Abstract
Members of the genus Flavobacterium occur in a variety of ecological niches and represent an interesting diversity of lifestyles. Flavobacterium branchiophilum is the main causative agent of bacterial gill disease, a severe condition affecting various cultured freshwater fish species worldwide, in particular salmonids in Canada and Japan. We report here the complete genome sequence of strain FL-15 isolated from a diseased sheatfish (Silurus glanis) in Hungary. The analysis of the F. branchiophilum genome revealed putative mechanisms of pathogenicity strikingly different from those of the other, closely related fish pathogen Flavobacterium psychrophilum, including the first cholera-like toxin in a non-Proteobacteria and a wealth of adhesins. The comparison with available genomes of other Flavobacterium species revealed a small genome size, large differences in chromosome organization, and fewer rRNA and tRNA genes, in line with its more fastidious growth. In addition, horizontal gene transfer shaped the evolution of F. branchiophilum, as evidenced by its virulence factors, genomic islands, and CRISPR (clustered regularly interspaced short palindromic repeats) systems. Further functional analysis should help in the understanding of host-pathogen interactions and in the development of rational diagnostic tools and control strategies in fish farms.
INTRODUCTION
The genus Flavobacterium, thus far encompassing 65 validly named species, is the type genus of the family Flavobacteriaceae, phylum Bacteroidetes (3). Representatives of the genus Flavobacterium have colonized a wide variety of temperate and polar habitats in terrestrial, freshwater, and marine environments. They are likely of high importance for the turnover/degradation of organic matter in these ecosystems (4). Hence, members of the genus Flavobacterium have recently acquired important ecological interest. In addition, some strains may have biotechnological applications owing to the production of cold-adapted enzymes (40) and to their potential for bioremediation (24) or wastewater treatment (44, 47). Although most members of the genus are environmental bacteria, three Flavobacterium species are serious fish pathogens that severely impact freshwater aquaculture worldwide (4).
Bacterial gill disease (BGD) is characterized by the presence of numerous bacteria on the surface of the gill epithelium that severely affect the respiratory function of infected fish. Although Flavobacterium psychrophilum and Flavobacterium columnare may cause gill necrosis (4), Flavobacterium branchiophilum (62) is actually the main causative agent of this condition. First recognized on salmonid fish in Japan and Oregon (28, 61), BGD caused by F. branchiophilum has been one of the most important conditions affecting the salmonid industry in Ontario, Canada, for 2 decades (43). The disease has also been reported on salmonid as well as nonsalmonid fish in Hungary and The Netherlands (12) and in South Korea (12, 29).
In contrast with F. psychrophilum and F. columnare, F. branchiophilum is a fastidious, nongliding organism with a unique tropism for the gill epithelium and is usually not isolated from internal organs. The disease is characterized by explosive morbidity and mortality attributable to massive bacterial colonization of gill lamellar surfaces, causing irritation and fusion of gill filaments and lamellae. The subsequent necrosis of the gills rapidly impairs the respiratory and osmoregulatory functions (53, 63). So far, no commercial vaccine is available, and the control of BGD by bath treatments using various chemotherapeutics has met with various degrees of success (51).
Only scarce information has been available so far on the pathogenesis of BGD and on the virulence mechanisms of F. branchiophilum, making it difficult to adopt preventive approaches to combat this pathogen. In order to get insight into the molecular determinants, with particular emphasis on pathogenicity, we determined and analyzed the complete genome sequence of F. branchiophilum FL-15 (CIP 109950), isolated in 1983 from a sheatfish (Silurus glanis) fingerling with BGD in a Hungarian fish farm (12). Strain FL-15 was compared to isolates from Japan and Oregon and included in the original description of F. branchiophilum (62).
Analysis of the whole genome of F. psychrophilum JIP02/86 has revealed sets of genes related to colonization, invasion, and destruction of the host tissues as well as particular metabolic properties related to stress response and long-term survival outside the host (9). In contrast, the genome of Flavobacterium johnsoniae UW101T has revealed unique sets of genes in relation to its environmental lifestyle, in particular, genes encoding polysaccharide utilization proteins, gliding motility, and novel biochemical features (35). By sequencing the whole genome of F. branchiophilum and performing comparative genomic studies, we aimed at understanding the adaptation of this poorly studied organism and the evolution of virulence in the genus Flavobacterium.
MATERIALS AND METHODS
F. branchiophilum genome sequencing.
To sequence the complete genome of strain FL-15, a shotgun sequencing strategy based on three different clone libraries and capillary Sanger sequencing was used to obtain a 14-fold coverage of the complete genome. The genomic DNA was fragmented by mechanical shearing. The 3-kb (library A) and 10-kb (library B) inserts were, respectively, cloned into the pcdna2.1 (Invitrogen) and pCNS (pSU18 derived) plasmid vectors while large inserts (40 kb; library C) were cloned into the fosmid vector pCC1Fos. Vector DNAs were purified and end sequenced (library A, 37,632 reads; library B, 13,056 reads; and library C, 5,568 reads) using dye terminator chemistry on ABI 3730 sequencers. The reads were assembled using the whole-genome shotgun assembler Arachne (http://www.broadinstitute.org), and the assembly was visualized by the interface Consed (CodonCode Corp., Dedham, MA). For the finishing phase, we used primer walking of clones, PCRs, and in vitro transposition technology (Template Generation System II Kit; Finnzyme, Espoo, Finland), corresponding to 108, 44, and 1,152 reads, respectively. All frameshift sequences were checked, and the assembly was validated by optical mapping (31).
