Abstract
Investigation of viruses in the environment often requires the amplification of viral DNA before sequencing of viral metagenomes. In this study, two of the most widely used amplification methods, the linker amplified shotgun library (LASL) and multiple displacement amplification (MDA) methods, were applied to a sample from the seawater surface. Viral DNA was extracted from viruses concentrated by tangential flow filtration and amplified by these two methods. 454 pyrosequencing was used to read the metagenomic sequences from different libraries. The resulting taxonomic classifications of the viruses, their functional assignments, and assembly patterns differed substantially depending on the amplification method. Only double-stranded DNA viruses were retrieved from the LASL, whereas most sequences in the MDA library were from single-stranded DNA viruses, and double-stranded DNA viral sequences were minorities. Thus, the two amplification methods reveal different aspects of viral diversity.
INTRODUCTION
Viruses are the most numerous biological entities, existing in almost every ecosystem, including aquatic, terrestrial, and symbiotic environments, in which they outnumber bacteria by over 10 times (for a review, see reference 37). Viruses significantly influence their environments; for example, in the sea, they control the mortalities of the bacterial hosts (35). Because there are no genes common to all viruses to serve as molecular markers, it is impossible to take an overall view of viruses in a defined ecosystem by using PCR-based molecular methods (12). Metagenomic approaches have been adapted to overcome this limitation; viral metagenomics has become crucial to the understanding of the environmental ecology of viruses (12). Recent studies of viral metagenomes, known as viromes, have revealed an as-yet-unexplored diversity of viruses in environments as different as seawater, soil, and human feces (2–8, 10, 11, 13, 20, 25, 41). To conduct a metagenomic study, viruses are usually separated from bacteria by filtration and/or ultracentrifugation (22, 38) and their nucleic acids are extracted. Because viral genomes are smaller and relatively shorter than those of bacteria, the amount of DNA obtained from environmental samples is frequently insufficient to conduct analyses such as the construction of cloning libraries or 454 pyrosequencing. Thus, viral DNA must be amplified for most metagenomic studies (2–8, 10, 11, 13, 20, 25, 26, 41).
Breitbart and collaborators (8) have developed a method known as the linker amplified shotgun library (LASL) to amplify marine viral DNA. In the method, viral DNA is sheared into small fragments (several hundred base pairs), polished, and ligated with an adapter (linker). The adapter is ligated to the fragments using a single orientation due to the presence of an overhanging 3′ end and phosphates at the 5′ end of the oligonucleotides. Finally, the ligated DNA fragments are PCR amplified with a single primer specific to the adapter. Because the adapter can be ligated only to double-stranded DNA (dsDNA), this method is restricted to the amplification of dsDNA. The LASL method has been used in viral metagenomic studies conducted in various environments, such as the sea (3, 8), human feces (6), infant gut (5), marine sediment (4), blood (7), stromatolites and thrombolites (11), soil (13), hot springs (33), and fermented foods (26). It was also applied to investigate RNA viral diversity to amplify double-stranded cDNA obtained from the genomes of RNA viruses by reverse transcription (10).
Another technique, known as multiple displacement amplification (MDA), has been used to amplify viral DNA (2). The prominent feature of MDA is that random hexamers and phi29 DNA polymerase are used to amplify DNA isothermally. In addition to its use in various microbiology applications, such as single-cell DNA amplification (40), environmental DNA amplification (1), and microarray construction (9), MDA has also been used for the amplification of viromes from a variety of environments, such as surface seawater (2), an Antarctic lake (24), reclaimed water (30), human feces (28), and water ponds with various salinity levels (29). In a previous study, we demonstrated that MDA amplifies circular DNA more efficiently than linear DNA and that the genomes of single-stranded DNA (ssDNA) viruses could be selectively amplified in viral assemblages from soil samples using MDA (20).
The advantages and disadvantages of these two amplification methods were discussed in a recent paper (27). The LASL method is time-consuming and requires a relatively high initial DNA concentration, whereas MDA is easily performed and requires lower initial DNA concentrations (27). However, MDA has been associated with the production of biases and artifacts, such as the formation of chimeras (21), quantitative biases (1, 39, 40), and the preferential amplification of ssDNA viruses (20). Yet to our knowledge, there have been no studies comparing the results produced by the employment of these two methods in the same sample.
