of our environmental strains. We therefore analyzed a subset of the genes developed to type Vibrio species (65), which successfully amplified loci from all collected isolates. We included recA and pyrH because these genes differentiate closely related Vibrio species (61), especially V. cholerae and V. mimicus (64). We also included gapA, gyrB, and topA, which are less valuable for discriminating between species of Vibrio due to their relatively high divergence but could potentially reveal more differences between strains from the same population. A neighbor-joining tree based on concatenated sequences showed a diverse population with two major clades and several clonal complexes, designated a to i (Fig. 1A). All O1 clinical reference strains grouped within the larger and more diverse clade 1 and most closely with isolates 1114 and 1173, which grouped closely within the classical and El Tor biotypes subclade (Fig. 1A). We further characterized 1114 and 1173 by standard methods that differentiate between El Tor and classical biotypes and compared them with V. cholerae N16961, C6706 (both El Tor), O395 (classical), and three GBE strains outside the El Tor/classical subclade (428, 658, and 1068). The five GBE strains share multiple traits with the two El Tor biotype strains, that is, they are all polymyxin B resistant, HapR positive, can use citrate as a sole carbon source, and do not autoagglutinate on LB (pH 6.5) at 30°C. However, none of the strains contained marker genes or products for VPI-1, VPI-2, ctxφ phage, VSP-I, or VSP-II. The only exception was 428, which encodes NanA and is able to utilize sialic acid as a sole carbon source, indicating the presence of the nan-nag cluster, which is typically encoded within VPI-2, a pathogenicity island whose canonical form is confined to cholerogenic strains of V. cholerae (2).
Fig. 1.
Consensus neighbor-joining tree of GBE isolates. (A) A tree was constructed from concatenated sequences of 5 loci, including gapA, gyrB, pyrH, recA, and topA sequences (2,792 bp), using the Jukes-Cantor model of substitution for 31 GBE isolates from Nannie Island (circles) and Oyster River (squares) collected in 2008 (open symbols) and 2009 (filled symbols), as well as five serotype O1 (four El Tor biotypes and one classical biotype) reference strains and eight clinical non-O1 reference strains. V. mimicus VM603 and V. parahaemolyticus RIMD 2210633 are included as tree roots for comparison, but the V. parahaemolyticus branch was shortened in the figure (dashed line). Bootstrap values are from 1,000 replicates. Bar, 0.2% divergence. (B) MLST clonal complexes are labeled with an italic letter to differentiate the clones in the PARS tree, which is based on the discrete levels of the most variable eight phenotypes tested.
Five GBE isolates, all from Nannie Island, formed clade 2. This clade is more distant from the clinical isolates, less diverse than clade 1, but nevertheless shares 100% identity in the V2-V3 region of 16S rRNA genes with type strains of V. cholerae. Moreover, these isolates did not cluster with environmental strains of V. mimicus (Fig. 1A) (64). Phylogenies of each individual gene mirrored the concatenated tree, except for gyrB, which indicates either unique patterns of recombination or atypical divergence and selective pressures on this locus. All combinations of four out of five concatenated genes produced the same topology as the five-gene analysis (data not shown), indicating that the relationships between strains were not biased by the inclusion of any one locus.
Although our analysis revealed high diversity within this geographically confined population, it was unclear whether it was typical of natural ecosystems. We compared our small collection with a geographically broader collection of 156 environmental isolates from central California and Hawaii (referred to here as CA) at two loci that overlapped with our MLSA scheme (recA and gyrB) and found similar diversity. For recA, the 31 GBE isolates were equally diverse (maximum pairwise distance, CA = 0.077, GBE = 0.062; average pairwise distance, CA = 0.019, GBE = 0.032), whereas for gyrB, the CA collection was slightly more diverse (maximum pairwise distance, CA = 0.063, GBE = 0.045; average pairwise distance, CA = 0.016, GBE = 0.01). When both collections were combined to construct a phylogeny based on these two loci, each major clade from the CA collection paired with at least one GBE isolate (data not shown), further demonstrating that this presumably small population is diverse.
We also noticed that some MLSA clones appeared in both years and some associated with site (Fig. 1A). To evaluate these associations statistically, we used adaptML (23), which correlates phylogenetic clusters with habitat. This analysis confirmed that over 75% of isolates correlate with site, but no single isolate or phylogenetic branch correlated with year (data not shown), suggesting that the population is geographically structured.
