Abstract
Growth is one of the basic attributes of any living organism. Surprisingly, the growth rates of marine bacterioplankton are only poorly known. Current data suggest that marine bacteria grow relatively slowly, having generation times of several days. However, some bacterial groups, such as the aerobic anoxygenic phototrophic (AAP) bacteria, have been shown to grow much faster. Two manipulation experiments, in which grazing, viruses, and resource competition were reduced, were conducted in the coastal Mediterranean Sea (Blanes Bay Microbial Observatory). The growth rates of AAP bacteria and of several important phylogenetic groups (the Bacteroidetes, the alphaproteobacterial groups Roseobacter and SAR11, and the Gammaproteobacteria group and its subgroups the Alteromonadaceae and the NOR5/OM60 clade) were calculated from changes in cell numbers in the manipulation treatments. In addition, we examined the role that top-down (mortality due to grazers and viruses) and bottom-up (resource availability) factors play in determining the growth rates of these groups. Manipulations resulted in an increase of the growth rates of all groups studied, but its extent differed largely among the individual treatments and among the different groups. Interestingly, higher growth rates were found for the AAP bacteria (up to 3.71 day−1) and for the Alteromonadaceae (up to 5.44 day−1), in spite of the fact that these bacterial groups represented only a very low percentage of the total prokaryotic community. In contrast, the SAR11 clade, which was the most abundant group, was the slower grower in all treatments. Our results show that, in general, the least abundant groups exhibited the highest rates, whereas the most abundant groups were those growing more slowly, indicating that some minor groups, such the AAP bacteria, very likely contribute much more to the recycling of organic matter in the ocean than what their abundances alone would predict.
INTRODUCTION
The structure of bacterioplankton communities is defined by the type of organisms and by their relative proportions. Marine surface waters are typically composed of a few abundant groups, generally members of the Alphaproteobacteria and Gammaproteobacteria and the phylum Bacteroidetes (17), and many low-abundant taxa (32). The sizes of the different populations depend to a large extent on their growth rates, which can range from organisms that are almost inactive or dormant to cells growing very rapidly (12). In addition to the differences in activity between individual cells (40), recent evidence suggests that variability among different bacterioplankton groups also occurs (42, 44, 45). Since actively growing bacteria are responsible for major carbon and nutrient transformations in the ocean, determining the growth rates of individual groups is critical to understand their ecological roles and specific contributions to marine biogeochemical cycles.
One of the groups that has been reported to grow at high rates in the ocean is the aerobic anoxygenic phototrophic (AAP) bacteria, which are photoheterotrophic organisms containing bacteriochlorophyll a (BChl a). These bacteria require organic substrates for their metabolism and growth but can derive a portion of their energy requirements using light, an ability that could provide an ecological advantage (28). Data from the Atlantic Ocean and the Baltic Sea show that AAP bacteria can grow at rates much higher than those of the total community (26, 27). In spite of their high growth rate, AAP cells typically account for less than 10% of the total bacterial abundance (9, 30, 38). A possible explanation for this paradox is the large cell size of AAP bacteria (25, 38), which may make them more vulnerable to flagellate attack (34). However, the role that grazing and other factors such as viral attack or resource availability plays in constraining the growth of this functional group as well as that of other bacterial groups remains largely unexplored. In fact, there are very few reports in which the growth rates of specific bacterial groups have been estimated (see references 22 and 39 for freshwaters and references 7, 42, 44, and 45 for the ocean). Furthermore, in most of these reports, growth rates were calculated only in either dilution or grazer-free experiments, a strategy that allows the estimation of the gross but not the net growth rates of the individual groups. In addition, reports from marine systems published to date have focused mainly on an understanding of the link between growth and resource availability, typically known as bottom-up control, and less attention has been given to studying top-down processes, i.e., the effect of grazing and viral lysis.
In previous studies, we established that the microbial community structure at the Blanes Bay Microbial Observatory (BBMO) (Northwest Mediterranean) is dominated by the Alphaproteobacteria, particularly by the SAR11 clade; the Gammaproteobacteria; and the Bacteroidetes (1). Bulk community growth rates at this coastal site, based on the [3H]leucine incorporation method, are on average low (6-year monthly growth rate of 0.17 ± 0.05 day−1 [average ± standard error] [2; J. M. Gasol, unpublished data]), which is consistent with data from other reports of similar oligotrophic environments (13). In contrast, based on BChl a diel changes, the AAP community was found to grow at rates of 1.15 to 1.42 day−1 (E. Hojerová et al., unpublished results). To understand such differences, we designed manipulation experiments with Blanes Bay seawater and measured the net and gross growth rates of AAP bacteria compared to those of several phylogenetic groups of bacteria. Additionally, we examined the roles that top-down and bottom-up control processes play in constraining the growth of these bacterioplankton groups.
MATERIALS AND METHODS
Sample collection and basic data.
Samples were collected from the Blanes Bay Microbial Observatory (BBMO) (41°40′N, 2°48′E), which is a shallow (∼20-m) coastal site about 1 km offshore on the Mediterranean coast, approximately 70 km north of Barcelona, Spain. Two experiments were performed, starting on 9 June 2009 (experiment 1) and 7 July 2009 (experiment 2). Samples (∼50 liters) were sieved through a 200-μm mesh and transported to the laboratory within 2 h. Water temperature and salinity were measured in situ with a CTD (conductivity, temperature, and depth) probe, and light penetration was estimated by using a Secchi disk (36). Underwater profiles of photosynthetically active radiation (PAR) at the sampling site were measured with a multichannel filter radiometer (PUV-2500; Biospherical Instruments Inc.). The concentration of inorganic nutrients was determined spectrophotometrically by using an Alliance Evolution II autoanalyzer according to standard procedures (19). The chlorophyll a (Chl a) concentration was measured from acetone extracts by fluorometry, and the abundances of heterotrophic bacteria and photosynthetic phytoplankton were measured by flow cytometry as described elsewhere (1).