In this study, we amplified viral DNA from a single sample from the seawater surface using both the LASL and MDA methods. The amplified metagenomes were then pyrosequenced. Consequently, we compared the effects of the amplification methods and the diversities of ssDNA and dsDNA viruses from the same marine environment.
MATERIALS AND METHODS
Sampling and concentration.
A sample from the seawater surface was collected near a breakwater (35°35.15′N, 126°37.11′E) in November 2007. Samples were collected with aseptic water-collecting bottles, transferred to the lab within 2 h, and stored at 4°C. The sample was filtered through 0.22-μm-pore-size polyvinylidene difluoride (PVDF) Durapore membrane filters (Millipore, Billerica, MA). The filtrated seawater was concentrated using the Labscale tangential flow filter system (Millipore) equipped with a 100-kDa polyethersulfone tangential flow filter cartridge (Pellicon XL filter; Millipore) (20). Sixteen liters of seawater was concentrated to 50 ml. The concentrated seawater was filtered three times using a 0.22-μm syringe filter.
DNA extraction.
Before DNA extraction, DNase I (final concentration of 20 U/ml at 37°C for 30 min) was added to the filtrate to degrade exogenous DNA. DNA was purified from 10 ml of the concentrated viruses using proteinase K and phenol-chloroform/isoamyl alcohol as described previously (31).
Construction of the LASL.
The DNA was sheared into small fragments by sonication for 3 s on ice to prevent heat denaturation (32). The fragmented DNA was polished using T4 DNA polymerase (Takara, Shiga, Japan) and T4 alkaline phosphatase supplied with deoxynucleoside triphosphate (dNTP) at 37°C for 30 min. The NotI linker (adapter) (long strand, 5′-AATTCGCGGCCGCGTCGAC-3′; short strand, 5′-phosphate-GTCGACGCGGCCGCG-3′) was ligated to the sheared DNA fragments using T4 DNA ligase for 1.5 h. The ligated DNA was PCR amplified with a primer (5′-CGGCCGCGTCGAC-3′) specific to the linker. Taq DNA polymerase was used for the PCR amplification (Bioneer, Daejeon, South Korea). PCR comprised an initial denaturation at 94°C for 5 min; 30 amplifying cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 3 min; and a final extension at 72°C for 10 min. The DNA amplified in this way was designated LASL.
Construction of the MDA library.
DNA was amplified with the GenomiPhi V2 DNA amplification kit (GE Healthcare, Buckinghamshire, United Kingdom). First, 1 μl of DNA was mixed with 19 μl of the sample buffer of the kit and heated at 95°C for 3 min. Next, 18 μl of the reaction buffer of the kit and 2 μl phi29 DNA polymerase were added and incubated for 1.5 h at 30°C. This amplification method was designated MDAH. A modified MDA method (referred to as MDAX) that did not include the heat-denaturing step described above (but, instead, placing the tube in ice for 3 min as opposed to heating it at 95°C) was also used to amplify the viral DNA. DNAs amplified by MDAH and MDAX were digested with 5 U/μl S1 nuclease (Takara) in 1× buffer at 30°C for 1 h and purified by ethanol precipitation. Both MDAH and MDAX libraries were collectively designated MDA libraries.
454 pyrosequencing and preprocessing of metagenome sequences.
The DNA libraries produced (LASL, MDAH, and MDAX) were sequenced through 454 pyrosequencing by a commercial sequencing company (Macrogen, Seoul, South Korea). Briefly, DNA was randomly sheared in the case of MDAH and MDAX libraries but was not sheared in the case of LASL. DNA of the three libraries, ranging in size from 500 to 1,000 bp, was purified by agarose gel purification before pyrosequencing. A 1/16 fraction of a PicoTiterPlate device was used to sequence each sample with the Genome Sequencer FLX system (GS FLX chemistry; Roche, Mannheim, Germany). Duplicate sequences or those with poor quality were discarded using the QC filter (minimum average score of 21 and minimum read length of 65 bp) and 454 duplicate clustering workflows (sequence identity threshold of 0.96 and word length of 10) based on the CDHIT program (19) of the CAMERA 2.0 website (34). About 10% of the sequences were identified as duplicates. Adapter sequences were removed from LASL sequences: if the adapter was inserted into the middle of a read, the read was split at the adapter sequence and the larger part of the read was used. Those sequences shorter than 100 nucleotides (nt) and containing any ambiguous base N were also discarded.
