Abstract
The early detection of microbial contamination is crucial to avoid process failure and costly delays in fermentation industries. However, traditional detection methods such as plate counting and microscopy are labor-intensive, insensitive, and time-consuming. Modern techniques that can detect microbial contamination rapidly and cost-effectively are therefore sought. In the present study, we propose gas chromatography-mass spectrometry (GC-MS)-based metabolic footprint analysis as a rapid and reliable method for the detection of microbial contamination in fermentation processes. Our metabolic footprint analysis detected statistically significant differences in metabolite profiles of axenic and contaminated batch cultures of microalgae as early as 3 h after contamination was introduced, while classical detection methods could detect contamination only after 24 h. The data were analyzed by discriminant function analysis and were validated by leave-one-out cross-validation. We obtained a 97% success rate in correctly classifying samples coming from contaminated or axenic cultures. Therefore, metabolic footprint analysis combined with discriminant function analysis presents a rapid and cost-effective approach to monitor microbial contamination in industrial fermentation processes.
INTRODUCTION
Fermentation processes are an important technology in biotechnological industries, contributing to the production of high-value goods including food, beverages, bulk chemicals, and pharmaceuticals (3). Industrial fermentations are typically large-scale processes, requiring several scale-up steps over time and consuming often expensive raw materials (3). A loss of sterility of fermentation has serious consequences, impacting production schedules, costs, and time and affecting product quality and quantity (9). Contamination is a predominant cause of process failure (9). It is therefore crucial that industrial fermentations are carefully monitored to detect microbial contamination as early as possible after it occurs.
While bioreactors are expensive to operate and maintain, closed-batch or fed-batch bioreactor systems are favored for industrial microbial growth due to their ability to control environmental parameters such as pH, temperature, and gas transfer. Consequently, closed-system fermentations result in higher productivity and improved product consistency and quality. Microalgal biotechnology relies heavily upon fermentation. Microalgae are rich sources of commercially valuable products including amino acids, vitamins, lipids, and pigments (2, 7, 15); hence, microalgal biotechnology is emerging as a promising and exciting alternative for new natural products and applications. The risk of contamination by fast-growing microorganisms is increased in microalgal fermentations due to the comparatively slow growth of microalgae and the nutrient-rich nature of their industrial fermentation media. Therefore, the industry is eager to develop a sensitive, rapid, and cost-effective method for the detection of microbial contamination in fermentation broth.
Traditional methods for the detection of microbial contamination are generally insensitive, labor-intensive, and time-consuming, mainly because they require access to a reasonable amount of contaminant cells via sampling. Plate counting and fluorescence microscopy are the most commonly used methods, and these methods rely on the presence of contaminant cells in the samples. Consequently, the detection of cells is often delayed until the number of contaminant cells increases above the detection limit, and it is often limited to particular types of microorganisms. These traditional techniques usually exclude viable but nonculturable (VBNC) contaminant cells, slow-growing cells, as well as cells that cannot grow in the selective media and/or under the incubation conditions applied (13). On the other hand, DNA-based techniques are expensive and also require access to a reasonable amount of contaminant cells, which delays the time to detection. False-negative results are not uncommon due to the poor representation of the cell population usually caused by poor sampling procedures (8, 13). Additionally, detection sensitivity is insufficient in early stages of contamination, when the proportion of contaminant cells to cultured cells is minimal (1). Delays required for plate incubation and colony enumeration also result in the contamination being detected only several days after contamination occurred, which often precludes the safe recovery of fermentation products.
On the other hand, microorganisms are known to rapidly adjust their metabolism in response to environmental changes such as the presence of a contaminant organism (10, 11). Changes in microbial metabolism are quickly detected through changes in their metabolic footprint profile. The metabolic footprint is the profile of extracellular metabolites resulting from nutrient uptake, extracellular biochemical reactions, and the secretion of metabolic products performed by a population of cells in the environment where they grew (10, 20). This metabolic footprint is highly specific to the genetic background of the cells and the environmental conditions under which they grew (10, 20). Thus, through metabolic footprint analysis (exometabolomics), we are able to distinguish different metabolic states of the cells, different microbial species, and even different strains or mutants from the same species (10, 19, 20). Metabolic footprinting therefore has the potential to reveal unique differences in the profiles of metabolites of fermentation broths that result from the presence of contaminant cells.