Experimental setup.
Seawater was subjected to four experimental treatments: (i) whole unfiltered seawater (control [CT]), (ii) seawater prefiltered with a 1-μm filter to remove large predators while maintaining most bacteria (predator reduced [PR]), (iii) a 1:4 dilution of whole seawater with 0.2-μm-filtered seawater to reduce both predators and resource competition among bacteria (diluted [DL]), and (iv) a 1:4 dilution of whole seawater with seawater filtered through a 30-kDa VivaFlow cartridge to reduce viruses, predators, and resource competition (virus reduced [VR]). The samples were subjected to these manipulations and distributed into 2-liter Nalgene bottles that were incubated in duplicate for 3 days in a large water bath (200 liters) with circulating seawater to maintain the temperature close to in situ conditions. The water bath was maintained under natural light conditions (15-h–9-h light-dark cycles), except for the exclusion of UV using two layers of an Ultraphan URUV Farblos filter and a net that reduced in situ light intensity to roughly mimic the light conditions of a water depth of 3 m, with the transparency measured in situ at the sampling time. PAR radiation was monitored with a radiometer placed inside the incubation water bath. Samples were collected regularly during 3 days for the enumeration of total prokaryotes and AAP bacteria, measurements of leucine incorporation, BChl a measurements, and fluorescence in situ hybridization (FISH).
Enumeration of total prokaryotes and AAP bacteria by epifluorescence microscopy.
Subsamples were collected daily, fixed with 2% formaldehyde, and filtered on a 0.2-μm polycarbonate filter. Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) and counted by using an Olympus BX51TF fluorescence microscope as described previously (30). Briefly, three fluorescence images were captured for each frame. First, total DAPI-stained bacteria were recorded in the blue part of the spectrum; Chl a autofluorescence was then recorded in the red part of the spectrum; and finally, both BChl a- and Chl a-containing organisms were recorded in the infrared part of the spectrum (>850 nm). For each sample, at least 8 frames (400 to 600 DAPI-stained cells) were recorded and analyzed semimanually with AnalySiS software (Soft Imaging Systems) to distinguish between heterotrophic bacteria, picocyanobacteria, and AAP bacteria. To obtain net AAP bacterial counts, the contribution of Chl a-containing organisms to the infrared image was subtracted.
Bacterial biomass production.
Bacterial biomass production was estimated by using the [3H]leucine incorporation method (24), modified as described previously by Smith and Azam (41). Leucine incorporation was measured twice a day by incubating three replicates plus one trichloroacetic acid-killed control for each treatment in the dark with [3H]leucine (40 nM final concentration) for 2 h at an in situ temperature. Activity was converted to bacterial production using the theoretical conversion factor of 1.55 kg C mol−1 Leu, which is similar to the average empirical conversion factor measured throughout a year in our study site (3).
Bacteriochlorophyll a turnover.
The measurement of bacteriochlorophyll a turnover allows a simple assessment of mortality rates (loss term) of AAP bacteria from diel changes of the BChl a concentration (26, 27). Changes in pigment concentrations in the incubation mixtures were monitored by using infrared kinetic fluorometry, as described previously (26), at least 4 times a day, usually starting about 1 h after sunrise and finishing before dusk. BChl a turnover was computed by single exponential decay fitting.
CARD-FISH.
For the determination of bacterial community composition, samples were fixed in paraformaldehyde (2% final concentration), and catalyzed reporter deposition FISH (CARD-FISH) was performed as described previously by Pernthaler et al. (33). Samples were hybridized with the following probes: a mixture of Eub338-I, -II, and -III (4, 10) for Eubacteria; CF319a (4) for Bacteroidetes; Gam42a (4) for Gammaproteobacteria; Alt1413 (14) for Alteromonadaceae; NOR5-730 (14) for NOR5/OM60; Ros537 (14) for Roseobacter; and SAR11-441R (31) for SAR11 (for details, see reference 1).
Calculation of specific growth rates.
Growth rates of the individual bacterial groups were calculated based on the time course measurements of cell abundances. Growth rates were calculated as μ = ln[(PΔt/B0) + 1]/Δt, where PΔt is the change in cell abundance after Δt, B0 is the cell abundance at time zero, and Δt is the difference between the final time and time zero (35). To minimize the potential effects of prolonged incubation, the growth rates presented here were calculated considering only the first 18 h of incubation in experiment 1 and the first 20 h in experiment 2. Additionally, we used bacterial production data derived from leucine incorporation to estimate bulk community growth rates, as described previously by Kirchman (23), using 12 fg C per cell as a conversion factor (18).
RESULTS
To estimate the effects of top-down and bottom-up controls on different bacterioplankton groups, we conducted two manipulation experiments in which the growth rates of individual groups were determined in a control treatment and when the pressure of grazers, resource competition, and viruses was largely reduced. The initial seawater sample for both experiments was collected from the Blanes Bay Microbial Observatory in the coastal Northwestern Mediterranean Sea. Most bacterial predators were removed by filtering the water through a 1-μm filter (PR treatment), resource competition was reduced by diluting the original sample 1:4 with seawater filtered with a 0.2-μm filter (DL treatment), and lysis by viruses was reduced by diluting the original sample 1:4 with virus-free seawater (VR treatment)(see Materials and Methods for details). Abundances of picoeukaryotes and Synechococcus were measured in the initial incubation mixtures to check whether the filtration and dilution treatments had reduced cell numbers as expected. Numbers of Synechococcus and picoeukaryotes were reduced about one-half in the PR treatment and about 25% in the DL and VR treatments, as theoretically expected (data not shown).