Although they formed two diverse clades, most isolates collected were apparently clones of at least one other isolate based on MLSA, which reflects relatedness but may improperly define some isolates as clones because only limited sequence information is considered. Indeed, further genotyping and phenotyping (Table 1 and Fig. 1B) revealed diversity even within these clonal complexes. Phenotypic diversity among the strains was highest for antibiotic resistance, methyl red reactivity, and hemolysin activity. In contrast, the GBE isolates were homogeneous in other phenotypes used to differentiate V. cholerae strains in previous studies, including arginine and lysine oxidation, the Voges-Proskauer test, and growth in various NaCl concentrations (7). The lack of correlation of any particular phenotype or clone with substrate further emphasizes that the traits examined have limited utility as a reflection of ecologically relevant characteristics, with perhaps one exception: the GBE strains grew in all concentrations of NaCl tested, even those that are typically inhibitory to V. cholerae (8%), which may reflect adaptation to large variations in salinity in the GBE.
Comparison of a phenogram constructed from the eight most variable phenotypes (Fig. 1B) with the phylogeny constructed from concatenated MLSA loci (Fig. 1A) indicated differences in topology. The phenogram often grouped strains of different multilocus types, and MLSA clones were often phenotypically dissimilar. Antibiotic resistance and methyl red reactivity were most often the phenotypes that distinguished among the complexes. Thus, MLSA alone did not capture the true diversity of V. cholerae in the GBE, and no specific phenotype or set of phenotypes grouped strains by genetic relatedness. Likely because these phenotypes are not discrete genetic characters, this phenotypic profile offered a complementary but distinct profile of the diversity of V. cholerae in the GBE.
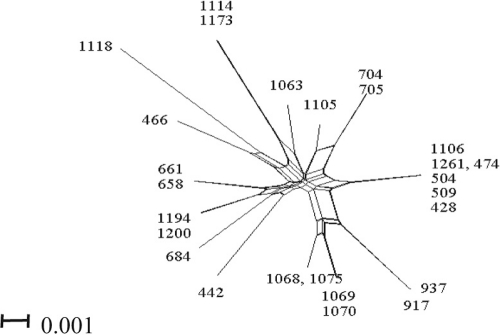
We also evaluated the MLSA data for evidence of recombination by two statistical tests of sequence diversity and association among alleles. Sequence diversity of the entire group of 31 environmental isolates, as measured by the Phi test (24), demonstrated no significant recombination (P = 0.196). However, when the divergent clade 2 was excluded from this analysis, we observed significant recombination among isolates in clade 1 (Phi test P < 0.001) (Fig. 2). We further scrutinized the relationships within clade 1 to determine whether certain loci were more prone to recombine by sequentially removing individual loci from the analysis. Although gyrB accounted for the most recombination observed, each gene displayed significant evidence of recombination. We noted that toxigenic serotype O1 strains also group within clade 1 as well as other clinical non-O1 strains (Fig. 1A) and may be capable of similar levels of recombination. Similar results were obtained using the LIAN test of recombination, which is based on allelic profiles; it failed to reject the null hypothesis of linkage equilibrium (IA = 0.3204; P < 0.001). We recorded one recombination event (r) for every mutation (m) from the ClonalFrame analysis (r/m, 1.33; 95% confidence range, 0.33 to 3.12). This suggests that diversity in GBE strains is equally explained by recombination and mutation, which is consistent with both the previous evidence of recombination and the divergence (rather than convergence) observed in the gene trees.
Fig. 2.
Splits tree of GBE clade 1 isolates. Shown is a concatenated split network tree based on five gene loci: gapA, gyrB, pyrH, recA, and topA sequences (2,792 bp). Sequences were concatenated and reconstructed using the SplitsTree4 program (see Materials and Methods for details).