Gas chromatography coupled to mass spectrometry (GC-MS) is the most commonly used analytical approach to obtain comprehensive metabolite profiles of biological samples (10, 21). GC-MS is a sensitive analytical instrument that presents the best chromatographic resolution in metabolite analysis (21). In addition, it is one of the most cost-effective mass spectrometry-based instruments available today and is easy to operate. GC-MS produces reproducible results and has been shown to present less than 6% variability in biological samples (18). However, GC-MS analyses depend on the analytes being volatile. Most metabolites are not sufficiently volatile to be analyzed directly by GC-MS, so chemical derivatization is a crucial step in sample preparation (18). Our platform for metabolite analysis using GC-MS is based on an alkylation reaction of amino and non-amino organic acids and makes use of a rapid and efficient derivatization procedure targeting important metabolic intermediates from the central carbon metabolism (14). This procedure is ideal for the routine analysis of metabolites in fermentation broths, as the derivatization reagents are cost-effective, and the reaction is rapid (∼1 min), is carried out at room temperature and in aqueous medium, and results in stable volatile derivatives with high reproducibility (21). Also, the derivatization reactions can be automated by using robotic autosamplers for GC-MS systems (e.g., the CTC Combi PAL or Gerstel MultiPurpose sampler instrument).
In this study, we applied metabolic footprint analysis using GC-MS to detect microbial contamination in microalgal fermentation broths. Microalgal fermentation broths were inoculated with contaminant microbial cells and were monitored to determine when significant changes in the metabolic footprint profile of the broth could be observed.
MATERIALS AND METHODS
Chemicals.
Methanol, sodium hydroxide, chloroform, and sodium sulfate used for chemical derivatization were all of analytical grade and purchased from different suppliers. The derivatization reagent methyl chloroformate (MCF), pyridine, and the isotope-labeled internal standard l-alanine-2,3,3,3-d4 were obtained from Sigma-Aldrich (St. Louis, MO).
Microbial strains.
The UTEX 2047 strain of the pennate diatom Nitzschia laevis (University of Texas Microalgal Collection) was used in all microalgal fermentations, and a strain of Pseudomonas aeruginosa (School of Biological Sciences collection) and a strain of Bacillus subtilis (School of Biological Sciences collection) were used to mimic bacterial contamination in microalgal cultures by deliberate contamination experiments.
Microalgal fermentation.
A total of 55 batch fermentations were carried out under aerobic conditions (not concomitantly) in 500-ml shake flasks and incubated at 20°C under continuous agitation (200 rpm). Each flask contained 200 ml of defined culture medium (pH 8.0), prepared in several separated batches according to Photonz' specifications, supplemented with glucose (20 g/liter), vitamins, and trace metals.
Microbial contamination experiments.
A flow chart summarizing all contamination experiments carried out in this work can be found in Fig. S1 in the supplemental material. We carried out three independent contamination experiments as a proof of concept that metabolic footprint analysis is able to detect early microbial contamination in microalgal fermentation broth. First, 1-day-old N. laevis culture flasks (n = 3) were opened on the laboratory bench for 2 h to allow naturally occurring airborne microorganisms to contaminate the liquid cultures. Two-milliliter samples were harvested when the flasks were opened (time zero) and every 24 h for 4 days. Samples were filtered using 0.2-μm filter membranes to remove microbial biomass. Filtrates were stored at −20°C until chemical derivatization and metabolite analysis were performed.