The physicochemical and biological parameters of the original seawater samples in the two experiments are listed in Table 1. The Chl a concentration, inorganic nutrient concentration, and bacterial and picophytoplankton abundances were higher in experiment 1 (June) than in experiment 2 (July). The level of bacterial heterotrophic production in the original seawater was over 2-fold higher in June than in July. Based on infrared epifluorescence microscopy counts, aerobic anoxygenic phototrophic (AAP) bacteria accounted for 5% of the total DAPI counts in experiment 1 and 7% in experiment 2. In terms of phylogenetic groups, both samples were dominated by the alphaproteobacterial group SAR11, followed by the Bacteroidetes and the Gammaproteobacteria. Roseobacter, the NOR5/OM60 clade, and Alteromonadaceae were found in low abundances. In general, the percentages of the different phylogenetic groups were similar between the two starting samples, except for the SAR11 group, which accounted for 45% of the total prokaryotic community in experiment 2, whereas it represented 33% in experiment 1 (Table 1).
Table 1.
Physicochemical and biological parameters of initial sample in both experiments
| Variable | Value for expt: |
|
|---|---|---|
| 1 | 2 | |
| Temp (°C) | 16.5 | 21 |
| Secchi disk (m) | 13 | 18 |
| PAR subsurface (μmol photons m−2 s−1) | 557 | 561 |
| Chlorophyll a (μg liter−1) | 0.3 | 0.18 |
| [PO4] (μM) | 0.069 | 0.038 |
| [NH4] (μM) | 0.478 | 0.385 |
| [NO2] (μM) | 0.022 | 0.001 |
| [NO3] (μM) | 0.113 | 0.086 |
| [Si] (μM) | 1.464 | 0.556 |
| Bacterial activity (pmol Leu h−1 liter−1) | 295 | 133 |
| Bacterial abundance (103 cells ml−1) | 1.140 | 765 |
| Synechococcus abundance (103 cells ml−1) | 29.0 | 18.8 |
| Picoeukaryote abundance (103 cells ml−1) | 4.74 | 1.49 |
| Avg abundance (%) ± SDa | ||
| Eubacteria | 87 ± 7 | 72 ± 5 |
| Bacteroidetes | 17 ± 3 | 12 ± 1 |
| Gammaproteobacteria | 12 ± 3 | 10 ± 1 |
| Alteromonadaceae | 2 ± 1 | 1 ± 1 |
| NOR5/OM60 | 3 ± 2 | 3 ± 1 |
| SAR11 | 33 ± 5 | 45 ± 5 |
| Roseobacter | 6 ± 2 | 4 ± 2 |
| AAP bacteria | 5 ± 1 | 7 ± 1 |
In situ contributions to the bacterial abundances of the different bacterioplankton groups are represented as percentages of DAPI-positive cells.
Effect of manipulations on abundances of total bacteria and AAP bacteria.
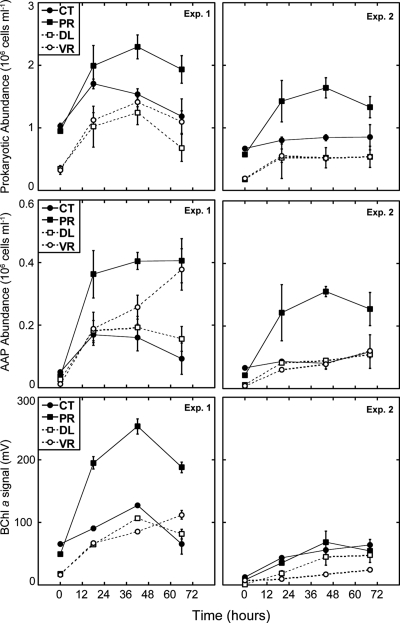
The manipulation of top-down and bottom-up factors caused more than a 2-fold increase in the total bacterial abundance during the first 2 days of incubation in experiment 1 and during the first day in experiment 2 (Fig. 1, top). There was also an increase in bacterial abundance in the controls, especially in experiment 1, but that increase was lower than that for the other treatments. Interestingly, the removal of predators led to a very rapid growth of AAP bacteria during the first day of incubation (Fig. 1, middle), and their relative abundances changed from the initial 5% to 18% in experiment 1 and from 7% to 15% in experiment 2. A reduction in resource competition (DL treatment) also promoted an increase in the percentages of AAP bacteria up to 15% and 17% in both experiments. For the VR treatment, the percentages of AAP bacteria increased up to 17% and 11% in each experiment, respectively. There was a minor increase in the abundance of AAP bacteria in the control treatment of experiment 1 and almost no change in experiment 2. In addition to absolute counts of AAP bacteria, we monitored BChl a signals throughout the experiments and found that, in general, the day-to-day accumulation of the pigment measured shortly after sunrise followed the same trends as those of AAP bacterial abundance (Fig. 1, bottom).
Fig. 1.
Changes in abundances of total prokaryotes (top), aerobic anoxygenic phototrophic (AAP) bacteria (middle), and bacteriochlorophyll a signals (lower) during incubations in the two experiments. CT, control; PR, predator reduced; DL, diluted; VR, virus reduced. Data shown represent averages ± standard deviations for two replicated bottles.
Effect of manipulations on bacterial biomass production.
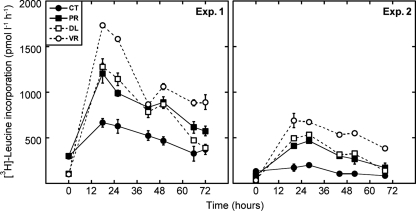
Despite the differences in the initial characteristics of the sample, similar changes in bacterial biomass production were observed in both experiments (Fig. 2). Both prefiltration and dilution treatments resulted in an immediate increase in leucine uptake rates within the first day of incubation. Diluting seawater had greater effects on bacterial production than did the removal of predators by filtration. In particular, the greatest increase was observed when samples were diluted with virus-depleted seawater. There was also a small increase of leucine uptake rates in the controls that could be explained by the “bottle effects” of the incubation (water in the controls was neither filtered nor treated by any means). After the increase on the first day, leucine incorporation rates decreased toward the end of the incubation in all treatments (Fig. 2).
Fig. 2.
Bacterial heterotrophic production measured as the rate of leucine incorporation during incubations in the two experiments. Data shown represent averages ± standard deviations for two replicated bottles.