Open reading frame (ORF) prediction and annotation.
The prediction of coding sequences was generated using the self-training gene detection software SHOW (38) based on hidden Markov models ([HMMs] http://genome.jouy.inra.fr/ssb/SHOW/). The ribosome-binding sites and transcriptional terminators were detected using the SHOW and Petrin software programs (7), respectively, while the tRNA- and rRNA-encoding genes were detected using the tRNA-scan (32) and RNammer (30) software programs, respectively. Genome annotation including manual validation was performed using the AGMIAL annotation platform (6). Insertion sequences (ISs) were identified using the procedure described in Touchon and Rocha (56) and annotated using IS finder (http://www-is.biotoul.fr/) (52).
ABC transporters and proteases were classified using the ABCISSE (http://www.pasteur.fr/recherche/unites/pmtg/abc/) and MEROPS (http://merops.sanger.ac.uk/) databases, respectively. Laterally transferred regions were predicted by Alien Hunter (http://www.sanger.ac.uk/resources/software/alien_hunter/) (58). Protein localization was predicted using PSORTb, version 3.0 (64). Putative CRISPR (clustered regularly interspaced short palindromic repeats) elements were identified using CRISPRFinder (17). The hidden Markov models for the 45 CRISPR-associated (Cas) protein families described in Haft et al. (19) were obtained from the TIGRFAM database, version 6.0 (http://www.tigr.org/TIGRFAMs/). The cas genes were identified with these Cas HMM profiles using hmmpfam (10) with the thresholds of an E value of <0.001 and a positive score. Blastn was used for similarity searches between CRISPR spacer sequences and the 834 complete prokaryote genomes, 1,725 complete plasmid genomes, and 522 virus genomes available in GenBank. Only matches showing an E value of <1 × 10−5 and less than 10% difference in sequence length were retained; matches to sequences found within CRISPR loci were ignored. Prophages were identified using PhageFinder (14). Genes with homology to the transduction-like gene transfer agent (GTA) described in McDaniel et al. (36) were identified using standard Blastp search.
Assignment of orthology.
Orthologs were defined by identifying unique pairwise reciprocal best hits, with at least 50% similarity in amino acid sequence and less than 20% difference in protein length. The analysis of orthology was made for every pair of Flavobacterium or Flavobacteriaceae genomes available in GenBank. The core genome consists of genes found in all genomes analyzed and was defined as the intersection of pairwise lists. Thus, the core genome of the genus Flavobacterium is around 1,400 genes. The core genome of the family Flavobacteriaceae is comprised of 595 genes detected in all 11 genomes.
Phylogenetic analyses.
The reference phylogenetic tree of the family Flavobacteriaceae was reconstructed from the concatenated alignments of 595 proteins of the core genome obtained with MUSCLE, version 3.6 (11), and then back-translated to DNA, as is standard usage. We used Tree-Puzzle (49) to compute the distance matrix between all genomes using maximum likelihood under the HKY+G(8)+I (Hasegawa-Kishino-Yano model of nucleotide substitution with gamma distribution allowing 8 categories and a proportion of invariant sites) model. The tree of the core genome was built from the distance matrix using BioNJ (16). We made 1,000 bootstrap experiments on the concatenated sequences to assess the robustness of the topology. The topology of this tree is congruent with previous phylogenetic analyses based on 16S rRNA (33).
Homologs to FBFL15_0919 were searched using Blastp (with an E value of <10−10) in the 249 metagenomes of the Integrated Microbial Genomes with Microbiome Samples (IMG/M) system (34). The molecular phylogeny of these putative enterotoxin proteins has been explored by the construction of multiple sequence alignments with MUSCLE, version 3.6 (11). The phylogenetic tree was reconstructed using the maximum-likelihood method implemented in the PhyML program (version 3.0, with approximate likelihood ratio test [aLRT]) with the WAG (Wheelan and Goldman) matrix and a gamma correction for variable evolutionary rates (18). Reliability for the internal branch was assessed using the aLRT (2).
Nucleotide sequence accession numbers.
The genomic sequences reported in this paper have been deposited in the EMBL database under the accession number FQ859183 for the bacterial chromosome and FQ859182 for the plasmid.
RESULTS AND DISCUSSION
General genome features.
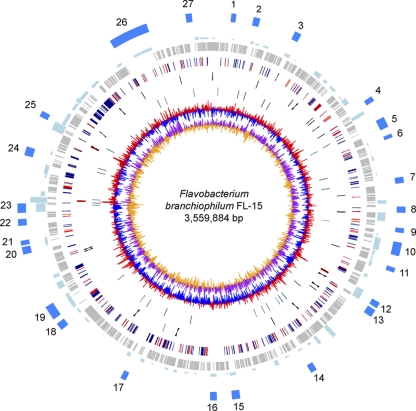
The complete genome of F. branchiophilum FL-15 consists of a circular chromosome of 3,559,884 bp (Fig. 1 and Table 1) and one small plasmid, pFB1, of 3,408 bp. The average G+C content is 33% for both the chromosome and the pFB1 plasmid. The chromosome is predicted to contain 2,867 protein-coding genes. We identified three rRNA operons and 44 tRNA genes. Therefore, F. branchiophilum contains one of the smallest subset of these genes among the sequenced genomes of the family Flavobacteriaceae. This is in accordance with the fastidious growth of F. branchiophilum strains as slow growers tend to have fewer such genes (59). The FL-15 genome encodes 36 insertion sequences, of which 8 appear incomplete (see Table S1 in the supplemental material). The pFB1 plasmid considerably differs from the pCP1 plasmid of F. psychrophilum JIP02/86 although they share the same size. The pFB1 plasmid is predicted to contain five genes encoding proteins including (i) a plasmid replication initiation protein, (ii) a mobilization protein similar to those previously identified on plasmids from different members of the phylum Bacteroidetes, and (iii) a toxin-antitoxin module.