DISCUSSION
As part of a larger survey of human pathogenic Vibrio spp. in the GBE of New Hampshire, we cultured and characterized 31 V. cholerae isolates. The analysis revealed an incongruence between physical traits and heritage and also highlighted the challenges of applying typing schemes developed with clinical strains to more diverse wild relatives. MLSA revealed a diverse population of V. cholerae, with some isolates related to pathogenic lineages and others related to more diverse clones from environmental sources. Strains collected worldwide all group within the phylogeny of GBE isolates that were collected in a relatively isolated northern estuary. As observed in other studies of Vibrio populations (19, 31, 61, 69), this population displayed a history of recombination; however, the extent observed here is noteworthy because most prior analyses of this species have focused on epidemic (and hence clonal) lineages (5, 18, 37). Even so, the influence of recombination on population structure was modest (r/m, 1.33) compared to a parallel study with environmental V. cholerae samples isolated from central coastal California and Hawaii (r/m, 4 to 8) (31). Mutation may contribute more to diversity in the GBE population, perhaps because of genetic or ecological barriers to recombination. Our analysis also suggests that pathogenic members of clade 1 (Fig. 1A) could exchange homologous alleles with other nonpathogenic strains from this clade. Whether transient residency of toxigenic V. cholerae could lead to recombination and evolution of infective biovars is debatable, but genetic transfer between avirulent and virulent strains is believed to have led to disease emergence in Bangladesh (16), and the emergence of environmental non-O1/non-O139 strains that laterally acquired the ctxAB via the CTX prophage have been reported in India, Malaysia, and California (25, 45, 59).
Non-O1/non-O139 endemic populations in the United States rarely cause infections and are thought to have little clinical opportunity for outbreaks (35, 43). However, current climate change models predict increasing surface water temperatures and rainfall events in some areas, including New Hampshire (51). These conditions have been associated with V. cholerae abundance in this and other studies (10, 41, 42) and could increase endemic populations and promote niche expansion, both of which could facilitate emergence of new biovars. Indeed, outbreaks of cholera related to unprecedented increases in surface water temperatures and invasion by Asiatic strains have occurred in regions such as Peru, where cholera had not been a problem for over a century (10, 39, 46). The strongest correlating factor in our study was major rainfall events, which was only apparent when data over several days prior to collection were used in the analysis, whereas reduced salinity and increased temperature were only weakly correlated with abundance. This apparent contradiction with previous studies could reflect a true adaptation of northern temperate strains that may experience greater variations in both salinity and temperature. Regardless, this observation may prove useful in developing predictive models for temperate regions on the cusp of niche expansion.
Vibrio species have been a focus for study of New Hamsphire's Great Bay Estuary since the 1960s, and although V. parahaemolyticus and V. vulnificus have been isolated with regularity (4, 27, 28, 52), only during the last 2 years of intense study of this ecosystem have we recovered V. cholerae. It is unclear whether our isolation of V. cholerae resulted from a recent niche expansion event and/or population growth or whether the population was simply overlooked due to its presumed improbability. Even so, it is unlikely that a recently established community would display such high diversity, and the detection of MLSA clones in both years of this study suggests the strains can overwinter and reemerge in the spring. This discovery presents an opportunity to study this potentially pathogenic species in a highly variable estuary at the northern range limit for this species. Due to the GBE's relative isolation and the extreme environmental conditions that it experiences relative to the greater Gulf of Maine and other known endemic populations, here we may explore the ecological and evolutionary dynamics of resident vibrios in the context of a natural and potentially changing ecosystem.
ACKNOWLEDGMENTS
We thank C. Edwards, R. Desy, J. Mahoney, E. Jones, A. Lamb, A. Urbano, B. Morris, and S. McLean for assistance with the collection of environmental V. cholerae, R. Taylor for helpful suggestions, protocols, and reference strains, A. DePaola for reference strains, and F. Thompson for discussions and protocols.
This work was funded by Sea Grant number R/CE-137, National Institutes of Health grant R03AI081102, and a fellowship from the Great Bay Stewards. Partial funding was provided by the New Hampshire Agricultural Experiment Station. This is Scientific Contribution number 2463.