Effect of manipulations on bacteriochlorophyll a turnover rates.
Diel changes in BChl a concentrations were used to calculate mortality rates for AAP bacteria as demonstrated in previous work (26, 27). The method is based on the fact that BChl a pigment synthesis in AAP bacteria is naturally inhibited by light (21, 26, 46), and therefore, the mortality of AAP bacteria results in a decrease of the BChl a concentration during daylight that is proportional to the rate of mortality for AAP bacteria. The validity of this method relies on two main assumptions: that pigment synthesis is fully inhibited by light and that pigment loss is due to cell mortality only (26). The incomplete inhibition of BChl a synthesis (i.e., due to low-light conditions) would result in underestimated mortality rates, whereas an unspecific pigment loss (such as photobleaching) would cause an overestimation of real values.
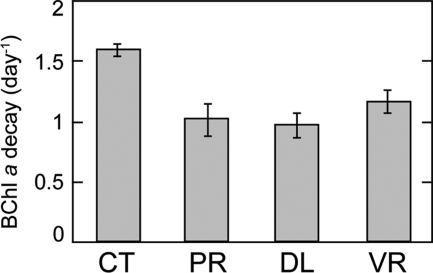
We estimated the mortality rates of AAP bacteria by measuring turnover rates in the control treatment. BChl a decay rates were measured during day 2 of experiment 1 (water samples were placed in the incubators on day 1 at around 4 p.m.). Unfortunately, we could not measure BChl a turnover rates in experiment 2 since BChl a synthesis was not fully inhibited, probably due to low-light conditions (overcast). The average PAR during the light hours of the 3-day incubation in experiment 1 was 600 μmol photons m−2 s−1, whereas during experiment 2, it was 360 μmol photons m−2 s−1. Furthermore, the variability in irradiance was much greater during experiment 2 because of alternating sunny skies and overcast and rainy weather conditions. The BChl a signal in the control treatment of experiment 1 decayed at a rate of 1.60 ± 0.04 day−1 (mean of 2 replicates ± standard deviation). The manipulation of top-down and bottom-up controls resulted in a reduction of the decay rate in all treatments (Fig. 3). The removal of grazers by filtration (PR treatment) reduced the rate of decay to 1.01 ± 0.18 day−1 (∼37% reduction), whereas the dilution treatments reduced the decay rates to 0.97 ± 0.18 day−1 (DL) and to 1.16 ± 0.18 day−1 (VR), which were approximately 39% and 27% reductions compared to the control rate, respectively.
Fig. 3.
Bacteriochlorophyll a turnover rate measured from diel changes of pigment concentrations in the different treatments of experiment 1. Error bars represent standard deviations for two replicated bottles.
Effect of manipulations on growth rates.
Growth rates measured for the control treatment are considered net growth rates, since predators and viruses were present in the sample and would represent in situ growth rates. Using changes in abundance over time in the control treatment, we estimated that the total prokaryotic community (as estimated from DAPI counts) grew at net rates of 0.66 ± 0.07 day−1 and 0.24 ± 0.03 day−1 in each experiment, respectively. The whole bacterial community (FISH-determined Eubacteria-positive [Eub+] cells) grew at about the same rate as the total prokaryotes (0.72 ± 0.05 and 0.44 ± 0.10 day−1), which is consistent with the fact that the percentage of cells hybridized with the Eubacteria probe was very high (87% ± 7% and 72% ± 5% of total DAPI counts in each experiment). In addition, we calculated the growth rate of the whole community using leucine incorporation rates and standard conversion factors. In experiment 1, the growth rate in the control experiment derived from bacterial production data was 1.82 day−1, and in experiment 2, it was 1.22 day−1, which are higher than the growth rates based on changes in cell abundance. However, using production data, growth estimates should be close to gross growth rates, since the loss is minimized by short sample incubation times (1 to 2 h), and therefore, the mortality that occurs on a longer temporal scale is not taken into account. Aerobic anoxygenic phototrophic bacteria displayed different growth rates between experiments: in June, they were growing at a rate of 1.62 ± 0.20 day−1 in the control treatment, whereas in July, their growth rate was 0.32 ± 0.09 day−1.
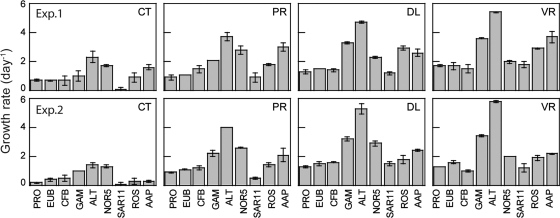
In addition to measurements of heterotrophic and AAP bacteria, we determined the growth rates of six distinct phylogenetic groups (Fig. 4, CT panels) that were identified by CARD-FISH. In general, the results were similar in the two experiments. Bacteroidetes grew at about the same rate as the total community (0.71 ± 0.32 day−1 in experiment 1 and 0.48 ± 0.20 day−1 in experiment 2). As expected, the two groups analyzed that belong to the Alphaproteobacteria subclass presented contrasting behaviors: Roseobacter grew much faster than SAR11, which displayed the lowest growth rates in both experiments (∼0.07 day−1). The maximal growth rates corresponded to the Gammaproteobacteria (1.05 ± 0.30 day−1 in experiment 1 and 1.01 ± 0.02 day−1 in experiment 2) and its subgroups. In particular, we examined the gammaproteobacterial NOR5/OM60 clade, which includes cultured AAP bacterium representatives of the Gammaproteobacteria (16) and the Alteromonadaceae, which can bloom under certain conditions and, thus, are expected to grow at high rates. Indeed, both NOR5/OM60 and Alteromonadaceae presented high growth rates (1.74 ± 0.07 day−1 and 2.35 ± 0.39 day−1 in experiment 1 and 1.28 ± 0.10 day−1 and 1.39 ± 0.21 day−1 in experiment 2, respectively).
Fig. 4.