Classification of metagenome sequences based on database comparison.
BLASTX analysis was performed against NCBI nonredundant protein (NR) sequences in the CAMERA databases (E value of 0.001; October 2010 version). The viral protein database (E value of 0.001; October 2010 version) in the CAMERA server, consisting of proteins from publicly available viral genomes, was also used to identify the viral sequences in the metagenomes. The NCBI taxonomy identifier number of the hit was used to classify the sequences. The NCBI viral taxonomic system is based on the taxonomy of the International Committee on Taxonomy of Viruses. The taxonomic information corresponding to the hit with the highest bit score was used.
Functional assignment of metagenome sequences.
Metagenome sequences were analyzed with the RAMMCAP (23) workflow available on the CAMERA server. In the workflow, open reading frames (ORFs) predicted from the metagenomes were compared to the COG (36), TigrFAM (15), and PFAM (14) protein family databases. A detailed description of the methods employed is provided in the workflow manual (January 2010 version), available on the CAMERA site.
Assembly of metagenome sequences.
Sequences were assembled with the meta-assembler in the CAMERA workflows (default parameters of the July 2010 version were used). Sequences are available at the MG-RAST repository under identifiers 4464802.3, 4464804.3, and 4464805.3.
RESULTS AND DISCUSSION
Pyrosequencing of viral metagenomes from seawater.
A total of 16,796, 15,410, and 17,412 reads were obtained in the LASL, MDAH, and MDAX libraries, respectively. Of these, a total of 10,166, 12,788, and 14,031 reads, in the same respective order, remained in the libraries following the preprocessing of the sequences (Table 1). The number of the discarded sequences was larger in the LASL than in the MDA library, because much shorter sequences were obtained from the LASL during the removal of adapters and preparation of the DNA for 454 pyrosequencing (average nucleotide lengths of 184 bp, 233 bp, and 246 bp for LASL, MDAH, and MDAX, respectively). Sequences with ambiguous bases were more abundant in the LASL (19% was removed) than in the other libraries (3% was removed). Most (70%) of the ambiguous bases were positioned nearby the terminus (within 20 bp) of the sequence, which implies that adapter ligation might be one of the reasons for the abundance of ambiguous sequences in LASL.
Table 1.
Description of sequences from each library and results of assembly from each metagenomic sequence
| Library | No. of sequences | Mean length (bp) | % of sequence assembled | No. of contigs | Length of the longest contig (bp) | No. of sequences per contig (all)a | No. of sequences per contig (first quarter)b |
|---|---|---|---|---|---|---|---|
| LASL | 10,166 | 184 | 3.4 | 55 | 790 | 7 | 12 |
| MDAH | 12,788 | 233 | 66.0 | 384 | 4,724 | 44 | 146 |
| MDAX | 15,410 | 246 | 73.2 | 484 | 4,215 | 47 | 151 |
Average number of sequences per contig, considering all contigs.
Average number of sequences per contig, considering only the first quarter of contigs when the contigs were sorted in descending order according to the number of sequences assembled in a contig.
TBLASTX comparison of each library.
The three libraries were compared by TBLASTX analysis, with an E value of 0.001 as a criterion. Of the LASL sequences, 4.5% and 1.9% matched the sequences from the MDAH and MDAX libraries, respectively, whereas 83.9% of the MDAH sequences were also found in the MDAX library. Although each library was amplified from the same environmental DNA, LASL and MDA libraries had very few common sequences, and most of the hits between the LASL and MDA libraries showed much higher E values (low similarities) than between MDAH and MDAX (see Fig. S1 in the supplemental material), indicating that significant bias was introduced during the amplification process, as discussed below.
Taxonomic classification of sequences.