In a second experiment, liquid cultures of specific microbial contaminants were prepared by inoculating single colonies of either P. aeruginosa or B. subtilis in 100 ml nutrient broth containing yeast extract (3 g/liter), peptone (5 g/liter), and dextrose (1 g/liter) at pH 6.5. Cultures were incubated in 250-ml shake flasks at 25°C and 200 rpm. Bacterial cells were harvested by centrifugation after approximately 19 h of growth (while the cells were still at the exponential growth phase). The concentration of bacterial biomass was estimated by determining the optical density at 600 nm, and the cells were diluted in the microalgal medium to obtain approximately 1 CFU per μl of cell suspension. One microliter of the diluted bacterial cell suspension was then used to inoculate 5-day-old N. laevis cultures (n = 3) with either B. subtilis or P. aeruginosa. Two-milliliter broth samples were harvested every 6 h, starting from time zero (at the moment of contaminant inoculation) until 12 h after contamination. A further sample was taken 24 h after contamination. The samples were filtered to remove microbial biomass (pore size, 0.2 μm), and the filtrates were stored at −20°C until chemical derivatization and metabolite analysis were performed.
Lastly, to build a discriminant model for the automatic prediction of microbial contamination based on the metabolic footprint profile of microalgal cultures, the second experiment described above was repeated using 40 culture replicates. Twenty flasks were maintained axenic (control), and the other 20 were contaminated with 1 CFU of P. aeruginosa as described above. The 40 cultures were incubated on different days as well as using medium prepared in different batches in an attempt to include the natural variation in the extracellular metabolite composition of axenic cultures that might occur due to technical variation. Two-milliliter broth samples were harvested every 3 h, starting from time zero (at the moment of contaminant inoculation) until 9 h after contamination. The samples were filtered to remove microbial biomass (pore size, 0.2 μm), and the filtrates were stored at −20°C until chemical derivatization and metabolite analysis were performed.
Biomass quantification.
An 8-ml broth sample was harvested at every sampling time point for the quantification of total microbial biomass (dry weight).
Traditional methods for controlling contamination.
Before membrane filtration, all samples harvested for metabolic footprint analysis were examined by light microscopy (wet mount), and 1 ml was spread over plate count agar (PCA) plates for the counting of CFU of contaminants present in the samples. The PCA plates were incubated at 30°C for 48 h.
Sample derivatization.
The MCF derivatization was performed according to a method described previously by Smart et al. (14). In summary, 150 μl of filtered broth in triplicate was mixed with 30 μl of sodium hydroxide solution (3 M) and 20 μl of internal standard (l-alanine-2,3,3,3-d4 [10 mM]). Two hundred microliters of the alkalinized broth sample was then mixed with 34 μl of pyridine and 167 μl of methanol. Twenty microliters of MCF was added to the reagent mixture, followed by vigorous mixing for 30 s using a vortex. Another 20 μl of MCF was added to the reactive mixture, followed again by vigorous mixing for another 30s. To separate the MCF derivatives from the reactive mixture, 400 μl of chloroform was added to the mixture and then mixed vigorously for 10 s, followed by the addition of 400 μl of sodium bicarbonate solution (50 mM) and vigorous mixing for an additional 10 s. The upper aqueous layer was discarded, and the chloroform phase was subjected to GC-MS analysis.
GC-MS analysis.
GC-MS analysis was performed with an Agilent GC7890 gas chromatograph coupled to an MSD5975 mass selective detector. The column used for all analyses was a ZB1701 column (Zebron; Phenomenex), with a 30-m by 250-μm internal diameter (i.d.) and a 0.15-μm film thickness with a 5-m guard column. The mass spectrometer was operated in scan mode (start after 4.5 min, with a mass range of 40 to 650 atomic mass units [amu] at a rate of 0.15 s/scan). The parameters for the separation and analysis of MCF derivatives were described previously by Smart et al. (14).
Data analysis.
Data were analyzed according to methods described previously by Smart et al. (14). Metabolites were identified by using our in-house MCF-MS library of metabolite standards and Automated Mass Spectral Deconvolution and Identification System (AMDIS) software, which relies on both the chromatographic retention times of the analytes and their respective mass spectra. Metabolite levels were determined by the intensity of the base peak normalized by the internal standard and the biomass concentration in the respective sample. Data mining was performed by using an in-house R script using R software (version 2.10.0; r-project) loaded with the following packages: xcms, KEGG.db, gplots, reshape, plotrix, KEGGSOAP, and keggorth.
