Abstract
Cryptosporidiosis is one of the most common waterborne diseases reported worldwide. Outbreaks of this gastrointestinal disease, which is caused by the Cryptosporidium parasite, are often attributed to public swimming pools and municipal water supplies. Between the months of January and April in 2009, New South Wales, Australia, experienced the largest waterborne cryptosporidiosis outbreak reported in Australia to date. Through the course of the contamination event, 1,141 individuals became infected with Cryptosporidium. Health authorities in New South Wales indicated that public swimming pool use was a contributing factor in the outbreak. To identify the Cryptosporidium species responsible for the outbreak, fecal samples from infected patients were collected from hospitals and pathology companies throughout New South Wales for genetic analyses. Genetic characterization of Cryptosporidium oocysts from the fecal samples identified the anthroponotic Cryptosporidium hominis IbA10G2 subtype as the causative parasite. Equal proportions of infections were found in males and females, and an increased susceptibility was observed in the 0- to 4-year age group. Spatiotemporal analysis indicated that the outbreak was primarily confined to the densely populated coastal cities of Sydney and Newcastle.
INTRODUCTION
Emerging infectious diseases (EIDs) cause significant impacts on human and animal health. Protozoan parasites are the third most common cause of EIDs, with 10.7% of EID events over the last 64 years attributed to them (15). Protozoan parasites of the genus Cryptosporidium, which are responsible for the gastrointestinal illness cryptosporidiosis, are one of the most important disease-causing agents in humans. Although 22 Cryptosporidium species have been described, over 90% of all reported human cryptosporidiosis infections are attributed to only two species, the anthroponotic C. hominis and the zoonotic C. parvum (18). Cryptosporidium meleagridis, C. canis, C. felis, C. suis, C. muris, C. fayeri, C. ubiquitum, and C. cuniculus are also considered human pathogens (5, 9, 22, 26). Cryptosporidium is transmitted between hosts via the oocyst, an environmentally robust endogenous life cycle stage that is resistant to the disinfectants used in water treatment. The oocyst is excreted in a fully infective form, and transmission is completed through the fecal-oral route. Chemical resistance and immediate infectivity coupled with a low infective dose make Cryptosporidium a significant threat to human health.
Waterborne disease transmission is the most common pathway for the spread of cryptosporidiosis (6). The majority of waterborne outbreaks have been attributed to both C. hominis and C. parvum, but a recent outbreak in Northamptonshire, England, was caused by C. cuniculus (4, 28). Species identification requires molecular tools due to morphological similarities exhibited within the Cryptosporidium genus. Numerous genetic markers are used for species-specific Cryptosporidium identification. However, the gp60 locus, which encodes glycoprotein 60, a highly variable surface antigen, is preferentially utilized in epidemiological investigations. The extensive sequence variation in gp60 enables grouping of Cryptosporidium into subtype families. For C. hominis, 6 subtype families have been identified, and for C. parvum, there are 11 subtype families (28). Analysis of a microsatellite region that encodes serine repeats is used to further characterize C. hominis and C. parvum to what is termed the subtype level.
Differences in virulence and clinical manifestations have been observed between Cryptosporidium species and the subtype families. Cryptosporidium hominis infections, which are commonly associated with nonintestinal sequelae, are more virulent than those from C. parvum (12). All C. hominis infections cause diarrhea; however, subtype family Ib is the most virulent and is associated with nausea, vomiting, and general malaise (3). The number of waterborne outbreaks caused by C. hominis Ib, particularly the IbA10G2 subtype, support the high virulence of this subtype family (28).
Despite good hygiene, sanitary living conditions, safe food and water supplies, and access to medicine (immunizations and antibiotics), Australia's east coast has been identified as an EID hot spot (15). A myriad of bacterial, viral, and protozoal infectious agents have contributed to this hot spot status. New South Wales, Australia, has experienced six waterborne outbreaks of cryptosporidiosis, all of which have been attributed to public swimming pool use (2, 16, 17, 21, 23). Cryptosporidiosis has been a notifiable disease in New South Wales since 1996 (19), and the data show that infections are increasing, particularly during the warmer months from November to March (http://www.health.nsw.gov.au/data/diseases/cryptosporidiosis.asp). Between January and April 2009, 1,141 cryptosporidiosis cases were reported to New South Wales Health Communicable Disease Branch, representing a 313% increase in incidence compared to the same period the previous year. Contaminated public swimming pools were identified as the source of the outbreak, and up to 19 pools were hyperchlorinated as a result (Jeremy McAnulty New South Wales Health, personal communication). The aim of this research was to undertake a genetic analysis to identify the Cryptosporidium species and subtypes responsible for the 2009 outbreak and to relate this to patient demographics. A spatially based approach was included in the study to investigate the geographical extent of the outbreak.