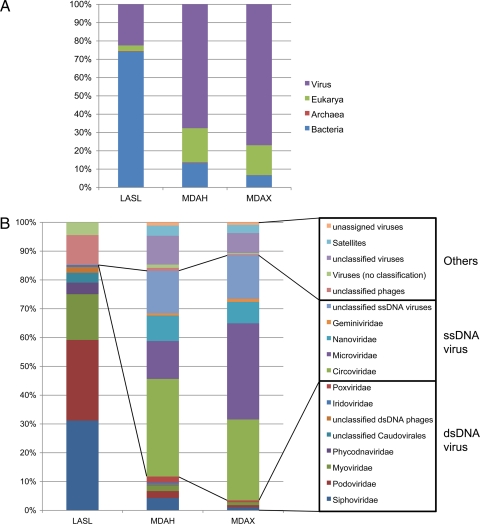
Respectively, 14.2%, 6.0%, and 9.3% of the LASL, MDAH, and MDAX sequences showed similarities with sequences in the NCBI nonredundant protein database, with an E value lower than 0.001. Among these known sequences, 74.2%, 13.5%, and 6.6%, respectively, were related to bacteria; 3.1%, 18.6%, and 16.3% to Eukarya; and 22.4%, 67.7%, and 76.9% to viruses (Fig. 1A). A large proportion of the sequences in the LASL showed homology to bacterial but not to viral sequences. Those sequences might have originated from contaminated bacterial DNA during the preparation of metagenomic libraries, but this phenomenon is more likely to be due to the lack of viral sequences or genomes in public databases and the inclusion of prophage sequences in bacterial genomes, as discussed in previous studies of viral metagenomics (6, 8), which means that the sequence databases need more genes and genomes from bacteria and viruses to investigate the viral metagenome.
Fig. 1.
Classification of metagenomic sequences from the three libraries based on biological groups (A) and viral genome types and viral families (B) from BLASTX analysis. Among total reads analyzed in the LASL, MDAH, and MDAX libraries, only 14.2%, 6.0%, and 9.3%, respectively, showed hits against the NCBI nonredundant protein database (A) and only 9.8%, 6.2%, and 9.8%, respectively, against the CAMERA viral protein database (B).
BLASTX analyses against the CAMERA viral protein database were performed to investigate viral diversity. A total of 9.8%, 6.2%, and 9.8% of the sequences in the LASL, MDAH, and MDAX libraries, respectively, matched a sequence from the viral database. Figure 1B depicts a classification of the sequences based on their viral types. The patterns were very different between the LASL and MDA libraries. In the former case, all classified sequences belonged to dsDNA viruses, including Siphoviridae (31.1%), Podoviridae (27.9%), Myoviridae (15.8%), Phycodnaviridae (4.0%), and Iridoviridae (0.7%). The former three families are bacteriophages, whereas the latter two represent groups of viruses that infect marine and freshwater eukaryotic algae (Phycodnaviridae) and invertebrates and vertebrate species (Iridoviridae). The viral groups comprising more than 0.7% of abundance are listed in Table S1 in the supplemental material. In the LASL, no sequence associated with ssDNA viruses was isolated, confirming the observation that only dsDNA can in principle be amplified by this method.
In the MDAH and MDAX libraries, most virus-related sequences were associated with ssDNA viruses, including members of the Microviridae (13.1% and 33.4%), Circoviridae (33.8% and 28.1%), Nanoviridae (8.8% and 7.4%), and Geminiviridae (0.8% and 1.1%) families. The first family represents a bacteriophage group, whereas the remaining families represent viruses that infect eukaryotes: Circoviridae viruses infect animals, and Nanoviridae and Geminiviridae viruses infect plants.