The difference in the metabolite profiles of the different samples was assessed by discriminant function analysis (DFA). First, we listed the relative normalized level of each identified metabolite under all conditions (see Table S1 in the supplemental material). Conditions for which the level of a metabolite was below the detection limit of the method were assigned a value of zero. To avoid violating the independent-sample assumption, three separate paired-sample DFAs were implemented for each time point. Therefore, it was not the actual expression level of metabolites (relative abundance) at all time points but the change compared to the baseline at time zero that was used. In other words, the metabolite abundances at time zero were subtracted from the abundances of the same metabolites at a time of 3 h. The difference in metabolite abundance was used as the input for DFA for prediction.
Using a stepwise procedure (12), we selected 19 metabolites that contribute the most to the discrimination between the two conditions (Table 1). From the list of all metabolites, variables were added one by one to the model until the classification result did not increase more than 0.5%. Following this, two variables were fitted into the model, one previously selected and another that gave the best classification, and 3 variables were then fitted, two of those used before and a third one that gave the best classification, and so on.
Table 1.
List of identified extracellular metabolites analyzed by GC-MS and used as variables for DFA
| Metabolite |
|---|
| Amino acids |
| Valine |
| Leucine |
| tert-Leucine |
| 4-Aminobutyrate |
| Proline |
| Asparagine |
| Phenylalanine |
| Lysine |
| Simple organic acids |
| 2-Oxoglutarate |
| Citrate |
| Malate |
| Succinate |
| Lactate |
| Nicotinate |
| Fatty acids |
| 10,13-Dimethyltetradecanoate |
| 14-Methylpentadecanoate |
| Stearate |
| Caproate |
| Other |
| EDTA |
The projections of samples on three dimensions were computed by the dicrcoord function of the fpc package (http://www.homepages.ucl.ac.uk/∼ucakche/). Visual clustering was achieved by plotting the first three DFA projections. The lda function of the MASS package was used to classify samples into two experimental categories. The results were validated by using a leave-one-out cross-validation technique, using a single observation as the testing data for the DFA and the remaining data as the training set. The procedure was repeated until all observations were used as the testing sample. The data were log transformed to fit the normal distribution criteria.
RESULTS
Metabolic footprint analysis of axenic versus contaminated microalgal cultures.
Approximately 53 different metabolites that play important metabolic roles in the central carbon metabolism and lipid and amino acid biosynthesis were identified among hundreds of detected compounds (see Table S1 in the supplemental material). To distinguish samples among classes, we projected the identified metabolite level data from each sample to a lower-dimensional space. Two often-used data projection methods are PCA (principal component analysis) and DFA (discriminant function analysis). PCA maximizes variation in the reduced dimensions, whereas DFA maximizes separation between classes (19). For this reason, we applied DFA to visualize samples in an attempt to distinguish them among classes, which revealed a very clear separation, as shown in Fig. 1 and 2. Our metabolomics data successfully demonstrated that each data class (axenic and contaminated samples) presents distinct metabolite profiles, with samples from the same data class clustering closer to each other despite being sampled at different time points. This proves that the changes in the metabolic footprint profiles of contaminated cultures exceed natural changes in metabolite profiles of axenic cultures over time.
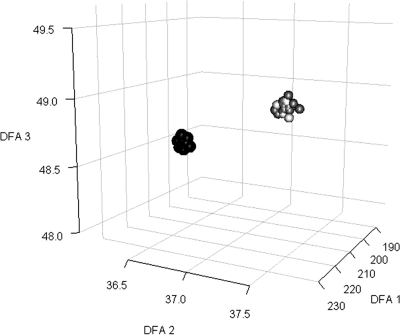
Fig. 1.
Discriminant function analysis (DFA) for sample visualization (uncontrolled contamination). GC-MS metabolite data successfully distinguished samples from contaminated versus noncontaminated microalgal flasks. Projections of the log-transformed intracellular metabolite data from 36 samples into three-dimensional (3D) space show two very distinct clusters of the two data classes (contaminated versus noncontaminated). For each sample, the projection values were calculated as the linear combination of metabolite values determined by DFA. Only metabolites that were detected in more than 25% of the samples for each data class were used for the analysis. Black, samples from contaminated flasks (time [t] = 24 to t = 96 h); dark gray, samples from noncontaminated flaks (t = 24 to t = 96 h); light gray, samples from contaminated flasks at time zero.