MATERIALS AND METHODS
Sample sources, parasite enumeration, and DNA extraction.
Five hundred eighty-nine fecal samples positive for Cryptosporidium were obtained from hospitals and pathology companies in New South Wales, Australia. Patient names were removed from samples to maintain privacy. All specimens were identified as Cryptosporidium positive by the pathology companies, using the Remel ProSpecT Giardia/Cryptosporidium microplate assay. Oocysts were purified from feces using a sucrose flotation gradient (25), and DNA was extracted using PrepGem (Zygem Corporation Ltd., Hamilton, New Zealand) (10). Oocysts were stained with the Cryptosporidium-specific antibody CRY104 labeled with fluorescein isothiocyanate (FITC; Biotech Frontiers, Sydney, Australia) and were enumerated by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences, Sydney, Australia) (1).
Identification of Cryptosporidium species by PCR-RFLP analysis.
Cryptosporidium species were identified by restriction fragment length polymorphism (RFLP) analysis of a diagnostic fragment of the 18S rRNA. The fragment was amplified using a previously described nested PCR (29). The PCR mixtures contained 6 mM MgCl2, 200 μM deoxynucleoside triphosphates (dNTPs), 200 nM each primer, and 1 U Red Hot Taq DNA polymerase (Thermo Scientific, Australia) with 2 μl of DNA template for the primary reaction and 1 μl of the primary PCR product for the secondary reaction. A total of 35 cycles, each consisting of 94°C for 45 s, 56°C for 45 s, and 72°C for 1 min, with an initial denaturation of 94°C for 3 min and a final extension step of 72°C for 7 min, were performed for both primary and secondary reactions. THe PCR controls included a negative sample containing PCR water only and a positive sample containing C. parvum DNA. Reactions were run on Eppendorf Mastercycler personal instruments (Eppendorf, North Ryde, Australia). The amplicons were resolved by electrophoresis on 2% (wt/vol) agarose gel containing Sybr Safe (Invitrogen, Mulgrave, Australia) and visualized under UV light. Secondary products of the correct size (832 to 835 bp depending on species) were purified using the QIAquick PCR purification kit (Qiagen, Melbourne, Australia) following the manufacturer's instructions.
RFLP analysis of the 18S rRNA amplicon was performed using a previously described protocol with the restriction enzyme VspI (10 units/μl; New England BioLabs) (29). Digested fragments were resolved on 3.5% (wt/vol) agarose gels at 100 V for 50 min. The RFLP patterns were visualized under UV light after prestaining with Sybr Safe according to the manufacturer's instructions.
Identification of Cryptosporidium subtype families and subtypes.
A previously described nested PCR targeting the hypervariable gp60 gene was performed to identify Cryptosporidium subtype families and subtypes (27). The PCR mixtures contained 4 mM MgCl2, 200 nM dNTPs, 200 nM each forward and reverse primer, and 1 U of Red Hot Taq polymerase with 2 μl of DNA template for the primary reaction and 1 μl of the primary PCR product for the secondary reaction. The reaction conditions comprised an initial denaturation at 94°C for 3 min followed by 35 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 1 min 30 s, with a final extension at 72°C for 7 min. The amplicons were separated on 2% (wt/vol) agarose gel containing Sybr safe and were visualized under UV light. Reaction mixtures containing the correct size fragment (∼1,000 bp) were purified as described above and sequenced using an ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, CA) with a BigDye Terminator kit (Applied Biosystems).
Nucleotide sequences were analyzed using Geneious version 4.8.2 (Biomatters Ltd., Auckland, New Zealand). Isolates were assigned a subtype according to the nomenclature system described by Sulaiman et al. (24).
Patient data and spatial analysis.
Patient information, such as age, gender, and residential postal code, was obtained for each sample from New South Wales Health. To examine the spatial distribution of the outbreak, the numbers of patients infected with the various Cryptosporidium subtypes were mapped. Analysis of the distribution of the outbreak over the 4-month period was achieved by using the pathology sample date. Samples were divided into their respective months and mapped with their corresponding postal code. Geographical mapping was completed using Esri ArcGIS version 10.0 (http://esriaustralia.com.au/esri/default.html) in conjunction with New South Wales digital postal boundary postcodes 2006 (http://www.abs.gov.au/AUSSTATS/abs@.nsf/DetailsPage/2923.0.30.0012006).