Fig. 1.
Circular representation of the F. branchiophilum FL-15 genome. Circles represent the following (from the inside out): 1, GC skew [(G − C)/(G + C) using a 2-kbp sliding window] (purple, positive GC skew; orange, negative GC skew); 2, G+C deviation (difference between the average G+C content in a 2-kbp window and the genomic average G+C), where red areas indicate that the deviation is greater than 2 standard deviations; 3, location of ISs; 4, location of tRNA genes (black) and rRNA operons (red); 5, genes with orthologs in F. johnsoniae (blue) or F. psychrophilum (red); 6, core Flavobacterium genome, i.e., genes with orthologs in F. johnsoniae UW101T and F. psychrophilum JIP02/86 (gray); 7, putative horizontal gene transfer regions detected by Alien Hunter (58), where the height corresponds to the score prediction (the higher the score, the more significant the detected region); and 8, regions specific to F. branchiophilum FL-15 (blue). In the outermost circle, a specific region corresponds to at least 10 consecutive noncore genes and has less than 40% of the genes present in F. johnsoniae UW101T or F. psychrophilum JIP02/86. Characteristics of each region (defined by number) are indicated in Table S2 in the supplemental material.
Table 1.
Chromosome features of F. branchiophilum FL-15 compared with those of F. psychrophilum JIP02/86 and F. johnsoniae UW101T
| Chromosome feature | Value for the parameter in: |
||
|---|---|---|---|
| F. branchiophilum FL-15 | F. psychrophilum JIP02/86 | F. johnsoniae UW101T | |
| Genome size (bp) | 3,559,884 | 2,861,988 | 6,096,872 |
| Plasmid size (bp) | 3,408 | 3,407 | No |
| G+C content (%) | 33 | 32.5 | 34.1 |
| No. of rRNA operons | 3 | 6 | 6 |
| No. of tRNA genes | 44 | 49 | 62 |
| Total no. of protein-coding genes | 2,867 | 2,432 | 5,056 |
| Protein coding density (%) | 82.9 | 84.5 | 87.3 |
| Avg gene length (bp) | 1,030 | 1,003 | 1,061 |
| No. of complete IS elements (pseudogene[s]) | 28 (8) | 28 (21) | 16 (1) |
Genome comparison.
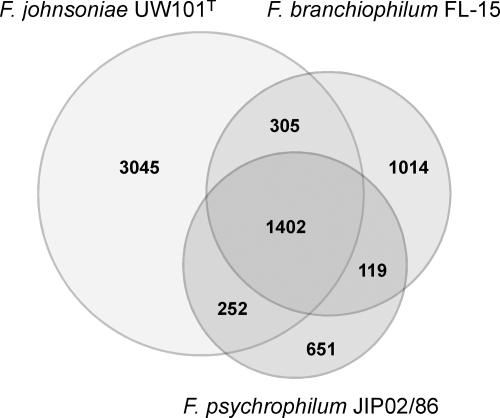
Pairwise reciprocal best hits were used to identify core protein-encoding genes, i.e., the genes shared by F. branchiophilum FL-15, F. psychrophilum JIP02/86, and F. johnsoniae UW101T. Using a threshold of 50% similarity in amino acid sequence and less than 20% of difference in protein length, 1,402 core genome genes were identified (Fig. 2). The bulk of the core proteins share 70% to 90% sequence similarity (see Fig. S1 in the supplemental material). This core genome of about 1,400 genes, about half of the F. branchiophilum FL-15 genome, is involved in central metabolism and transcription and translation machinery. Little conservation of the gene order was observed between the genomes (see Fig. S2 in the supplemental material) although the orthologs do tend to remain at similar relative positions on the chromosome. This observed X-plot shape likely derives from the accumulation of rearrangements that tend to be symmetrical relative to the replication origin (55), which appears to be conserved in the genus Flavobacterium. In contrast with F. johnsoniae and F. psychrophilum, we found weak GC skew deviations (Fig. 1), suggesting extensive genome shuffling in the lineage leading to F. branchiophilum. Chromosomal rearrangements may have resulted from homologous or other types of recombination between DNA repeats in the genome. Apart from the above-mentioned IS, we found a large number of Rhs (for rearrangement hot spot) elements in the genome. Rhs elements are complex genetic composites ubiquitous within the family Enterobacteriaceae (22). Although commonly found within accessory regions and thought to be implicated in genomic rearrangements in Escherichia coli, their functions are poorly understood (23). The FL-15 genome contains 65 rhs genes, of which 59 appear incomplete (gene remnants), and five cognate vgr genes, two of which are gene remnants. Most of the rhs and vgr genes are located in the predicted genomic islands (see below).
Fig. 2.
Venn diagram illustrating the overlap of gene repertoires of F. branchiophilum FL-15, F. johnsoniae UW101T, and F. psychrophilum JIP02/86. The core Flavobacterium genome contains 1,402 genes, i.e., ∼50% of the F. branchiophilum gene repertoire and ∼21% of the pan-Flavobacterium genome (6,788 genes).