The dsDNA viruses shown in the LASL were also detected in the MDAH and MDAX libraries in relatively low numbers. Although dsDNA viruses also can be studied with the MDA method, the bias toward the amplification of circular genomes by this method resulted in the amplification of a much larger number of ssDNA than dsDNA viral sequences (85.9% and 96.1% ssDNA viruses among total dsDNA and ssDNA viruses in the MDAH and MDAX libraries, respectively). These are thought to be the highest ratios compared to other viral metagenome studies amplified by MDA. We calculated the ratios of ssDNA viruses among the total ssDNA and dsDNA viruses based on the previously determined sequences (2) and obtained percentages of 0.7%, 1.1%, 10.9%, and 25.0% for sequences from the Arctic Ocean, Gulf of Mexico, British Columbia, and Sargasso Sea, respectively. A high ratio (78.8%) was obtained in the case of the Antarctic lake in spring but not in summer (9.6%) (30). Those differences might have reflected various factors, such as a real abundance of ssDNA viruses in each environment, sampling sites, initial concentration of DNA, types of MDA kits, amplification time, and unknown factors. It is important to know that the ratio between ssDNA and dsDNA viruses in the viral metagenome amplified by MDA did not reflect the real ratio between them. The previous study of a rice paddy soil environment (20) reported that the proportion of ssDNA viral sequences in the unamplified viral metagenome was lower than those in the amplified viral metagenome by at least 2 to 3 orders of magnitude. Other methods, such as real-time PCR, unamplified metagenome library, or unbiased amplification methods are needed to quantitatively compare these two virus types. The small numbers of sequences (79 and 39, respectively) related to dsDNA viruses in the MDAH and MDAX libraries made it statistically unreliable to predict the composition between dsDNA viral family types. The prediction might be more reliable if a larger-scale sequencing effort had been performed.
Functional assignment of metagenome sequences.
The comparison of the LASL, MDAH, and MDAX metagenomic sequences against the COG, TigrFAM, and PFAM databases showed that the patterns of functional assignment were also quite different between the LASL and the MDA libraries (Tables 2 and 3; see also Tables S2 and S3 in the supplemental material). The majority of hits with the PFAM database in the MDA libraries were due to the sequences related to two viral protein families, a putative viral replication protein (PF02407) and a capsid protein F (PF02305), which originated from ssDNA viruses (Tables 3). The COG and the TigrFAM databases showed few hits in the MDA libraries, which means that the two databases did not detect the ssDNA viral genes, including the two major protein families in the PFAM databases. In all databases, less diverse kinds of protein families were detected in the MDA libraries than in the LASL (Table 2), because the majority of genes in the MDA libraries originated from ssDNA viruses that have small and simple genomes (17). The protein families detected in the LASL were not detected in the MDA libraries and vice versa (Table 3; see also Tables S2 and S3 in the supplemental material).
Table 2.
Results of metagenome analyses against the protein family databases COG, TigrFAM, and PFAM
| Database | No. of hits (no. of the kinds of protein families) |
||
|---|---|---|---|
| LASL | MDAH | MDAX | |
| COG | 283 (17) | 38 (9) | 34 (11) |
| TigrFAM | 170 (86) | 24 (22) | 28 (18) |
| PFAM | 205 (101) | 198 (25) | 446 (25) |
Table 3.
PFAM protein families detected most frequently in each library
| Parameter | Family identifier | Description | Result |
||
|---|---|---|---|---|---|
| LASL | MDAH | MDAX | |||
| Top 10 hits in the LASL (no. of hits) | PF04586 | Caudovirus prohead protease | 10 | 0 | 0 |
| PF01555 | DNA methylase | 8 | 2 | 0 | |
| PF03354 | Phage terminase | 7 | 1 | 1 | |
| PF05876 | Phage terminase large subunit (GpA) | 7 | 0 | 0 | |
| PF08291 | Peptidase M15 | 7 | 0 | 1 | |
| PF03796 | DnaB-like helicase C-terminal domain | 6 | 0 | 0 | |
| PF09588 | YqaJ viral recombinase family | 6 | 0 | 0 | |
| PF00476 | DNA polymerase family A | 5 | 1 | 0 | |
| PF00940 | DNA-dependent RNA polymerase | 5 | 0 | 0 | |
| PF09374 | Predicted peptidoglycan domain | 5 | 0 | 0 | |
| % of top 10 hits among total hits | 32.2 | 2.0 | 0.4 | ||
| Top 5 hits in the MDA library (no. of hits) | PF02407 | Putative viral replication protein | 0 | 110 | 160 |
| PF02305 | Capsid protein (F protein) | 0 | 47 | 227 | |
| PF06803 | Protein of unknown function (DUF1232) | 0 | 11 | 10 | |
| PF00910 | RNA helicase | 0 | 5 | 13 | |
| PF00799 | Geminivirus Rep catalytic domain | 0 | 3 | 14 | |
| % of top 5 hits among total hits | 0.0 | 88.9 | 95.1 | ||
| Total no. of hits for PFAM | 205 | 198 | 446 | ||
Assembly of metagenomes.