Growth rates of individual groups derived from changes in abundance during the incubations in experiment 1 (top) and experiment 2 (bottom). Error bars represent the standard deviations for two replicated incubations. PRO, total prokaryotes as measured by DAPI staining; EUB, Eubacteria; AAP, aerobic anoxygenic phototrophic bacteria; GAM, Gammaproteobacteria; ALT, Alteromonadaceae; NOR5, NOR5/OM60 clade; SAR11, SAR11 clade; ROS, Roseobacter; CFB, Bacteroidetes.
Manipulation treatments resulted in increases in the growth rates of all groups studied; however, the extent of the increase differed largely among the individual treatments and among the different groups (Fig. 4, PR, DL, and VR panels). The growth rates of AAP bacteria increased substantially in all treatments compared to the controls and grew at maximum growth rates of 3.71 ± 0.38 day−1 in the VR treatment in experiment 1 and 2.38 ± 0.07 day−1 in the DL treatment in experiment 2 (Fig. 4). However, again, the Gammaproteobacteria and their subgroups were the faster growers in all cases. In particular, the Alteromonadaceae displayed the highest rates, growing at rates of up to 3.66 ± 0.28 day−1 (experiment 1) and 3.97 ± 0.03 day−1 (experiment 2) when predators were removed, 4.70 ± 0.08 and 5.32 ± 0.34 day−1 as a response to dilution, and 5.44 ± 0.05 and 5.81 ± 0.07 day−1 in the virus-depleted treatment.
DISCUSSION
Growth rates of marine bacterioplankton are routinely estimated from radiolabeled substrate uptake rates (23). In the last few decades, much has been learned about bacterial production and growth in the ocean (11, 13, 23), but most of those studies addressed bacterioplankton as a homogeneous assemblage, and only very few studies determined growth rates of individual bacterial groups. In this work, we measured the growth rates of aerobic anoxygenic phototrophic bacteria and of several important phylogenetic groups in the coastal Mediterranean Sea and used experimental manipulations to ascertain which are the key factors controlling their growth.
Our results demonstrate that some bacterioplankton groups such as the AAP bacteria and the Alteromonadaceae are capable of growing at much higher rates than others (Fig. 4). The rapid growth of AAP bacteria is consistent with previous observations that, based on bacteriochlorophyll a decay measurements, suggested that these organisms grow at high rates in the ocean (27). The estimated growth rates of AAP bacteria in our experiments with the BChl a decay method and with changes in cell abundance were within the same range. These results confirm that the decay approach is a valid and quick method to monitor in situ growth rates of AAP bacteria.
As expected from previous reports (14, 15, 44) (Table 2), the Gammaproteobacteria were also growing at high rates. Despite being present in very low abundances in the original sample, both individual gammaproteobacterial groups targeted, the NOR5/OM60 clade and the Alteromonadaceae, displayed high growth rates. The ability to grow at such rates may explain why, despite generally representing a small percentage of marine bacterioplankton, these groups can be abundant or even dominate microbial communities under certain conditions (1, 5, 43). In contrast, the Betaproteobacteria and Alphaproteobacteria are typically the fastest growers in freshwater lakes (22) and in estuaries (15, 45), respectively. We did not measure the growth rates of the Betaproteobacteria because they are not usually present in our study site (1), nor did we study those of the Alphaproteobacteria, since this broad phylogenetic group includes subclusters with very different lifestyles and ecological traits (17). We instead measured the growth rates of two relevant alphaproteobacterial subgroups, SAR11 and Roseobacter, which are present in significant numbers in diverse marine habitats (8, 31). As expected, they displayed different rates of growth: SAR11 grew at very low rates (∼0.07 day−1 in both experiments), in agreement with previous observations from other coastal systems (20, 42), whereas Roseobacter grew at higher rates (0.90 and 0.30 day−1). The Bacteroidetes grew at rates similar to those of the bulk community and within the same range as those of previous reports (Table 2). Interestingly, phylogenetic groups that include members of the AAP bacteria, i.e., Roseobacter and the NOR5/OM60 clade, displayed rates of growth comparable to those measured for the AAP bacteria, which may be an indication that the AAP bacterial community in our study site is composed of both gamma- and alphaproteobacterial phylotypes, as reported previously for other areas of the Mediterranean Sea (29).
Table 2.
Summary of minimal and maximal growth rates of specific bacterioplankton groups from marine systemsa
| Marine system | Treatment | Growth rate (day−1) for group |
Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PRO | Eubacteria | Bacteroidetes | Gammaproteobacteria | Alteromonadaceae | NOR5 | SAR11 | Roseobacter | Alphaproteobacteria | AAP bacteria | |||
| NW Mediterranean | Untreated | 0.2–0.7 | 0.4–0.7 | 0.5–0.7 | 1.0 | 1.4–2.3 | 1.3–1.7 | 0.1 | 0.3–0.9 | 0.3–1.6 | This work | |
| Filtration (1-μm filter) | 0.9 | 1.1 | 1.2–1.7 | 2.1–2.2 | 3.7–4 | 2.6–2.8 | 0.5–0.9 | 1.4–1.8 | 2.1–3.0 | This work | ||
| Dilution | 1.3 | 1.5 | 1.4–1.6 | 3.2–3.3 | 4.7–5.4 | 2.3–2.9 | 1.2–1.5 | 1.8–2.4 | 2.4–2.6 | This work | ||
| Dilution and virus reduction | 1.3–1.7 | 1.6–1.7 | 1.0–1.5 | 3.4–3.6 | 5.4–5.8 | 2.0 | 1.2–1.8 | 1.9–2.9 | 2.2–3.7 | This work | ||
| Baltic Sea | Untreated | 0.7–2.2b | 26 | |||||||||
| Atlantic Ocean | Untreated | 0.7–2.1b | 27 | |||||||||
| Ría de Vigo (NW Spain) | Dilution | 0.4–0.5 | 0.5–0.8 | 1.1–0.8 | 0.9–1.8 | 0.5 | 1–1.4 | 42 | ||||
| NW Pacific | Dilution | 0.3–3.2 | 0.3–4.2 | 0.2–3.1 | 44 | |||||||
| North Sea | Filtration (1.2-μm filter) | 0.5 | 1.0 | 0.5 | 1.1 | 1.0 | 14c | |||||
| Filtration (1.2-μm filter) + nutrients added | 0.6 | 1.1 | 0.9 | 1.8 | 0.5 | 14c | ||||||
| Dilution | 1.9 | 2.5–2.9 | 5c | |||||||||
| Delaware Bay | Dilution | 0.4–3.5 | 0.2–3.2 | 1.4–2.3 | 0.0–4.3 | 0–6.1 | 45 | |||||
| Plymouth Sound and English Channel | Dilution | 2.3 | 3.2–3.8 | 1.0–5.1 | 4.0–4.6 | 2.6–3.9 | 15c | |||||
PRO, total prokaryotes; NOR5, NOR5/OM60 clade; AAP bacteria, aerobic anoxygenic phototrophic bacteria.