We identified 1,014 protein-encoding genes in the F. branchiophilum FL-15 genome that are absent from the published F. psychrophilum and F. johnsoniae genomes. Of these, 42% are randomly distributed along the genome in small clusters of one to three genes. Taking into account the regions encompassing at least 10 noncore-genome genes, we identified 27 large genomic regions specific to F. branchiophilum FL-15 (Fig. 1; see also Table S2 in the supplemental material). These regions encompass 526 protein-encoding genes (52%) out of the 1,014. Regions 1, 10, 11, 18, and 20 contain genes likely involved in the detoxification of/resistance to various compounds, including heavy metals, antibiotics, and hydrogen peroxide; regions 15, 17, and 26 contain genes encoding retron-type reverse transcriptases suggesting a foreign origin; regions 8, 13, 22, and 27 contain genes encoding specific carbohydrate metabolism pathways likely involved in exopolysaccharide biosynthesis; region 16 contains phenylacetic acid degradation pathway-encoding genes; regions 5 and 24 contain sugar import- and metabolism-encoding genes; region 9 contains lipid metabolism-encoding genes; and regions 3, 19, and 26 are rich in rhs fragments (7, 11, and 33 fragments, respectively). Most of these genomic regions (60%) are predicted to be horizontally transferred by the Alien Hunter program, suggesting that they are indeed genomic islands. The FL-15 genome appears devoid of known prophages, gene transfer agents (GTAs), and integrative conjugative element (ICEs). Located in region 23, the FBFL15_2297 gene probably encodes an integrase, and the FBFL15_2295 and FBFL15_2299 genes probably encode excisionases; however, the neighboring genes are unrelated to prophage (i.e., phage structural phage genes) or ICE (i.e., conjugation-related genes).
CRISPR (clustered regularly interspaced short palindromic repeats) loci.
The CRISPR system is thought to be an adaptative hereditary immune system of prokaryotes that allows them to cope with foreign genetic elements (26). The FL-15 genome contains three CRISPR loci with 28, 31, and 39 direct repeats (DR), respectively. While the DR length is 36 bp in all cases, the sequence is different in each CRISPR locus. The third CRISPR locus is not canonical, with three interruptions in the modular pattern. Only the first CRISPR locus appears to be located near two known CRISPR-associated (cas) genes (FBFL15_1617 and FBFL15_1622 encode Cas2 and Csn1 proteins, respectively). However, this CRISPR Nmeni subtype system (which is complete in the genome of F. psychrophilum JIP02/86) seems incomplete in the FL-15 genome as the cas1 gene is absent, and the cas2 gene is rearranged (i.e., cas2 is downstream of the CRISPR locus and is no longer near csn1). Since cas1 is probably essential for the correct functioning of CRISPR, these results suggest that these CRISPR loci are no longer functional, at least in strain FL-15.
Using nonstringent criteria (Blastn E value of <1 × 10−5 and less than 10% difference in sequence length), no homologous sequence (i.e., proto-spacers) has been identified in any of the plasmid/phage/bacterial complete genomes available in GenBank. This is likely the consequence of the very low number of sequenced mobile genetic elements of members of the family Flavobacteriaceae available thus far (one phage and seven plasmids). The lack of CRISPR in F. johnsoniae UW101T and the presence of ISs in the direct vicinity of CRISPR in F. psychrophilum JIP02/86 suggest that CRISPR are poorly suited for the typing of Flavobacterium strains.
Toxins.
Strikingly, the genome of strain FL-15 is devoid of most of the predicted toxins previously identified in the F. psychrophilum JIP02/86 genome (9). Nevertheless, it contains a peculiar set of genes likely to be involved in virulence. The FBFL15_0919 gene probably encodes a preprotein 51% similar to the heat-labile toxin (LTA) expressed by enterotoxigenic Escherichia coli strains. The presence of a signal peptide and the conservation of the amino acid residues at the active site and at the NAD binding site support the functional homology between the FBFL15_0919 protein and LTA. LTA shows high sequence similarity (89%) with the cholera toxin (CTA), and both have been intensively studied as virulence factors in mammalian species (8). The LTA and CTA toxins function similarly by stimulating adenylate cyclase and provoking massive loss of water and electrolytes through the intestinal epithelium cells of the host (57). It is therefore tempting to speculate that FBFL15_0919 has a similar mode of action that disturbs the osmoregulatory function of the epithelial cells of the gills. Indeed, these cells are of utmost importance not only for the oxygen uptake of fish but also for the excretion of urea and for the active import of salts that compensate passive salt lost in freshwater (20). To our knowledge, it is the first time a gene similar to LTA and CTA has been detected outside the phylum Proteobacteria. The molecular phylogeny of these proteins argues against a direct lateral gene transfer between proteobacteria and strain FL-15 (see Fig. S3 in the supplemental material).
The FBFL15_0520 gene encodes a preprotein 47% similar to streptopain, an important streptococcal virulence factor playing a role in bacterial colonization, invasion, and inhibition of wound healing (25). In F. psychrophilum, this gene is absent from the genome of strain JIP02/86 (9) but present on a genomic island in the genome of strain THC02/90 (E. Duchaud, unpublished data).