Footnotes
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Albert M. J., et al. 1993. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet 341:704. [DOI] [PubMed] [Google Scholar]
- 2. Almagro-Moreno S., Boyd E. F. 2009. Sialic acid catabolism confers a competitive advantage to pathogenic Vibrio cholerae in the mouse intestine. Infect. Immun. 77:3807–3816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ausubel F., et al. 1990. Current protocols in molecular biology. Wiley and Sons, Inc., New York, NY [Google Scholar]
- 4. Bartley C. H., Slanetz L. W. 1971. Occurrence of Vibrio parahaemolyticus in estuarine waters and oysters of New Hampshire. Appl. Environ. Microbiol. 21:965–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Byun R., Elbourne L. D. H., Lan R., Reeves P. R. 1999. Evolutionary relationships of pathogenic clones of Vibrio cholerae by sequence analysis of four housekeeping genes. Infect. Immun. 67:1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chin C.-S., et al. 2011. The origin of the Haitian cholera outbreak strain. N. Engl. J. Med. 364:33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choopun N., Louis V., Huq A., Colwell R. R. 2002. Simple procedure for rapid identification of Vibrio cholerae from the aquatic environment. Appl. Environ. Microbiol. 68:995–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chow K. H., Ng T. K., Yuen K. Y., Yam W. C. 2001. Detection of RTX toxin gene in Vibrio cholerae by PCR. J. Clin. Microbiol. 39:2594–2597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chun J., et al. 2009. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 106:15442–15447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Colwell R. R. 2004. Infectious disease and environment: cholera as a paradigm for waterborne disease. Int. Microbiol. 7:285–289 [PubMed] [Google Scholar]
- 11. Colwell R. R., Spira W. M. 1992. Cholera. Plenum, New York, NY [Google Scholar]
- 12. Constantin de Magny G., Colwell R. R. 2009. Cholera and climate: a demonstrated relationship. Trans. Am. Clin. Climatol. Assoc. 2009:119–128 [PMC free article] [PubMed] [Google Scholar]
- 13. Didelot X., Falush D. 2007. Inference of bacterial microevolution using multilocus sequence data. Genetics 175:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Farfan M., Minana-Galbis D., Fuste M. C., Loren J. G. 2002. Allelic diversity and population structure in Vibrio cholerae O139 Bengal based on nucleotide sequence analysis. J. Bacteriol. 184:1304–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Faruque S. M., Albert M. J., Mekalanos J. J. 1998. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Faruque S. M., et al. 2003. Reemergence of epidemic Vibrio cholerae O139, Bangladesh. Emerg. Infect. Dis. 9:1116–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Felsenstein J. 1989. PHYLIP: phylogeny inference package, version 3.2. Cladistics 5:164–166 [Google Scholar]
- 18. Garg P., et al. 2003. Molecular epidemiology of O139 Vibrio cholerae: mutation, lateral gene transfer, and founder flush. Emerg. Infect. Dis. 9:810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gonzalez-Escalona N., et al. 2008. Determination of molecular phylogenetics of Vibrio parahaemolyticus strains by multilocus sequence typing. J. Bacteriol. 190:2831–2840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hall R. H., Drasar B. S. 1990. Vibrio cholerae HlyA hemolysin is processed by proteolysis. Infect. Immun. 58:3375–3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haubold B., Hudson R. R. 2000. LIAN 3.0: detecting linkage disequilibrium in multilocus data. Bioinformatics 16:847–849 [DOI] [PubMed] [Google Scholar]
- 22. Huhulescu S., et al. 2007. Occurrence of Vibrio cholerae; serogroups other than O1 and O139 in Austria. Wien. Klin. Wochenschr. 119:235–241 [DOI] [PubMed] [Google Scholar]
- 23. Hunt D. E., et al. 2008. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320:1081–1085 [DOI] [PubMed] [Google Scholar]
- 24. Huson D. H., Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254–267 [DOI] [PubMed] [Google Scholar]
- 25. Jiang S., Chu W., Fu W. 2003. Prevalence of cholera toxin genes (ctxA and zot) among non-O1/O139 Vibrio cholerae strains from Newport Bay, California. Appl. Environ. Microbiol. 69:7541–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jolley K. A., Feil E. J., Chan M.-S., Maiden M. C. J. 2001. Sequence type analysis and recombinational tests (START). Bioinformatics 17:1230–1231 [DOI] [PubMed] [Google Scholar]