A chi-squared statistical analysis was performed on the relative numbers of males and females, as well as the various age groups, affected throughout the outbreak.
Nucleotide sequence accession numbers.
The gp60 sequences generated from this study were submitted to GenBank under accession numbers JF727750 to JF727762.
RESULTS
Cryptosporidium species and subtypes and parasite enumeration.
Positive results for PCR of the 18S rRNA were obtained for 521/589 (88%) isolates. RFLP analysis showed that 502/521 (96%) patients were infected with C. hominis and 19/521 (4%) were infected with C. parvum.
Amplification of gp60 was successful for 473/521 (91%) isolates typed by 18S rRNA (Table 1). Sequence analysis of the C. hominis isolates identified four subtype families, Ia (1/473), Ib (453/473), Id (2/473), and Ie (1/473). The remaining isolates were identified to C. parvum subtype families IIa (15/473) and IId (1/473). Analysis of the microsatellite serine region identified 13 subtypes, 5 in C. hominis and 8 in C. parvum. The most common subtype, C. hominis IbA10G2, was identified in 449/473 (95%) isolates. The next most frequently detected subtype was C. parvum IIaA18G3R1, which was identified in 8/473 (2%) isolates.
Table 1.
Summary of oocyst numbers in feces and the Cryptosporidium species and subtypes causing human illness in the outbreak from January to April of 2009 in New South Wales, Australia
| Cryptosporidium species | gp60 subtype | No. of isolates | Oocysts/g feces |
|||
|---|---|---|---|---|---|---|
| ≤102 | 103–104 | 105–106 | ≥107 | |||
| C. hominis | IaA26 | 1 | 1 | 0 | 0 | 0 |
| IbA9G3 | 4 | 0 | 0 | 3 | 1 | |
| IbA10G2 | 449 | 36 | 118 | 223 | 72 | |
| IdA17G1 | 2 | 0 | 0 | 2 | 0 | |
| IeA12G3T2 | 1 | 0 | 0 | 1 | 0 | |
| Subtotal | 5 | 457 | 37 | 118 | 229 | 73 |
| C. parvum | IIaA15G2R1 | 1 | 0 | 0 | 1 | 0 |
| IIaA17G2R1 | 2 | 0 | 1 | 1 | 0 | |
| IIaA17G4R1 | 1 | 0 | 0 | 1 | 0 | |
| IIaA18G3R1 | 8 | 1 | 5 | 2 | 0 | |
| IIaA19G6R1 | 1 | 1 | 0 | 0 | 0 | |
| IIaA20G3R1 | 1 | 0 | 1 | 0 | 0 | |
| IIaA22G3R1 | 1 | 0 | 0 | 1 | 0 | |
| IIdA23G1 | 1 | 0 | 1 | 0 | 0 | |
| Subtotal | 8 | 16 | 2 | 8 | 6 | 0 |
| Total | 13 | 473 | 39 | 126 | 235 | 73 |
The number of oocysts/g in feces was determined using flow cytometry. The patients infected with C. hominis were shedding higher numbers of parasites than those infected with C. parvum. The oocyst counts ranged from 102 to 107/g in cases identified as shedding C. hominis and from 102 to 106 oocysts/g in C. parvum cases (Table 1). The majority of cases identified as C. hominis infections were shedding parasite numbers between 105 and 106, with an additional 73 patients shedding 107 or more oocysts (Table 1). Seventy-two of these cases were infected with the C. hominis IbA10G2 subtype. In contrast, the majority of C. parvum-infected patients were shedding parasite numbers of between 103 and 104 oocysts/g of feces.
Age and gender of patients infected with C. hominis IbA10G2.