F. branchiophilum has been reported to degrade gelatin and casein (62), and extracellular proteases of F. psychrophilum and Porphyromonas gingivalis (another pathogenic member of the phylum Bacteroidetes) have been suggested to play important roles in virulence (5, 41). Among the 11 putative secreted protease precursors identified in the FL-15 genome, two belong to the M1 family, one to the M36 fungalysin family, one to the M43 cytophagalysin family, one to the M12B family, and two to the S46 family. Together with five predicted peptide and amino acid importers, they might be involved in the breakdown and uptake of proteinaceous compounds during host tissue colonization.
Adhesion, motility, and secretion.
Bacterial attachment is essential in the initiation of mucosal infection, and previous reports have stressed the adherence properties of F. branchiophilum. Formalin-killed or acetone-killed bacterial cells retained part of their adherent nature, and adherence was never totally inhibited whatever the compound tested (42). In accordance with these findings, the FL-15 genome encodes 20 predicted adhesin precursors that could be implicated in cell-cell and cell-surface interactions.
Electron micrographs suggested that F. branchiophilum displays pili or fimbriae on the cell surface (43, 62), and pilus-like structures were indeed purified and partially characterized (21). However, no genes encoding known pilus or fimbrial proteins were identified in the FL-15 genome. If present, the corresponding genes may be hidden within those encoding hypothetical proteins; it is also possible that production of pilus-like structures is a strain-dependent feature absent from strain FL-15.
F. branchiophilum was reported to be devoid of gliding motility, the type of motility that occurs in many Flavobacterium species (3). Indeed, gliding motility of strain FL-15 was never observed in our hands. Two distinct groups of genes, gld and spr, are involved in gliding motility. The gld genes identified so far are thought to encode the gliding motor as they are mandatory for gliding motility. In addition, several spr genes encoding paralogous adhesins also have roles in motility (46). Intriguingly, the FL-15 genome contains all the gld genes and at least some of the spr genes. This suggests that F. branchiophilum may actually be motile, but experimental conditions used so far failed to mimic natural conditions where gliding motility is expressed.
A link between the Flavobacterium motility apparatus (i.e., Gld proteins) and a protein translocation system (referred to as the Por secretion system, or PorSS) has been described recently (48). The fact that the porP, porQ, porR, porS, and porT genes are present in the FL-15 genome suggests that F. branchiophilum is proficient in protein secretion through the PorSS. Moreover, 31 predicted secreted proteins, including proteases and adhesins, harbor a conserved C-terminal domain (CTD) (see Table S3 in the supplemental material). This CTD, also found in proteins of other members of the phylum Bacteroidetes (9, 27), is involved in their attachment to the outer membrane (50). Because the proteins that possess a CTD are likely secreted by the PorSS (48), they can be promising targets for vaccine development strategies.
Metabolism.
The elements of the central energy metabolism (glycolysis, tricarboxylic acid cycle, and oxidative phosphorylation) of F. branchiophilum are globally similar to those depicted in F. psychrophilum (9) and F. johnsoniae (35). Intriguingly, the F. branchiophilum genome encodes a class I fumarate hydratase (fumA) and an aconitate hydratase 2 (acnB), and the F. psychrophilum genome encodes a class II fumarate hydratase (fumC) and an aconitate hydratase 1 (acnA) while the F. johnsoniae genome encodes both type of enzymes.
In line with other Flavobacterium species, F. branchiophilum grows only by aerobic respiration and is unable to use fermentation or anaerobic respiration. Hence, the genes encoding menaquinone biosynthesis, cytochrome c, ATP synthase, and NADH dehydrogenases are all present. The genes actABCDEF and ccsBA encoding the recently described components of the alternative complex III menaquinol-cytochrome c oxidoreductase (45) and the components of system II cytochrome c synthetase and heme channel (15), respectively, are also present in the FL-15 genome.
The genomes of F. psychrophilum (9) and F. johnsoniae (35) both contain some amino acid catabolic pathway-encoding genes [i.e., glycine C-acetyltransferase (kbl), l-threonine 3-dehydrogenase (ltd), and phenylalanine 4-monooxygenase (phhA)] that are lacking in the FL-15 genome. Nevertheless, the latter contains a locus (absent in the genomes of F. psychrophilum and F. johnsoniae) of about 12 kb encompassing 10 genes (FBFL15_1502 to FBFL15_1511) similar to the paa gene cluster of some E. coli strains (13). These paa genes encode the only known aerobic degradation pathway for phenylacetate, which is the most common for phenylalanine (54). Moreover, this pathway occurs in various pathogens, and a connection with virulence through toxicity of reactive early intermediates was proposed (reference 54 and references therein).
Polysaccharide utilization.
Consistent with the strictly aerobic lifestyle of F. branchiophilum, its genome lacks phosphotransferase systems for sugar import. F. branchiophilum is able to hydrolyze starch and to produce acid from various carbohydrates under aerobic conditions (12, 43, 62). Indeed, analysis of the FL-15 genome identified 23 predicted glycoside hydrolases likely involved in carbohydrate catabolism, of which 17 contain a signal peptide (see Table S4 in the supplemental material). Among these, FBFL15_2407 is predicted to encode a levanase precursor, FBFL15_2433 encodes a cellulase precursor, and FBFL15_2455 encodes an arabinogalactan endo-1,4-β-galactosidase precursor. These genes are absent from the previously sequenced Flavobacterium genomes, but they were identified in members of the marine clade of the family Flavobacteriaceae.