The assembly pattern is another means to show diversity. The higher the abundance of an organism in a community, the greater the possibility that its genomic sequences are assembled after random sequencing of the metagenome (8). The assembly patterns observed in the LASL and MDA libraries were very different. The proportions of sequences assembled into contigs in the MDA libraries were 20 times more than those in the LASL (Table 1). The average number of sequences per contig was also much higher in the MDA libraries than in the LASL. Few contigs were assembled with a large number of sequences (more than several hundred sequences per contig and a maximum of 1,646 and 895 sequences in the MDAH and MDAX libraries, respectively) in the MDA libraries, and the distribution was similar to the power law distribution (18). Such patterns might be explained by the observation that many copies of (viral) small circular genomes are generated during the amplification step by the rolling circle mechanism (16, 20) in the MDA method.
Comparison of the diversity from the amplified metagenome sequences.
We calculated diversity estimators based on the PHACCS analysis and rank-abundance distributions to compare the LASL and MDA libraries (for methods, see the supplemental material). Results from both methods showed that the dsDNA viral assemblages represented in the LASL were distributed more evenly and showed a higher number of rare groups than the ssDNA viral assemblages represented by the MDA libraries (see Fig. S2 and Tables S4 and S5 in the supplemental material). The comparison of estimated diversity between LASL and MDA libraries must be interpreted with caution, since different amplification mechanisms were employed to acquire the libraries. It is currently still not known whether the MDA method reflects the real composition of ssDNA viral assemblages. Quantitative amplification bias by MDA has been reported in the studies about amplification of single-cell genomes (1) or environmental genomes (40). A recent report has shown in more detail that MDA leads to quantitative bias in the comparison of bacterial communities based on small subunit rRNA (39).
The results of this study imply that two different methods, LASL and MDA, which can be used to amplify viral metagenomic DNA, yield substantially different results with respect to the types and ratios of viral sequences. Therefore, the selection of the amplification method must be carefully made, and the effect of each amplification method must be recognized before its use. Two methods could be used to study uninvestigated and uncultured viral diversity in ssDNA and/or dsDNA viruses. Further studies should be performed to estimate the relative abundance of ssDNA and dsDNA by using unamplified viral metagenomes or by using unbiased amplification methods.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Basic Science Research Program (grant no. 2010-0002571) of the National Research Foundation of Korea (NRF) and the 21C Frontier Microbial Genomics and Application Center Program funded by the Ministry of Education, Science, and Technology.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Abulencia C. B., et al. 2006. Environmental whole-genome amplification to access microbial populations in contaminated sediments. Appl. Environ. Microbiol. 72:3291–3301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Angly F. E., et al. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bench S. R., et al. 2007. Metagenomic characterization of Chesapeake Bay virioplankton. Appl. Environ. Microbiol. 73:7629–7641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Breitbart M., et al. 2004. Diversity and population structure of a near-shore marine-sediment viral community. Proc. Biol. Sci. 271:565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breitbart M., et al. 2008. Viral diversity and dynamics in an infant gut. Res. Microbiol. 159:367–373 [DOI] [PubMed] [Google Scholar]
- 6. Breitbart M., et al. 2003. Metagenomic analyses of an uncultured viral community from human feces. J. Bacteriol. 185:6220–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Breitbart M., Rohwer F. 2005. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques 39:729–736 [DOI] [PubMed] [Google Scholar]
- 8. Breitbart M., et al. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 99:14250–14255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chang H. W., et al. 2008. Development of microbial genome-probing microarrays using digital multiple displacement amplification of uncultivated microbial single cells. Environ. Sci. Technol. 42:6058–6064 [DOI] [PubMed] [Google Scholar]
- 10. Culley A. I., Lang A. S., Suttle C. A. 2006. Metagenomic analysis of coastal RNA virus communities. Science 312:1795–1798 [DOI] [PubMed] [Google Scholar]
- 11. Desnues C., et al. 2008. Biodiversity and biogeography of phages in modern stromatolites and thrombolites. Nature 452:340–343 [DOI] [PubMed] [Google Scholar]