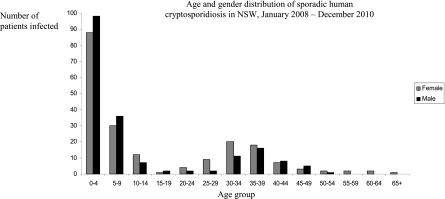
Information on patient age and gender were obtained for 387/521 (74%) samples. No gender bias was observed in outbreak cases (chi square = 0.26, df = 1, P = 0.61). The number of cases was 199/387 (51%) for females and 188/387 (49%) for males (Fig. 1). A bias in susceptibility by age was identified, with the 0- to 4-year year age category being the most commonly affected group for both males (chi square = 579.66, df = 1, P ≪ 0.01) and females (chi square = 468.79, df = 1, P ≪ 0.01). Of all cases, 88/199 (44%) female patients and 98/188 (52%) male patients were in the 0- to 4-year age group. Combined, the 0- to 4-year age category comprised 186/387 (48%) C. hominis IbA10G2 infections. Infections were also high for both genders in the 5- to 9-, 30- to 34-, and 35- to 39-year age groups. In the 0- to 4- and 5- to 9-year age groups, males were more affected than females. Females had a higher incidence than males in the 30- to 34- and 35- to 39-year age groups.
Fig. 1.
Age and gender distribution of patients infected with C. hominis IbA10G2 in the Cryptosporidium outbreak from January to April of 2009 in New South Wales (NSW), Australia.
Spatial distribution.
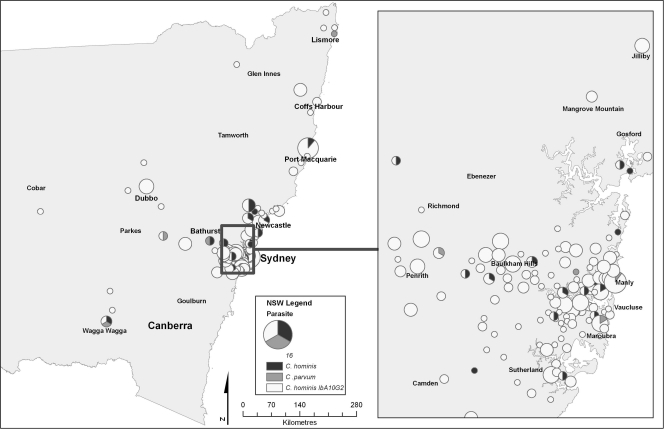
The postal codes for the locations from which isolates originated were obtained for 367/473 (78%) of the isolates successfully typed at the gp60 gene. Analysis of the New South Wales statewide map for the entire outbreak period (January to April 2009) showed a disease cluster centered in Sydney, with extensions 200 km north toward Newcastle (Fig. 2). Isolated clusters in two coastal communities north of Sydney, Port Macquarie and Coffs Harbor, were also observed, along with a cluster centered within the Orana rural community 400 km northwest of Sydney. An analysis of the greater Sydney region shows that the disease was widespread and not localized to any particular suburban area. However, clusters were seen through the inner west and northern suburbs of Sydney (Fig. 2).
Fig. 2.
Spatial distribution by postal-code areas of the patients infected with the C. hominis IbA10G2 subtype, other C. hominis subtypes, and C. parvum in New South Wales and Sydney between January and April 2009. The size of the circle represents the number of cases.
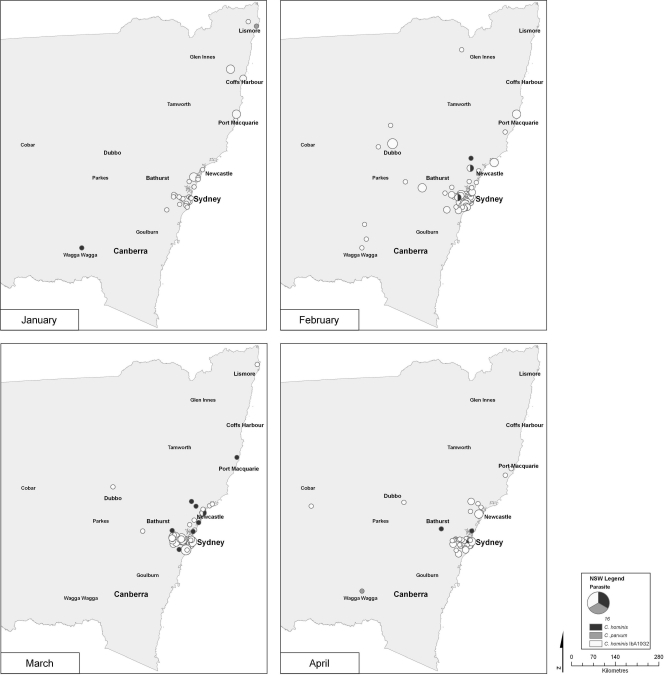
The spatiotemporal analysis depicts a monthly view of the spread of Cryptosporidium infections throughout the state during the January-to-April period (Fig. 3). In following the C. hominis IbA10G2 subtype (shown in white), analysis of the January infections showed that there were three disease clusters centered around the Sydney-to-Newcastle region, Port Macquarie, and Coffs Harbor. At this time, infections had not spread inland toward the rural areas. Infections peaked in February; the Sydney-to-Newcastle cluster increased, and the appearance of clusters in the rural areas northwest of Sydney occurred. The Port Macquarie incidence remained constant, while the third cluster previously present in Coffs Harbor had disappeared. By March, the outbreak was contracting back toward Sydney and Newcastle, with rural areas becoming less impacted. Decreased numbers of infections occurred in April, and the clusters dissipated, becoming centralized around the cities of Sydney and Newcastle. The remaining C. hominis subtypes (Fig. 3, shown in black) were located throughout the heavily populated urbanized regions of Sydney and Newcastle. Cryptosporidium parvum infections (shown in gray) were recorded sporadically in both urban and rural areas.