Commonly found in members of the phylum Bacteroidetes, polysaccharide utilization involves outer membrane oligomer transport systems encoded by susC-like and susD-like genes, referred to as polysaccharide utilization loci (PULs). This capacity to degrade complex polysaccharides has been shown to be of high importance for marine members of the family Flavobacteriaceae (37). In the FL-15 genome, four gene clusters encoding predicted PULs were identified, one of which may not be functional due to a frameshift mutation in the susC-like gene. These loci encompass susC-like, susD-like, and other adjacent genes likely involved in polysaccharide utilization (i.e., encoding glycoside hydrolases, polysaccharide lyases, and carbohydrate esterases). While F. psychrophilum is devoid of such systems, F. johnsoniae appears to have an arsenal of PULs for the digestion of carbohydrates, including plant cell wall polysaccharides, which may reflect its prevalence in soil and rhizosphere habitats (35). It was therefore rather surprising to identify PULs in the FL-15 genome, especially the FBFL15_2659-2677 locus resembling the Fjoh_4246-4265 locus predicted to be involved in pectin utilization (35). This may suggest that F. branchiophilum is able to use some plant carbohydrates or gill mucus mucopolysaccharides.
Pigments.
In contrast with F. johnsoniae and F. psychrophilum, F. branchiophilum cells do not produce flexirubin-type pigments (62). Indeed, the F. branchiophilum genome is devoid of the flexirubin biosynthesis gene cluster identified in the F. johnsoniae and F. psychrophilum genomes. Nevertheless, it contains the crtIBZY gene cluster predicted to be involved in the biosynthesis of carotenoid-type pigments, suggesting that carotenoids are the only pigments responsible for the light yellow appearance of the colonies.
Bacterial stress genes.
Most bacteria have to face many stresses, among which are UV exposure, cold/heat, starvation, and toxic chemicals (including heavy metals and antibiotics). In addition, pathogenic bacteria are confronted with a host response that includes the oxidative stress during infection and transmission.
The FL-15 genome is predicted to encode 37 proteins likely involved in detoxification. Among these, five TerZ/TerD, one TerC, and one TelA family proteins are predicted to be involved in tellurium resistance, two arsenate reductases and an arsenite efflux transporter are involved in resistance to arsenic compounds, two proteins (CopA and CopZ) are involved in resistance to copper compounds, four superoxide dismutases are involved in the elimination of superoxide radicals produced by the host oxidative burst during infection, and one DNA alkylation repair enzyme, AlkD, is involved in the repair of 7-methylguanine alkylation damage on DNA. Genes predicted to be involved in antibiotic resistance were also identified in the FL-15 genome, including one chloramphenicol acetyltransferase, two bleomycin resistance proteins, and two acetyltransferases highly similar to Vat, a staphylococcal protein inactivating the A-type compounds of virginiamycin-type antibiotics (1). Therefore, F. branchiophilum FL-15 seems well equipped to face various kinds of stress. This gene repertoire could be of importance for the survival of the bacterium and might provide a selective advantage during host colonization against competitors in the environment.
Summary and future directions.
We reported here the complete genome sequence of F. branchiophilum, a serious pathogen of freshwater fish in many geographic areas. Comparison with the available genomes of other Flavobacterium species has revealed striking differences in chromosome organization and gene content. F. branchiophilum is phylogenetically more closely related to F. psychrophilum than to F. johnsoniae on the basis of 16S rRNA genes (33) and concatenated core genome proteins (see Fig. S4 in the supplemental material). However, its biochemistry resembles more that of F. johnsoniae. In addition, its toxins are unrelated to those of F. psychrophilum and were originally described in widely distant taxa. These elements point to very different paths in the evolution of virulence in F. branchiophilum and F. psychrophilum. Intense gene transfer is suggested by the large number of genomic islands and by the presence of one plasmid; together with the accumulation of genomic rearrangements, gene transfer could have an important role in the diversification of the species. Indeed, Flavobacterium species are highly prone to homologous recombination (39, 60) and represent the major recipients of natural gene transfer agents in the oceans (36).
This genome sequence therefore provides insights into the lifestyle of this understudied pathogen and should help in the development of rational diagnostic tools and more efficient control strategies in fish farms. In addition, it should yield molecular markers for the development of population structure analysis and epidemiological survey using, e.g., multilocus sequence typing (MLST)-based or variable-number tandem repeat (VNTR) genotyping-based strategies. The availability of this genome sequence should also facilitate the development of functional genomic studies. As new sequencing methods now provide high-throughput short reads, the genome of strain FL-15 may also serve as a reference, allowing read mapping and scaffolding of the genome sequence of other F. branchiophilum strains for a better understanding of intraspecies diversity.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by grant 07-GMGE from the Agence Nationale de la Recherche of France. P.B. is a Université Evry Val d'Essonne Ph.D. fellowship.