- 12. Edwards R. A., Rohwer F. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3:504–510 [DOI] [PubMed] [Google Scholar]
- 13. Fierer N., et al. 2007. Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl. Environ. Microbiol. 73:7059–7066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Finn R. D., et al. 2006. Pfam: clans, Web tools and services. Nucleic Acids Res. 34:D247–D251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haft D. H., Selengut J. D., White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31:371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haible D., Kober S., Jeske H. 2006. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 135:9–16 [DOI] [PubMed] [Google Scholar]
- 17. Hino S. 2002. TTV, a new human virus with single stranded circular DNA genome. Rev. Med. Virol. 12:151–158 [DOI] [PubMed] [Google Scholar]
- 18. Hoffmann K. H., et al. 2007. Power law rank-abundance models for marine phage communities. FEMS Microbiol. Lett. 273:224–228 [DOI] [PubMed] [Google Scholar]
- 19. Huang Y., Niu B., Gao Y., Fu L., Li W. 2010. CD-HIT suite: a Web server for clustering and comparing biological sequences. Bioinformatics 26:680–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim K. H., et al. 2008. Amplification of uncultured single-stranded DNA viruses from rice paddy soil. Appl. Environ. Microbiol. 74:5975–5985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lasken R. S., Stockwell T. B. 2007. Mechanism of chimera formation during the multiple displacement amplification reaction. BMC Biotechnol. 7:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lawrence J. E., Steward G. F. 2010. Purification of viruses by centrifugation, p. 166–181 In Suttle C. A., Wilhelm S. W., Weinbauer M. G. (ed.), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX [Google Scholar]
- 23. Li W. 2009. Analysis and comparison of very large metagenomes with fast clustering and functional annotation. BMC Bioinformatics 10:359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lopez-Bueno A., et al. 2009. High diversity of the viral community from an Antarctic lake. Science 326:858–861 [DOI] [PubMed] [Google Scholar]
- 25. Ng T. F., et al. 2008. Discovery of a novel single-stranded DNA virus from a sea turtle fibropapilloma using viral metagenomics. J. Virol. 83:2500–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park E. J., et al. 2011. Metagenomic analysis of the viral communities in fermented foods. Appl. Environ. Microbiol. 77:1284–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Polson S. W., Wilhelm S. W., Wommack K. E. 2011. Unraveling the viral tapestry (from inside the capsid out). ISME J. 5:165–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reyes A., et al. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rodriguez-Brito B., et al. 2010. Viral and microbial community dynamics in four aquatic environments. ISME J. 4:739–751 [DOI] [PubMed] [Google Scholar]
- 30. Rosario K., Nilsson C., Lim Y. W., Ruan Y., Breitbart M. 2009. Metagenomic analysis of viruses in reclaimed water. Environ. Microbiol. 11:2806–2820 [DOI] [PubMed] [Google Scholar]
- 31. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 32. Sambrook J., Russell D. 2001. Appendix 8: commonly used techniques in molecular cloning. In Sambrook J., Russell D. (ed.), Molecular cloning, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 33. Schoenfeld T., et al. 2008. Assembly of viral metagenomes from Yellowstone hot springs. Appl. Environ. Microbiol. 74:4164–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seshadri R., Kravitz S. A., Smarr L., Gilna P., Frazier M. 2007. CAMERA: a community resource for metagenomics. PLoS Biol. 5:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suttle C. A. 2005. Viruses in the sea. Nature 437:356–361 [DOI] [PubMed] [Google Scholar]
- 36. Tatusov R. L., et al. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weinbauer M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127–181 [DOI] [PubMed] [Google Scholar]
- 38. Wommack K. E., Sime-Ngando T., Winget D. M., Jamindar S., Helton R. R. 2010. Filtration-based methods for the collection of viral concentrates from large water samples, p. 111–117 In Suttle C. A., Wilhelm S. W., Weinbauer M. G. (ed.), Manual of aquatic viral ecology. American Society of Limnology and Oceanography, Waco, TX [Google Scholar]
- 39. Yilmaz S., Allgaier M., Hugenholtz P. 2010. Multiple displacement amplification compromises quantitative analysis of metagenomes. Nat. Methods 7:943–944 [DOI] [PubMed] [Google Scholar]
- 40. Zhang K., et al. 2006. Sequencing genomes from single cells by polymerase cloning. Nat. Biotechnol. 24:680–686 [DOI] [PubMed] [Google Scholar]
- 41. Zhang T., et al. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4:e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.