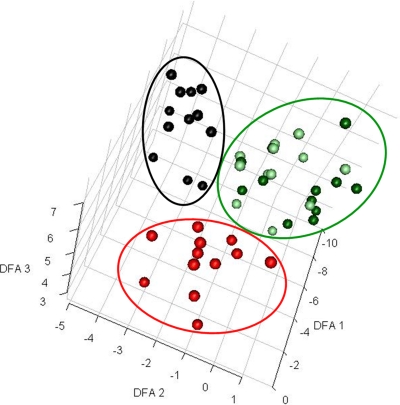
Fig. 2.
DFA for sample visualization (controlled contamination). GC-MS metabolite data successfully distinguished samples from contaminated versus noncontaminated microalgal flasks. Projections of the log-transformed intracellular metabolite data from 56 samples into 3D space show three distinct clusters of the three data classes. For each sample, the projection values were calculated as the linear combination of metabolite values determined by DFA. Only metabolites that were detected in more than 25% of the samples for each data class were used for the analysis. Black, samples from flasks contaminated by P. aeruginosa (t = 6 h to t = 24 h); red, samples from flasks contaminated by B. subtilis (t = 6 h to t = 24 h); dark green, samples from noncontaminated microalgal flasks; light green, samples collected from contaminated flasks at time zero.
Cultures exposed to uncontrolled contamination presented completely different metabolite profiles 24 h after the flasks were opened to induce contamination compared with their profiles at time zero or with the profile of noncontaminated flasks (Fig. 1). This shows that metabolic footprint profiling is capable of detecting microbial contamination in microalgal cultures within 24 h, while we could detect colonies of contaminant microbes only after 72 h (including incubation time).
In our second contamination experiment, we decided to harvest samples within 24 h of contamination in order to determine how early we could detect contamination in the microalgal fermentation broths using metabolic footprint analysis. Culture flasks contaminated with a single CFU of contaminant bacteria (P. aeruginosa or B. subtilis) presented distinct metabolite profiles as early as 6 h after contamination (Fig. 2). We could detect contaminating bacterial cells using light microscopy only at 24 h after the contamination was introduced, and we could detect bacterial CFU only in samples obtained 9 h after contamination was introduced. Considering that an additional 48 h of plate incubation is required to visualize contaminant bacterial colonies, the plating technique could detect bacterial contamination only after 57 h under the conditions tested.
Our preliminary contamination experiments clearly show that metabolic footprint profiling (extracellular metabolomics) is capable of detecting microbial contamination of microalgal batch fermentations and is certainly a more sensitive method than traditional techniques used for the detection of contamination in fermentation broth.
Classification model.
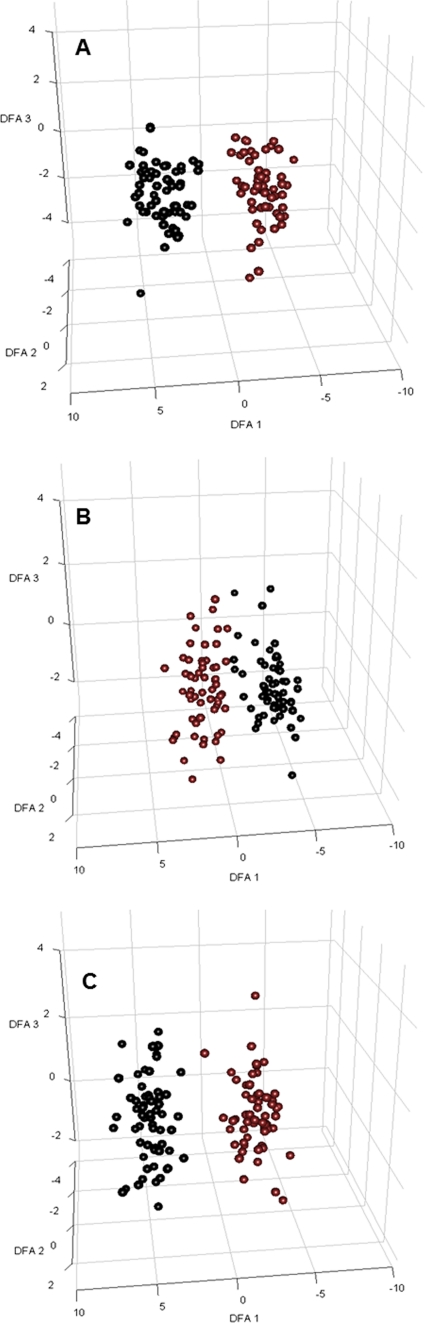
In order to validate our preliminary observations, a third contamination experiment using only P. aeruginosa was repeated in order to obtain a large number of replicate samples, which is necessary for creating a reliable classification model using DFA. A total of 60 replicates under each condition (20 biological replicates subdivided into three technical replicates each) were used for DFA. Comparative GC-MS chromatograms at different time points can be found in Fig. S2 in the supplemental material. DFA revealed a clear separation between all axenic and contaminated samples at different time points, as shown in Fig. 3. Once again, our metabolomics data successfully demonstrated that each data class (axenic and contaminated) presents distinct metabolite profiles, with samples from the same data class clustering very close to each other despite being sampled at different time points and coming from different fermentation batches. The classification rate of correctly classified samples as contaminated after 3 h of 1 CFU of contaminant being introduced into the flasks was 97%. A total of 0 out of 60 axenic samples was false positive, giving a classification rate of 100% accuracy (Table 2).