Fig. 3.
Temporal analysis by month of the patients infected with the C. hominis IbA10G2 subtype, other C. hominis subtypes, and C. parvum in New South Wales between January and April 2009. The size of the circle represents the number of cases.
DISCUSSION
A survey of global EID events indicated the east coast of Australia to be an emerging disease hot spot (15). Targeted surveillance and screening for potential disease outbreaks in hot spot zones is essential for the identification and protection of population groups most at risk of disease. New South Wales, Australia, has experienced six waterborne cryptosporidiosis outbreaks that have been linked to public swimming pools (2, 16, 17, 21, 23). This study analyzed the recent 2009 outbreak, which is the largest reported outbreak to date in Australia. A molecular and spatially based approach was applied to investigate the Cryptosporidium species and subtype responsible for this outbreak and to identify the population groups most affected.
One-third of the 22 million Australian residents live within New South Wales, making this state the most populated region in Australia. Within New South Wales, 63% of the population lives in the state's capital, Sydney. A growing city, Sydney encompasses both urban and rural localities and has a population of 4.6 million (2009 census data, Australian Bureau of Statistics [ABS], www.abs.gov.au). Newcastle is the second largest city within New South Wales, and similar to Sydney, it is a metropolis comprised of urban and rural areas and has a current population of 354,054 (ABS). Spatial analysis performed on isolates obtained during the outbreak period showed that the highest incidence of cryptosporidiosis was in the cities of Sydney and Newcastle. The higher incidence in these areas was likely associated with the higher population densities; this must be taken into consideration when interpreting the data. Although infections in Sydney were widespread, the densely populated inner west and northwest area were the most affected throughout the outbreak. Temporal analysis showed that the outbreak originated in the urban cities and expanded inland to the rural areas of New South Wales. These results are consistent with previous studies in that C. hominis infections are more common in urbanized city regions due to high population densities that provide stable anthroponotic transmission pathways (28, 30). Isolated clusters of C. hominis IbA10G2 infections in northern New South Wales were visualized in the early stage of the outbreak. With C. hominis IbA10G2 as the dominant parasite causing human cryptosporidiosis in New South Wales, it remains unclear whether these cases should be attributed to sporadic infections or if they were part of a separate outbreak.
The outbreak was caused by the anthroponotic C. hominis IbA10G2 subtype, which was identified in 95% of the disease cases. This subtype has a global distribution, and 44.5% of global human Cryptosporidium infections that have been studied to the subtype level have been attributed to it (13, 14, 20, 27). The C. hominis IbA10G2 subtype is also the most common cause of sporadic human infections in Australia (13, 14, 20, 27) and has been the agent responsible for large waterborne cryptosporidiosis outbreaks in the United States, Northern Ireland, and France (7, 11, 31). Most notably, C. hominis subtype family Ib caused the 1993 Milwaukee waterborne cryptosporidiosis outbreak which affected over 400,000 individuals and recorded a total outbreak cost of $96.2 million (8). Close to a decade later, this subtype was still the most common Cryptosporidium parasite detected in raw Milwaukee wastewater (32). To date, subtype analysis has not been conducted on Milwaukee C. hominis Ib isolates.