We are indebted to H. Wakabayashi for kindly providing strain FL-15. We also thank Stéphane Chaillou, Guillaume Achaz, and Pierre Nicolas for critical reading of the manuscript. We are grateful to the INRA MIGALE bioinformatics platform (http://migale.jouy.inra.fr) for providing computational resources.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Allignet J., el Solh N. 1995. Diversity among the gram-positive acetyltransferases inactivating streptogramin A and structurally related compounds and characterization of a new staphylococcal determinant, vatB. Antimicrob. Agents Chemother. 39:2027–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anisimova M., Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 3. Bernardet J.-F., Bowman J. P. 2011. Genus I. Flavobacterium, p. 112–154 In Whitman W.(ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 4 Williams and Wilkins, Baltimore, MD [Google Scholar]
- 4. Bernardet J.-F., Bowman J. P. 2006. The genus Flavobacterium, p. 481–531 In Dworkin M., Falkow S., Rosenberg E., Schleifer K. H., Stackebrandt E. (ed.), The prokaryotes, a handbook on the biology of bacteria, 3rd ed., vol. 7 Springer, New York, NY [Google Scholar]
- 5. Bertolini J. M., Wakabayashi H., Watral V. G., Whipple M. J., Rohovec J. S. 1994. Electrophoretic detection of proteases from selected strains of Flexibacter psychrophilus and assessment of their variability. J. Aquat. Anim. Health 6:224–233 [Google Scholar]
- 6. Bryson K., et al. 2006. AGMIAL: implementing an annotation strategy for prokaryote genomes as a distributed system. Nucleic Acids Res. 34:3533–3545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. d'Aubenton Carafa Y., Brody E., Thermes C. 1990. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol. 216:835–858 [DOI] [PubMed] [Google Scholar]
- 8. de Haan L., Hirst T. R. 2000. Cholera toxin and related enterotoxins: a cell biological and immunological perspective. J. Nat. Toxins 9:281–297 [PubMed] [Google Scholar]
- 9. Duchaud E., et al. 2007. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat. Biotechnol. 25:763–769 [DOI] [PubMed] [Google Scholar]
- 10. Eddy S. R. 1998. Profile hidden Markov models. Bioinformatics 14:755–763 [DOI] [PubMed] [Google Scholar]
- 11. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Farkas J. 1985. Filamentous Flavobacterium sp. isolated from fish with gill diseases in cold water. Aquaculture 44:1–10 [Google Scholar]
- 13. Ferrandez A., et al. 1998. Catabolism of phenylacetic acid in Escherichia coli. Characterization of a new aerobic hybrid pathway. J. Biol. Chem. 273:25974–25986 [DOI] [PubMed] [Google Scholar]
- 14. Fouts D. E. 2006. Phage Finder: automated identification and classification of prophage regions in complete bacterial genome sequences. Nucleic Acids Res. 34:5839–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frawley E. R., Kranz R. G. 2009. CcsBA is a cytochrome c synthetase that also functions in heme transport. Proc. Natl. Acad. Sci. U. S. A. 106:10201–10206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gascuel O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14:685–695 [DOI] [PubMed] [Google Scholar]
- 17. Grissa I., Vergnaud G., Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35:W52–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 19. Haft D. H., Selengut J., Mongodin E. F., Nelson K. E. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Helfman G. S., Collette B. B., Facey D. E., Bowen B. W. 2009. The diversity of fishes: biology, evolution, and ecology, 2nd ed Wiley-Blackwell, Oxford, United Kingdom [Google Scholar]
- 21. Heo G. J., Wakabayashi H., Watabe S. 1990. Purification and characterisation of pili from Flavobacterium branchiophila. Fish Pathol. 25:21–27 [Google Scholar]
- 22. Hill C. W., Sandt C. H., Vlazny D. A. 1994. Rhs elements of Escherichia coli: a family of genetic composites each encoding a large mosaic protein. Mol. Microbiol. 12:865–871 [DOI] [PubMed] [Google Scholar]
- 23. Jackson A. P., Thomas G. H., Parkhill J., Thomson N. R. 2009. Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genomics 10:584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jit S., Dadhwal M., Prakash O., Lal R. 2008. Flavobacterium lindanitolerans sp. nov., isolated from hexachlorocyclohexane-contaminated soil. Int. J. Syst. Evol. Microbiol. 58:1665–1669 [DOI] [PubMed] [Google Scholar]
- 25. Kapur V., et al. 1993. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb. Pathog. 15:327–346 [DOI] [PubMed] [Google Scholar]
- 26. Karginov F. V., Hannon G. J. 2010. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol. Cell 37:7–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Karlsson E. N., et al. 2004. The modular xylanase Xyn10A from Rhodothermus marinus is cell-attached, and its C-terminal domain has several putative homologues among cell-attached proteins within the phylum Bacteroidetes. FEMS Microbiol. Lett. 241:233–242 [DOI] [PubMed] [Google Scholar]
- 28. Kimura N., Wakabayashi H., Kudo S. 1978. Studies on bacterial gill disease in salmonids I. Selection of bacterium transmitting gill disease. Fish Pathol. 12:233–242 [Google Scholar]
- 29. Ko Y. M., Heo G. J. 1997. Characteristics of Flavobacterium branchiophilum isolated from rainbow trout in Korea. Fish Pathol. 32:97–102 [Google Scholar]
- 30. Lagesen K., et al. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35:3100–3108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Latreille P., et al. 2007. Optical mapping as a routine tool for bacterial genome sequence finishing. BMC Genomics 8:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lowe T. M., Eddy S. R. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Madhaiyan M., Poonguzhali S., Lee J. S., Lee K. C., Sundaram S. 2010. Flavobacterium glycines sp. nov., a facultative methylotroph isolated from the rhizosphere of soybean. Int. J. Syst. Evol. Microbiol. 60:2187–2192 [DOI] [PubMed] [Google Scholar]