- 27. Jones S. H., et al. 1991. The incidence and elimination of vibrios and fecal-borne bacteria from northern New England oysters. J. Shellfish Res. 60:163–167 [Google Scholar]
- 28. Jones S. H., Summer-Brason B. 1998. Incidence and detection of pathogenic Vibrio sp. in a northern New England estuary, USA. J. Shellfish Res. 17:1665–1669 [Google Scholar]
- 29. Karaolis D., Lan R., Reeves P. 1995. The sixth and seventh cholera pandemics are due to independent clones separately derived from environmental, nontoxigenic, non-O1 Vibrio cholerae. J. Bacteriol. 177:3191–3198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaysner C. A., DePaola A., Jr 2004. Bacteriological analytical manual online. Vibrio. U.S. Food and Drug Administration, Washington, DC: http://www.fda.gov/Food/ScienceResearch/LaboratoryMethods/BacteriologicalAnalyticalManualBAM/ucm070830.htm [Google Scholar]
- 31. Keymer D. P., Boehm A. B. 2011. Recombination shapes the structure of an environmental Vibrio cholerae population. Appl. Environ. Microbiol. 77:537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Keymer D. P., Lam L. H., Boehm A. B. 2009. Biogeographic patterns in genomic diversity among a large collection of Vibrio cholerae isolates. Appl. Environ. Microbiol. 75:1658–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Keymer D. P., Miller M. C., Schoolnik G. K., Boehm A. B. 2007. Genomic and phenotypic diversity of coastal Vibrio cholerae strains is linked to environmental factors. Appl. Environ. Microbiol. 73:3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kirschner A. K. T., et al. 2008. Rapid growth of planktonic Vibrio cholerae non-O1/non-O139 strains in a large alkaline lake in Austria: dependence on temperature and dissolved organic carbon quality. Appl. Environ. Microbiol. 74:2004–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Klontz K. C., Tauxe R. V., Cook W. L., Riley W. H., Wachsmuth I. K. 1987. Cholera after the consumption of raw oysters. Ann. Intern. Med. 107:846–848 [DOI] [PubMed] [Google Scholar]
- 36. Kotetishvili M., et al. 2003. Multilocus sequence typing has better discriminatory ability for typing Vibrio cholerae than does pulsed-field gel electrophoresis and provides a measure of phylogenetic relatedness. J. Clin. Microbiol. 41:2191–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lee J. H., et al. 2006. Multilocus sequence typing (MLST) analysis of Vibrio cholerae O1 El Tor isolates from Mozambique that harbour the classical CTX prophage. J. Med. Microbiol. 55:165–170 [DOI] [PubMed] [Google Scholar]
- 38. Lipp E. K., Huq A., Colwell R. R. 2002. Effects of global climate on infectious disease: the cholera model. Clin. Microbiol. Rev. 15:757–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lipp E. K., et al. 2003. Direct detection of Vibrio cholerae and ctxA in Peruvian coastal water and plankton by PCR. Appl. Environ. Microbiol. 69:3676–3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Z., DeSantis T. Z., Andersen G. L., Knight R. 2008. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res. 36:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lobitz B., et al. 2000. Climate and infectious disease: use of remote sensing for detection of Vibrio cholerae by indirect measurement. Proc. Natl. Acad. Sci. U. S. A. 97:1438–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Louis V. R., et al. 2003. Predictability of Vibrio cholerae in Chesapeake Bay. Appl. Environ. Microbiol. 69:2773–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. MacRae S. H., Clements T., Horan J. M. 1983. Non-O1 Vibrio cholerae gastroenteritis in New Hampshire: a case report. Am. J. Med. Technol. 49:2. [PubMed] [Google Scholar]
- 44. Mahoney J. C., Gerding M. J., Jones S. H., Whistler C. A. 2010. Comparison of the pathogenic potentials of environmental and clinical Vibrio parahaemolyticus strains indicates a role for temperature regulation in virulence. Appl. Environ. Microbiol. 76:7459–7465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maiti D., et al. 2006. Genetic organization of pre-CTX and CTX prophages in the genome of an environmental Vibrio cholerae non-O1, non-O139 strain. Microbiology 152:3633–3641 [DOI] [PubMed] [Google Scholar]
- 46. Martinez-Urtaza J., et al. 2008. Emergence of Asiatic Vibrio diseases in South America in phase with El Nino. Epidemiology 19:829–837 [DOI] [PubMed] [Google Scholar]