Growth rate measured from bacteriochlorophyll a turnover rates.
Growth rates reported in these papers were calculated and reported previously by Yokokawa et al. (45).
Net growth rates are dependent on the balance between the gross growth rate, determined largely by nutrient availability and other physicochemical variables (bottom-up factors), and the mortality rate, which is dictated by top-down processes (e.g., grazing and viral lysis). In addition to measuring net growth rates in the control treatment, we measured growth rates in treatments in which grazing, viruses, and resource competition were reduced. By comparing these growth rates, we can estimate the effects of top-down and bottom-up processes. Our estimations, however, indicate their relative effect, since the measurement of the net effect of top-down and bottom-up factors experimentally suffers from important methodological limitations. For example, in the predator-reduced treatment, grazing pressure was relieved by filtration, but the incomplete removal of predators, the removal of particle-attached bacteria, or carbon enrichment due to cell lysis during filtration may affect estimates of the net effect of predators. Moreover, by dilution, we decreased the competition for the available resources due to the reduction in bacterial cell numbers, but at the same time, we reduced bacterial grazing mortality by decreasing the encounter frequency between predator and prey. The extent of the reduction of grazing pressure was, however, different than that in the predator-reduced treatment. Moreover, in the virus-reduced treatment, we removed a high percentage of lytic viruses, but lysogenic viruses may still have an impact on the growth of the different bacterioplankton groups. Despite these methodological limitations, we could generate an estimation of the relative effect of grazing pressure by comparing the growth rates of the predator-reduced and the control treatments. The effect of resource availability was estimated by the difference between the dilution and predator-reduced treatment, and finally, by comparing the virus-depleted treatment with the diluted treatment, we estimated the mortality induced by viral lysis.
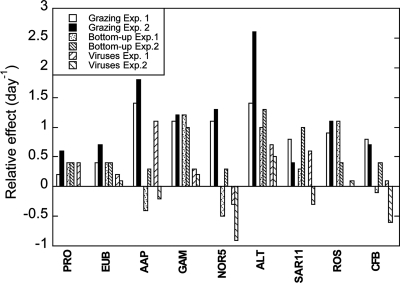
In general, we found that reductions of mortality and of resource competition had different effects on the growth rates of the individual bacterioplankton groups (Fig. 5). Differences between experiments may be explained by the different initial conditions, since organism abundances and nutrient availability were different (Table 1). In particular, growth rates of the total prokaryotic (DAPI counts) and bacterial (FISH-determined Eub+ cells) community increased when reducing mortality and competition for resources, which indicates that both top-down and bottom-up factors interact in controlling the community. However, we found some differences in the effects of grazing and viruses between experiments. Whereas in June, both viruses and grazers seemed to have similar effects on the growth of prokaryotic and bacterial communities, in July, the impact of grazers was stronger than that of viruses (Fig. 5). A 2-year study of bacterioplankton mortality in Blanes Bay (6) found that, in general, protists are the main cause of mortality, but during some periods, viruses can equal protists as agents of mortality. Our observations confirm that both forms of top-down processes may be important in controlling the microbial community in our study site.
Fig. 5.
Relative effects of top-down and bottom-up controls on the growth rates of the different groups. Differences in growth rates (day−1) between control (CT) and predator-reduced (PR) treatments indicate the effects of grazing, those between diluted (DL) and predator-reduced treatments indicate bottom-up control, and those between virus-reduced (VR) and diluted treatments indicate the effect of viruses. See Materials and Methods for details.
In contrast, grazing had a much stronger effect than other factors on the growth of AAP bacteria, supporting the hypothesis that their large size would make them more vulnerable to flagellate attack than other bacteria (27). Likewise, Bacteroidetes and NOR5/OM60 appeared to be more affected by predation than by other factors. The Alteromonadaceae, which were reported previously to be subjected to higher grazing pressure than other groups (5), responded positively to the reduction of grazers but also to the reduction of viruses and to resource availability. The alphaproteobacterial groups Roseobacter and SAR11 displayed different responses between experiments. The growth rate of Roseobacter increased significantly in June, when we reduced grazers and resource competition, whereas in July, the increase was mostly when grazing was reduced. In both cases, mortality due to viruses did not seem to be key in controlling the growth of Roseobacter populations. In contrast, viruses played an important role in controlling SAR11 growth, at least in one experiment, although SAR11 growth also increased when grazing was reduced. Despite the fact that the reported growth rates for SAR11 tend to be low (e.g., see references 37 and 42), we found that their turnover rates can be in some cases up to 1.82 day−1, which is much higher than the values previously reported.