Cryptosporidium hominis is a specialist pathogen, specifically adapted to the human host. Conversely, C. parvum is a generalist, capable of causing infections in humans and a variety of ruminant species. Specialist pathogens invoke more virulent infections than their generalist counterparts (12). Patients infected with C. hominis, particularly the IbA10G2 subtype, were shedding higher numbers of oocysts than those infected with C. parvum. For the majority of C. hominis IbA10G2 infections, oocyst shedding was between 105 to 107 oocysts/g. The high oocyst shedding intensity of C. hominis IbA10G2 may have facilitated the rapid spread of the parasite and contributed to the extent of the outbreak. Conversely, the majority of C. parvum cases in the same period were shedding between 103 to 104 oocysts/g of feces. Cryptosporidium parvum cases were considered to be sporadic and not associated with the outbreak. Human cryptosporidiosis infections typically last between 7 and 14 days, and the levels of oocyst shedding vary significantly during this period (5). The fecal samples analyzed in this study would have been obtained at various points in the respective infections, and hence, a range of numbers of oocysts being shed would be expected.
The increasing incidence of cryptosporidiosis in Australia indicates that this is an emerging disease. To reduce the risks of human illness, the implementation of Cryptosporidium-specific pool water treatments and monitoring systems is required. At this stage, these technologies are not available, and increased public education is the current resource that is important to preventing future cryptosporidiosis outbreaks. Based on the dominance, persistence, distribution, and virulence of the IbA10G2 subtype in human populations and its association with waterborne outbreaks, this subtype should be considered a significant threat to global human health.
ACKNOWLEDGMENTS
Funding for this research was provided through an Australian Research Council Linkage Grant in partnership with New South Wales Health and was approved by the Macquarie University Human Research Ethics Committee (approval reference no. HE23FEB2007-R05007).
We thank Douglas Hanley Moir of the SDS and Symbion Pathology companies in North Ryde and the Westmead, John Hunter of St. Vincent's, Blacktown, Gosford, and Davis Campbell of Wollongong and Seals Hospitals for providing the human fecal samples. Special acknowledgments to Jeremy McAnulty and Jennie Musto from the New South Wales Health Communicable Disease Branch for their assistance in sample collection, Borce Dimeski and Frank Siciliano for their assistance in spatial analyses, and Stephen Hoggard for his assistance in statistical analyses.
Footnotes
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Bennett J. W., Gauci M. R., Le Moenic S., Schaefer F. W., III, Lindquist H. D. A. 1999. A comparison of enumeration techniques for Cryptosporidium parvum oocysts. J. Parasitol. 85:1165–1168 [PubMed] [Google Scholar]
- 2. Black M., McAnulty J. M. 2006. The investigation of an outbreak of cryptosporidiosis in New South Wales in 2005. NSW Public Health Bull. 17:76–79 [DOI] [PubMed] [Google Scholar]
- 3. Cama V. A., et al. 2008. Cryptosporidium species and subtypes and clinical manifestations in children in Peru. Emerg. Infect. Dis. 14:1567–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chalmers R. M., et al. 2009. Cryptosporidium sp. rabbit genotype a newly identified human pathogen. Emerg. Infect. Dis. 15:829–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chalmers R. M., Davies A. P. 2010. Clinical cryptosporidiosis. Exp. Parasitol. 124:138–146 [DOI] [PubMed] [Google Scholar]
- 6. Chalmers R. M., et al. 2005. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int. J. Parasitol. 35:397–410 [DOI] [PubMed] [Google Scholar]
- 7. Cohen S., et al. 2006. Identification of Cpgp40/15 type Ib as the predominant allele in isolates of Cryptosporidium spp. from waterborne outbreak of gastroenteritis in South Burgundy, France. J. Clin. Microbiol. 44:589–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Corso P. S., et al. 2003. Cost of illness in the 1993 waterborne Cryptosporidium outbreak, Milwaukee, Wisconsin. Emerg. Infect. Dis. 9:426–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fayer R. 2010. Taxonomy and species delimitation in Cryptosporidium. Exp. Parasitol. 124:90–97 [DOI] [PubMed] [Google Scholar]