Fig. 3.
DFA for sample visualization of a large data set used for construction of the classification model (controlled contamination). GC-MS metabolite data successfully distinguished samples from contaminated versus noncontaminated microalgal flasks at different time points. Projections of the log-transformed intracellular metabolite data from 120 samples into 3D space show two distinct clusters of the two data classes. For each sample, the projection values were calculated as the linear combination of metabolite values determined by DFA. Only metabolites that were detected in all samples for each data class were used for the analysis. Black, samples from flasks contaminated by P. aeruginosa; red, samples from noncontaminated microalgal flasks. (A) Three hours after contamination; (B) 6 h after contamination; (C) 9 h after contamination.
Table 2.
Classification of contaminated culture samples (n = 60) and control samples for all time pointsa
| Contamination status | Time point (h) (no. of samples) | No. of incorrect classifications | Classification rate |
|---|---|---|---|
| Axenic | 0, control + contaminated (120) | 3 | 0.95 |
| 3, control (60) | 0 | 1.00 | |
| 6, control (60) | 8 | 0.86 | |
| 9, control (60) | 2 | 0.97 | |
| Overall | 13 | 0.96 | |
| Contaminated | 3 (60) | 2 | 0.97 |
| 6 (60) | 3 | 0.95 | |
| 9 (60) | 1 | 0.98 | |
| Overall | 6 | 0.97 | |
| Total overall | 0.96 |
Classification rate refers to the percentage of samples correctly classified by the DFA prediction model.
The monitoring of samples taken at 6 h and 9 h also showed high levels of accuracy (95% and 98% accuracy, respectively). This high success rate gives great statistical confidence that the model was highly effective in classifying metabolic footprints of unknown cultures as being axenic or contaminated.
From the total pool of samples, the rate of false-positive results was minimal (3% of the total samples were incorrectly classified). With an accuracy of 97%, the combination of metabolic footprinting and DFA was able to successfully detect bacterial contamination in microalgal fermentation broth within 3 h of a single CFU of a bacterium being introduced into the medium, validating our preliminary finding using lower numbers of replicates as well as emphasizing the greater sensitivity and reliability than those of traditional methods for the detection of microbial contamination in fermentation broths.
DISCUSSION
Metabolic footprint analysis examines low-molecular-weight compounds dissolved in liquid medium and does not rely on contaminant cells being present in a sample. This may explain why metabolic footprint analysis effectively detected microbial contamination in microalgal cultures as early as 3 h after contamination, while we could visually detect bacterial cell using light microscopy or bacterial CFU only after 24 and 57 h of the contamination taking place, respectively. Additionally, complications arising from inconsistent cell size, cell damage, and viability do not interfere with metabolite analysis.