We found that growth rates did not correlate with abundance under any conditions. The AAP bacterial community was growing at average rates of 1.6 day−1 in spite of accounting only for 5 to 7% of the total prokaryote abundance (Table 1). Besides the AAP bacteria, Alteromonadaceae, NOR5/OM60, and Roseobacter, which were also present in low abundances, were growing at high rates. The Bacteroidetes, which on average constitute ca. 11% of the bacterial community at the study site (1), presented intermediate growth rates. The slower grower in all treatments was the SAR11 group, which, however, dominates Mediterranean surface waters and seems to be the most abundant bacterial group in the world's oceans. Thus, the least abundant groups present were those that showed high growth rates, whereas contrarily, the most abundant groups showed lower growth rates. However, AAP bacteria and other fast-growing groups were subjected to high grazing pressure. This process might in part be responsible for the low population densities of these groups typically found in the marine environment.
In conclusion, our results indicate that some minor groups, such as the AAP bacteria, among others, are actively growing and therefore may play a much more important role in the recycling of organic matter in the ocean than what their abundances alone would predict. In oligotrophic systems, however, bacteria seem to maximize carbon utilization rather than bacterial growth efficiency (BGE) (11), and therefore, slow growers may also have a significant impact on marine biogeochemical cycles by having a particularly small BGE. Nevertheless, very little is known about intrinsic BGEs of different bacterioplankton groups, and further studies would be necessary to determine their real contribution to carbon processing in the ocean.
ACKNOWLEDGMENTS
We thank Clara Cardelús for her help with sampling, Irene Forn for helping with CARD-FISH, and Martí Galí for providing irradiance data. We thank two anonymous reviewers for their useful criticism and suggestions for improvement of the manuscript.
This research was supported by grants SUMMER (CTM2008-03309/MAR), STORM (CTM2009-09352/MAR), and ECOBAF (CTM2010-10462-E), funded by the Spanish Ministry of Science and Innovation, and by Czech projects GAČR P501/10/0221, AV ČR M200200903, Algatech (CZ.1.05/2.1.00/03.0110), and Inst. Research Concept AV0Z50200510. I.F. and M.S. were supported by Juan de la Cierva awards. M.K.'s stay in Barcelona was supported by a PIV fellowship from the Generalitat de Catalunya.
Footnotes
Published ahead of print on 1 July 2011.
REFERENCES
- 1. Alonso-Sáez L., et al. 2007. Seasonality in bacterial diversity in north-west Mediterranean coastal waters: assessment through clone libraries, fingerprinting and FISH. FEMS Microbiol. Ecol. 60:98–112 [DOI] [PubMed] [Google Scholar]
- 2. Alonso-Sáez L., et al. 2008. Factors controlling the year round variability in carbon flux through bacteria in a coastal marine system. Ecosystems 11:397–409 [Google Scholar]
- 3. Alonso-Sáez L., Pinhassi J., Pernthaler J., Gasol J. M. 2010. Leucine-to-carbon empirical conversion factor experiments: does bacterial community structure have an influence? Environ. Microbiol. 12:2988–2997 [DOI] [PubMed] [Google Scholar]
- 4. Amann R. I., et al. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beardsley C., Pernthaler J., Wosniok W., Amann R. 2003. Are readily culturable bacteria in coastal North Sea waters suppressed by selective grazing mortality? Appl. Environ. Microbiol. 69:2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boras J. A., Sala M. M., Vázquez-Domínguez E., Weinbauer M. G., Vaqué D. 2009. Annual changes of bacterial mortality due to viruses and protists in an oligotrophic coastal environment (NW Mediterranean). Environ. Microbiol. 11:1181–1193 [DOI] [PubMed] [Google Scholar]
- 7. Bouvier T., del Giorgio P. A. 2007. Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ. Microbiol. 9:287–297 [DOI] [PubMed] [Google Scholar]
- 8. Buchan A., González J. M., Moran M. A. 2005. Overview of the marine Roseobacter lineage. Appl. Environ. Microbiol. 71:5665–5677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cottrell M. T., Mannino A., Kirchman D. L. 2006. Aerobic anoxygenic phototrophic bacteria in the Mid-Atlantic Bight and the North Pacific Gyre. Appl. Environ. Microbiol. 72:557–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daims H., Bruhl A., Amann R., Schleifer K. H., Wagner M. 1999. The domain-specific probe EUB338 is insufficient for the detection of all bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434–444 [DOI] [PubMed] [Google Scholar]
- 11. del Giorgio P. A., Cole J. J. 1998. Bacterioplankton growth efficiency in aquatic systems Annu. Rev. Ecol. Syst. 29:503–541 [Google Scholar]
- 12. del Giorgio P. A., Gasol J. M. 2008. Physiological structure and single-cell activity in marine bacterioplankton, p. 243–298 In Kirchman D. L. (ed.), Microbial ecology of the oceans, 2nd ed John Wiley & Sons Inc., Hoboken, NJ [Google Scholar]
- 13. Ducklow H. W. 2000. Bacterial production and biomass in the oceans, p. 85–120 In Kirchman D. L. (ed.), Microbial ecology of the oceans, 2nd ed John Wiley & Sons Inc., Hoboken, NJ [Google Scholar]
- 14. Eilers H., Pernthaler J., Glöckner F. O., Amann R. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66:3044–3051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fuchs B. M., Zubkov M. V., Sahm K. M. V., Burkill P. H. K., Amann R. 2000. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometry and molecular biological techniques. Environ. Microbiol. 2:191–201 [DOI] [PubMed] [Google Scholar]
- 16. Fuchs B., et al. 2007. Characterization of a marine gammaproteobacterium capable of aerobic anoxygenic photosynthesis. Proc. Natl. Acad. Sci. U. S. A. 104:2891–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fuhrman J. A., Hagström A. 2008. Bacterial and archaeal community structure and its patterns, p. 45–90 In Kirchman D. L. (ed.), Microbial ecology of the oceans, 2nd ed John Wiley & Sons Inc., Hoboken, NJ [Google Scholar]