- 10. Ferrari B. C., Power M. L., Bergquist P. L. 2007. Closed-tube DNA extraction using a thermostable proteinase is highly sensitive, capable of single parasite detection. Biotechnol. Lett. 29:1831–1837 [DOI] [PubMed] [Google Scholar]
- 11. Glaberman S., et al. 2002. Three drinking water associated cryptosporidiosis outbreaks, Northern Ireland. Emerg. Infect. Dis. 8:631–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hunter P. R., et al. 2004. Health sequelae of human cryptosporidiosis in immunocompetent patients. Clin. Infect. Dis. 39:504–510 [DOI] [PubMed] [Google Scholar]
- 13. Jex A. R., Gasser R. B. 2010. Genetic richness and diversity in Cryptosporidium hominis and C. parvum reveals major knowledge gaps and a need for the application of “next generation” technologies. Biotechnol. Adv. 28:17–26 [DOI] [PubMed] [Google Scholar]
- 14. Jex A. R., et al. 2008. Classification of Cryptosporidium species from patients with sporadic cryptosporidiosis by use of sequence-based multilocus analysis following mutation scanning. J. Clin. Microbiol. 46:2252–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jones K. E., et al. 2008. Global trends in emerging infectious diseases. Nature 451:990–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lemmon J. M., McAnulty J. M., Bawden-Smith J. 1996. Outbreak of cryptosporidiosis linked to an indoor swimming pool. Med. J. Aust. 165:613–616 [DOI] [PubMed] [Google Scholar]
- 17. Menzies R. 2002. Cryptosporidiosis in NSW 1990-2000. NSW Public Health Bull. 13:54–57 [DOI] [PubMed] [Google Scholar]
- 18. Morgan-Ryan U. M., et al. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433–440 [DOI] [PubMed] [Google Scholar]
- 19. New South Wales Public Health Act of 1991 New South Wales Government. http://www.legislation.nsw.gov.au/viewtop/inforce/act+10+1991+FIRST+0+N
- 20. O'Brien E., McInnes L., Ryan U. 2008. Cryptosporidium gp60 genotypes from humans and domesticated animals in Australia, North America and Europe. Exp. Parasitol. 118:118–121 [DOI] [PubMed] [Google Scholar]
- 21. Peauch M. C., et al. 2001. A statewide outbreak of cryptosporidiosis in NSW associated with swimming at public pools. Epidemiol. Infect. 126:389–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robinson G., et al. 2010. Re-description of Cryptosporidium cuniculus Inman and Takeuchi, 1979 (Apicomplexa: Cryptosporidiidae): morphology, biology and phylogeny. Int. J. Parasitol. 40:1539–1548 [DOI] [PubMed] [Google Scholar]
- 23. Stafford R., Neville G., Towner C., McCall B. 2000. A community outbreak of Cryptosporidium infection associated with a swimming pool complex. Commun. Dis. Intell. 24:236–239 [PubMed] [Google Scholar]
- 24. Sulaiman I. M., et al. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 43:2805–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Truong Q., Ferrari B. C. 2006. Quantitative and qualitative comparisons of Cryptosporidium faecal purification procedures for the isolation of oocysts suitable for proteomic analysis. Int. J. Parasitol. 36:811–819 [DOI] [PubMed] [Google Scholar]
- 26. Waldron L. S., Cheung-Kwok-Sang C., Power M. L. 2010. Wildlife-associated Cryptosporidium fayeri in human, Australia. Emerg. Infect. Dis. 16:2006–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Waldron L. S., Ferrari B. C., Power M. L. 2009. Glycoprotein 60 diversity in C. hominis and C. parvum causing human cryptosporidiosis in NSW Australia. Exp. Parasitol. 122:124–127 [DOI] [PubMed] [Google Scholar]
- 28. Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80–89 [DOI] [PubMed] [Google Scholar]
- 29. Xiao L., et al. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species Appl. Environ. Microbiol. 65:3386–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xiao L., Ryan U. M. 2004. Cryptosporidiosis: an update in molecular epidemiology. Curr. Opin. Infect. Dis. 17:483–490 [DOI] [PubMed] [Google Scholar]
- 31. Xiao L., Ryan U. M. 2008. Molecular epidemiology, p. 119–171 In Fayer R., Xiao L. (ed.), Cryptosporidium and cryptosporidiosis. CRC Press and IWA Publishing, Boca Raton, FL [Google Scholar]
- 32. Zhou L., Singh A., Jiang J., Xiao L. 2003. Molecular surveillance of Cryptosporidium spp. in raw wastewater in Milwaukee: implications for understanding outbreak occurrence and transmission dynamics. J. Clin. Microbiol. 41:5254–5257 [DOI] [PMC free article] [PubMed] [Google Scholar]