The targeted analysis of signature metabolites is an approach that has been used in many studies for the detection of microorganisms in complex samples (4–6, 16). Certain metabolites have been considered indicative of the presence of particular microorganisms. These include muramic acid for all bacteria, myristic acid for Gram-negative bacteria, and glucosamine for fungi (4–6). However, these metabolic markers are not completely specific for a group of microorganisms, and it is known that the quantities of these metabolites vary from species to species, making it difficult to ascertain consistently the level of contamination based on metabolite levels. For example, myristic acid, proposed previously to be indicative of Gram-negative bacteria, is also synthesized by some Gram-positive bacteria (17) as well as fungi (17, 19). Therefore, it is safer to use the profile of metabolites instead of relying on single molecules as indicators.
Very few studies have examined the entire metabolite profile of contaminated cultures. Elmroth et al. (5, 6) previously compared the metabolite profiles of axenic cultures against those of contaminated cultures. Those authors detected changes in the metabolite profiles caused by the presence of microbial contaminants. However, their profiles were generated using only data representing targeted groups of intracellular metabolites. Intracellular metabolite analysis requires access to a considerably large number of contaminant cells, which is not ideal for the early detection of contamination. This approach makes use of spent culture media, which does not depend on having access to contaminant cells. Thus, to the best of our knowledge, this is the first time that metabolic footprinting analysis of extracellular metabolites has been proposed for the detection of microbial contamination.
Our proof-of-concept study clearly shows that metabolic footprint analysis provides rapid and conclusive detection of contaminated microalgal cultures. Although we have not tested other fermentation setups (e.g., bacterial or yeast fermentation), this approach has great potential to be used in industrial settings as a means of detecting microbial contamination in fermentations. As shown in Fig. 2, the metabolite profile of microalgal cultures contaminated by Pseudomonas was significantly different from that of microalgal cultures contaminated by Bacillus. It is already common for well-established fermentation companies to monitor the levels of some metabolites to ensure that production remains consistent over time. For example, beer production industries monitor the level of ethanol as a measure of consistency and quality. Our approach could therefore be easily adapted to an industrial routine. Since our approach uses the collective group of metabolites identified from metabolic footprints to generate metabolite profiles, it can be easily optimized and subsequently applied to a range of fermentation systems as long as a classification model (e.g., DFA) is trained with enough numbers of axenic samples covering the normal variability found in the fermentation culture media, such as batch-to-batch variations in medium compositions. In addition, raw GC-MS data can be used potentially to increase the data analysis throughput, and more samples would certainly increase the accuracy of the classification model, decreasing the error rate to below 3%. Nonetheless, our current GC-MS platform for metabolite analysis (14) would take no more than 1 h to classify a fermentation sample as being contaminated or axenic after a robust DFA model is built.
Supplementary Material
ACKNOWLEDGMENTS
We thank both the New Zealand Foundation for Research Science and Technology (FRST) and Photonz Corporation Limited for funding this research.