- 18. Fukuda R., Ogawa H., Nagata T., Koike I. 1998. Direct determination of carbon and nitrogen contents of natural bacterial assemblages in marine environments. Appl. Environ. Microbiol. 64:3352–3358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grasshoff K., Ehrhardt M., Kremling K. 1983. Methods on seawater analysis, 2nd ed Verlag Chemie, Weinheim, Germany [Google Scholar]
- 20. Hamasaki K., Taniguchi A., Tada Y., Long R. A., Azam F. 2007. Actively growing bacteria in the inland sea of Japan, identified by combined bromodeoxyuridine immunocapture and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 73:2787–2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iba K., Takami K. 1989. Action spectra for light-inhibition of bacteriochlorophyll and carotenoid accumulation during aerobic growth of photosynthetic bacteria. Plant Cell Physiol. 30:471–477 [Google Scholar]
- 22. Jürgens K., Pernthaler J., Schalla S., Amann R. 1999. Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl. Environ. Microbiol. 65:1241–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirchman D. 2001. Measuring bacterial biomass production and growth rates from leucine incorporation in natural aquatic environments, p. 227–237 In Paul J. H. (ed.), Methods in microbiology, vol. 30 Marine microbiology. Academic Press, San Diego, CA [Google Scholar]
- 24. Kirchman D. L., K'nees E., Hodson R. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic ecosystems. Appl. Environ. Microbiol. 49:599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koblížek M., Mlčoušková J., Kolber Z., Kopecký J. 2010. On the photosynthetic properties of marine bacterium COL2P belonging to Roseobacter clade. Arch. Microbiol. 192:41–49 [DOI] [PubMed] [Google Scholar]
- 26. Koblížek M., Stón-Egiert J., Sagan S., Kolber Z. 2005. Diel changes in bacteriochlorophyll a concentration suggest rapid bacterioplankton cycling in the Baltic Sea. FEMS Microbiol. Ecol. 51:353–361 [DOI] [PubMed] [Google Scholar]
- 27. Koblížek M., Mašín M., Ras J., Poulton A. J., Prášil O. 2007. Rapid growth rates of aerobic anoxygenic prototrophs in the ocean. Environ. Microbiol. 9:2401–2406 [DOI] [PubMed] [Google Scholar]
- 28. Koblížek M. 2011. Role of photoheterotrophic bacteria in the marine carbon cycle, p. 49–51 In Jiao N., Azam F., Sanders S. (ed.), Microbial carbon pump in the ocean. AAAS, Washington, DC [Google Scholar]
- 29. Lehours A. C., Cottrell M. T., Dahan O., Kirchman D. L., Jeanthon C. 2010. Summer distribution and diversity of aerobic anoxygenic phototrophic bacteria in the Mediterranean Sea in relation to environmental variables. FEMS Microbiol. Ecol. 74:397–409 [DOI] [PubMed] [Google Scholar]
- 30. Mašín M., et al. 2006. Seasonal changes and diversity of aerobic anoxygenic phototrophs in the Baltic Sea. Aquat. Microb. Ecol. 45:247–254 [Google Scholar]
- 31. Morris R. M., et al. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810 [DOI] [PubMed] [Google Scholar]
- 32. Pedrós-Alió C. 2006. Marine microbial diversity: can it be determined? Trends Microbiol. 14:257–263 [DOI] [PubMed] [Google Scholar]
- 33. Pernthaler A., Pernthaler J., Amann R. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Micriobiol. 68:3094–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3:537–546 [DOI] [PubMed] [Google Scholar]
- 35. Peters F. 2002. Overcoming linearisation errors in calculating bacterial growth rates. Mar. Ecol. Prog. Ser. 245:305–308 [Google Scholar]
- 36. Preisendorfer R. W. 1986. Secchi disk science: visual optics of natural waters. Limnol. Oceanogr. 31:909–926 [Google Scholar]
- 37. Rappé M. S., Connon S. A., Vergin K. L., Giovannoni S. J. 2002. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418:630–633 [DOI] [PubMed] [Google Scholar]
- 38. Sieracki M. E., Gilg I. C., Thier E. C., Poulton N. J., Goericke R. 2006. Distribution of planktonic aerobic anoxygenic photoheterotrophic bacteria in the northwest Atlantic. Limnol. Oceanogr. 51:38–46 [Google Scholar]
- 39. Šimek K., et al. 2006. Maximum growth rates and possible life strategies of different bacterioplankton groups in relation to phosphorous availability in a freshwater reservoir. Environ. Microbiol. 8:1613–1624 [DOI] [PubMed] [Google Scholar]
- 40. Sintes E., Herndl G. J. 2006. Quantifying substrate uptake by individual cells of marine bacterioplankton by catalyzed reporter deposition fluorescence in situ hybridization combined with microautoradiography. Appl. Environ. Microbiol. 72:7022–7028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smith D. C., Azam F. 1992. A simple, economical method for measuring bacteria protein synthesis rates in seawater using 3H-leucine. Mar. Microb. Food Webs 6:107–114 [Google Scholar]
- 42. Teira E., Martínez-García S., Lønborg C., Álvarez-Salgado X. A. 2009. Growth rates of different phylogenetic bacterioplankton groups in a coastal upwelling system. Environ. Microbiol. Rep. 1:545–554 [DOI] [PubMed] [Google Scholar]
- 43. Yan S., et al. 2009. Biogeography and phylogeny of the NOR5/OM60 clade of Gammaproteobacteria. Syst. Appl. Microbiol. 32:124–139 [DOI] [PubMed] [Google Scholar]
- 44. Yokokawa T., Nagata T. 2005. Growth and grazing rates of phylogenetic groups of bacterioplankton in coastal marine environments. Appl. Environ. Microbiol. 71:6799–6807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yokokawa T., Nagata T., Cottrell M. T., Kirchman D. L. 2004. Growth rate of the major phylogenetic bacterial groups in the Delaware estuary. Limnol. Oceanogr. 49:1620–1629 [Google Scholar]
- 46. Yurkov V. V., van Gemerden H. 1993. Impact of light/dark regimen on growth rate, biomass formation and bacteriochlorophyll synthesis in Erythromicrobium hydrolyticum. Arch. Microbiol. 159:84–89 [Google Scholar]