- 34. Markowitz V. M., et al. 2008. IMG/M: a data management and analysis system for metagenomes. Nucleic Acids Res. 36:D534–D538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McBride M. J., et al. 2009. Novel features of the polysaccharide-digesting gliding bacterium Flavobacterium johnsoniae as revealed by genome sequence analysis. Appl. Environ. Microbiol. 75:6864–6875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. McDaniel L. D., et al. 2010. High frequency of horizontal gene transfer in the oceans. Science 330:50. [DOI] [PubMed] [Google Scholar]
- 37. Michel G., Nyval-Collen P., Barbeyron T., Czjzek M., Helbert W. 2006. Bioconversion of red seaweed galactans: a focus on bacterial agarases and carrageenases. Appl. Microbiol. Biotechnol. 71:23–33 [DOI] [PubMed] [Google Scholar]
- 38. Nicolas P., et al. 2002. Mining Bacillus subtilis chromosome heterogeneities using hidden Markov models. Nucleic Acids Res. 30:1418–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nicolas P., et al. 2008. Population structure of the fish-pathogenic bacterium Flavobacterium psychrophilum. Appl. Environ. Microbiol. 74:3702–3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nogi Y., Soda K., Oikawa T. 2005. Flavobacterium frigidimaris sp. nov., isolated from Antarctic seawater. Syst. Appl. Microbiol. 28:310–315 [DOI] [PubMed] [Google Scholar]
- 41. O'Brien-Simpson N. M., et al. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527–7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ostland V. E., Lumsden J. S., MacPhee D. D., Derksen J. A., Ferguson H. W. 1997. Inhibition of the attachment of Flavobacterium branchiophilum to the gills of rainbow trout, Oncorhynchus mykiss (Walbaum). J. Fish Dis. 20:109–117 [Google Scholar]
- 43. Ostland V. E., Lumsden J. S., MacPhee D. D., Ferguson H. W. 1994. Characteristics of Flavobacterium branchiophilum, the cause of salmonid bacterial gill disease in Ontario. J. Aquat. Anim. Health 6:13–26 [Google Scholar]
- 44. Park M., et al. 2006. Flavobacterium croceum sp. nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 56:2443–2447 [DOI] [PubMed] [Google Scholar]
- 45. Refojo P. N., Sousa F. L., Teixeira M., Pereira M. M. 2010. The alternative complex III: a different architecture using known building modules. Biochim. Biophys. Acta 1797:1869–1876 [DOI] [PubMed] [Google Scholar]
- 46. Rhodes R. G., Nelson S. S., Pochiraju S., McBride M. J. 2011. Flavobacterium johnsoniae sprB is part of an operon spanning the additional gliding motility genes sprC, sprD, and sprF. J. Bacteriol. 193:599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ryu S. H., et al. 2007. Flavobacterium filum sp. nov., isolated from a wastewater treatment plant in Korea. Int. J. Syst. Evol. Microbiol. 57:2026–2030 [DOI] [PubMed] [Google Scholar]
- 48. Sato K., et al. 2010. A protein secretion system linked to Bacteroidetes gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 107:276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schmidt H. A., Strimmer K., Vingron M., von Haeseler A. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502–504 [DOI] [PubMed] [Google Scholar]
- 50. Seers C. A., et al. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 188:6376–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shotts E. B., Jr., Starliper C. E. 1999. Flavobacterial diseases: columnaris disease, cold-water disease and bacterial gill disease, p. 559–576 In Woo P. T. K., Bruno D. W. (ed.), Fish diseases and disorders, vol. 3 CABI Publishing, Oxford, United Kingdom [Google Scholar]
- 52. Siguier P., Perochon J., Lestrade L., Mahillon J., Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34:D32–D36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Spear D. J., Ferguson H. W., Beamish F. W. M., Yager J. A., Yamashiro S. 1991. Pathology of bacterial gill disease: sequential development of lesions during natural outbreaks of disease. J. Fish Dis. 14:21–32 [Google Scholar]
- 54. Teufel R., et al. 2010. Bacterial phenylalanine and phenylacetate catabolic pathway revealed. Proc. Natl. Acad. Sci. U. S. A. 107:14390–14395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tillier E. R., Collins R. A. 2000. Genome rearrangement by replication-directed translocation. Nat. Genet. 26:195–197 [DOI] [PubMed] [Google Scholar]
- 56. Touchon M., Rocha E. P. 2007. Causes of insertion sequences abundance in prokaryotic genomes. Mol. Biol. Evol. 24:969–981 [DOI] [PubMed] [Google Scholar]
- 57. Vanden Broeck D., Horvath C., De Wolf M. J. 2007. Vibrio cholerae: cholera toxin. Int. J. Biochem. Cell Biol. 39:1771–1775 [DOI] [PubMed] [Google Scholar]
- 58. Vernikos G. S., Parkhill J. 2006. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics 22:2196–2203 [DOI] [PubMed] [Google Scholar]
- 59. Vieira-Silva S., Rocha E. P. 2010. The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet. 6:1000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vos M., Didelot X. 2009. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 3:199–208 [DOI] [PubMed] [Google Scholar]
- 61. Wakabayashi H., Egusa S., Fryer J. L. 1980. Characteristics of filamentous bacteria isolated from a gill disease of salmonids. Can. J. Fisheries Aquatic Sci. 37:1499–1507 [Google Scholar]
- 62. Wakabayashi H., Hun G. J., Kimura N. 1989. Flavobacterium branchiophila sp. nov., a causative agent of bacterial gill disease of freshwater fishes. Int. J. Syst. Bacteriol. 39:213–216 [Google Scholar]
- 63. Wakabayashi H., Iwado T. 1985. Effects of a bacterial gill disease on the respiratory functions of juvenile rainbow trout, p. 153–160 In Ellis A. E. (ed.), Fish and shellfish pathology. Academic Press, London, United Kingdom [Google Scholar]
- 64. Yu N. Y., et al. 2007. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.