We also thank Raphael Aggio for assistance with data mining and figures in the supplemental material and Katya Ruggiero for fruitful statistical discussions.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Chaudhary S. C., Dodd P. W. 1994. Economic importance of sterility in industrial fermentation processes. Adv. Biochem. Eng. Biotechnol. 51:35–38 [Google Scholar]
- 2. Chen F. 1996. High cell density culture of microalgae in heterotrophic growth. Trends Biotechnol. 14:421–426 [Google Scholar]
- 3. Chisti Y. 1992. Assure bioreactor sterility. Chem. Eng. Prog. 88:80–85 [Google Scholar]
- 4. Elmroth I., Valeur A., Odham G., Larsson L. 1990. Detection of microbial contamination in fermentation processes: mass spectrometric determination of gram-negative bacteria in Leuconostoc mesenteroides cultures. Biotechnol. Bioeng. 35:787–792 [DOI] [PubMed] [Google Scholar]
- 5. Elmroth I., Sundin P., Valeur A., Larsson L., Odham G. 1992. Evaluation of chromatographic methods for the detection of bacterial contamination in biotechnical processes. J. Microbiol. Methods 15:215–228 [Google Scholar]
- 6. Elmroth I., Fox A., Holst O., Larsson L. 1993. Detection of bacterial contamination in cultures of eucaryotic cells by gas chromatography-mass spectrometry. Biotechnol. Bioeng. 42:421–429 [DOI] [PubMed] [Google Scholar]
- 7. Harun R., Singh M., Forde G. M., Danquah M. K. 2010. Bioprocess engineering of microalgae to produce a variety of consumer products. Renewable Sustainable Energy Rev. 14:1037–1047 [Google Scholar]
- 8. Hobson N. S., Tothill I., Turner A. P. F. 1996. Microbial detection. Biosens. Bioelectron. 11:455–477 [DOI] [PubMed] [Google Scholar]
- 9. Junker B., et al. 2006. Sustainable reduction of bioreactor contamination in an industrial fermentation pilot plant. J. Biosci. Bioeng. 102:251–268 [DOI] [PubMed] [Google Scholar]
- 10. Kell D. B., et al. 2005. Metabolic footprinting and systems biology: the medium is the message. Nat. Rev. Microbiol. 3:557–565 [DOI] [PubMed] [Google Scholar]
- 11. Mapelli V., Olsson L., Nielsen J. 2008. Metabolic footprinting in microbiology: methods and applications in functional genomics and biotechnology. Trends Biotechnol. 26:490–497 [DOI] [PubMed] [Google Scholar]
- 12. Martin F. L., et al. 2007. Identifying variables responsible for clustering in discriminant analysis of data from infrared microspectroscopy of a biological sample. J. Comput. Biol. 14:1176–1184 [DOI] [PubMed] [Google Scholar]
- 13. Maukonen J., et al. 2006. Suitability of the fluorescent techniques for the enumeration of probiotic bacteria in commercial non-dairy drinks and in pharmaceutical products. Food Res. Int. 39:22–32 [Google Scholar]
- 14. Smart K. F., Aggio R. B. M., Van Houtte J. R., Villas-Bôas S. G. 2010. Analytical platform for metabolome analysis of microbial cells using methyl chloroformate derivatization followed by gas chromatography-mass spectrometry. Nat. Protoc. 10:1711–1729 [DOI] [PubMed] [Google Scholar]
- 15. Spolaore P., Joannis-Cassan C., Duran E., Isambert A. 2006. Commercial applications of microalgae. J. Biosci. Bioeng. 101:87–96 [DOI] [PubMed] [Google Scholar]
- 16. Trifirò A., et al. 1997. Use of ion chromatography for monitoring microbial spoilage in the fruit juice industry. J. Chromatogr. A 770:243–252 [DOI] [PubMed] [Google Scholar]
- 17. Villas-Bôas S. G., Bruheim P. 2007. Cold glycerol-saline: the promising quenching solution for accurate intracellular metabolite analysis of microbial cells. Anal. Biochem. 370:87–97 [DOI] [PubMed] [Google Scholar]
- 18. Villas-Bôas S. G., Koulman A., Lane G. A. 2007. Analytical methods from the perspective of method standardization. Top. Curr. Genet. 18:11–52 [Google Scholar]
- 19. Villas-Bôas S. G., Moxley J. F., Åkesson M., Stephanopoulos G., Nielsen J. 2005. High-throughput metabolic state analysis: the missing link in integrated functional genomics of yeasts. Biochem. J. 388:669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Villas-Bôas S. G., Noel S., Lane G. A., Attwood G., Cookson A. 2006. Extracellular metabolomics: a metabolic footprinting approach to assess fiber degradation in complex media. Anal. Biochem. 349:297–305 [DOI] [PubMed] [Google Scholar]
- 21. Villas-Bôas S. G., Smart K. F., Sivakumaran S., Lane G. A. 2011. Alkylation or silylation for analysis of amino and non-amino organic acids by GC-MS? Metabolites 1:3–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.