- 47. Massad G., Oliver J. D. 1987. New selective and differential medium for Vibrio cholerae and Vibrio vulnificus. Appl. Environ. Microbiol. 53:2262–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. McCarthy S. A., Khambaty F. M. 1994. International dissemination of epidemic Vibrio cholerae by cargo ship ballast and other nonpotable waters. Appl. Environ. Microbiol. 60:2597–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mekalanos J. J., Rubin E. J., Waldor M. K. 1997. Cholera: molecular basis for emergence and pathogenesis. FEMS Immunol. Med. Microbiol. 18:241–248 [DOI] [PubMed] [Google Scholar]
- 50. Miller V. L., Mekalanos J. J. 1984. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc. Natl. Acad. Sci. U. S. A. 81:3471–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. New England Regional Assessment Group 2001. Preparing for a changing climate: the potential consequences of climate variability and change. New England Regional Overview. University of New Hampshire, Durham, NH [Google Scholar]
- 52. O'Neill K. R., Jones S. H., Grimes D. J. 1992. Seasonal incidence of Vibrio vulnificus in the Great Bay Estuary of New Hampshire and Maine. Appl. Environ. Microbiol. 58:3257–3262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. O'Shea Y. A., et al. 2004. The Vibrio seventh pandemic island-II is a 26.9 kb genomic island present in Vibrio cholerae El Tor and O139 serogroup isolates that shows homology to a 43.4 kb genomic island in V. vulnificus. Microbiology 150:4053–4063 [DOI] [PubMed] [Google Scholar]
- 54. O'Shea Y. A., Reen F. J., Quirke M., Boyd E. F. 2004. Evolutionary genetic analysis of the emergence of epidemic Vibrio cholerae isolates on the basis of comparative nucleotide sequence analysis and multilocus virulence gene profiles. J. Clin. Microbiol. 42:4657–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Palit A., Batabyal P. 2010. Toxigenic Vibrio cholerae from environmental sources associated with the cholera outbreak after ‘AILA’ cyclone in West Bengal, India. Lett. Appl. Microbiol. 51:241–243 [DOI] [PubMed] [Google Scholar]
- 56. Panicker G. 2004. Multiplex PCR detection of clinical and environmental strains of Vibrio vulnificus in shellfish. Can. J. Microbiol. 50:911–922 [DOI] [PubMed] [Google Scholar]
- 57. Pascual M., Bouma M. J., Dobson A. P. 2002. Cholera and climate: revisiting the quantitative evidence. Microbes Infect. 4:237–245 [DOI] [PubMed] [Google Scholar]
- 58. Preheim S. P., et al. 2011. Metapopulation structure of Vibrionaceae among coastal marine invertebrates. Environ. Microbiol. 13:265–275 [DOI] [PubMed] [Google Scholar]
- 59. Radu S., et al. 1999. Molecular characterization of Vibrio cholerae O1 and non-O1 from human and environmental sources in Malaysia. Epidemiol. Infect. 123:225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sack D. A., Sack R. B., Nair G. B., Siddique A. K. 2004. Cholera. Lancet 363:223–233 [DOI] [PubMed] [Google Scholar]
- 61. Sawabe T., Kita-Tsukamoto K., Thompson F. L. 2007. Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 189:7932–7936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Singh D. V., et al. 2001. Molecular analysis of Vibrio cholerae O1, O139, non-O1, and non-O139 strains: clonal relationships between clinical and environmental isolates. Appl. Environ. Microbiol. 67:910–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 [DOI] [PubMed] [Google Scholar]
- 64. Thompson C. C., Thompson F. L., Vicente A. C. P. 2008. Identification of Vibrio cholerae and Vibrio mimicus by multilocus sequence analysis (MLSA). Int. J. Syst. Evol. Microbiol. 58:617–621 [DOI] [PubMed] [Google Scholar]
- 65. Thompson F. L., et al. 2005. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol. 71:5107–5115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tobin-D'Angelo M., et al. 2008. Severe diarrhea aaused by cholera toxin-producing Vibrio cholerae serogroup O75 infections acquired in the southeastern United States. Clin. Infect. Dis. 47:1035–1040 [DOI] [PubMed] [Google Scholar]
- 67. Warner E., Oliver J. D. 2007. Refined medium for direct isolation of Vibrio vulnificus from oyster tissue and seawater. Appl. Environ. Microbiol. 73:3098–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wibbenmeyer J. A., Provenzano D., Landry C. F., Klose K. E., Delcour A. H. 2002. Vibrio cholerae OmpU and OmpT porins are differentially affected by bile. Infect. Immun. 70:121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yan Y., et al. 2011. Extended MLST-based population genetics and phylogeny of Vibrio parahaemolyticus with high levels of recombination. Int. J. Food Microbiol. 145:106–112 [DOI] [PubMed] [Google Scholar]