Abstract
Salmonellosis represents a worldwide health problem because it is one of the major causes of food-borne disease. Although motility is postulated as an important Salmonella virulence attribute, there is little information about variation in motility in natural isolates. Here we report the identification of a point mutation (T551 → G) in motA, a gene essential for flagellar rotation, in several Salmonella enterica serovar Enteritidis field isolates. This mutation results in bacteria that can biosynthesize structurally normal but paralyzed flagella and are impaired in their capacity to invade human intestinal epithelial cells. Introduction of a wild-type copy of motA into one of these isolates restored both motility and cell invasiveness. The motA mutant triggered higher proinflammatory transcriptional responses than an aflagellate isolate in differentiated Caco-2 cells, suggesting that the paralyzed flagella are able to signal through pattern recognition receptors. A specific PCR was designed to screen for the T551 → G mutation in a collection of 266 S. Enteritidis field isolates from a nationwide epidemic, comprising 194 from humans and 72 from other sources. We found that 72 of the 266 (27%) isolates were nonmotile, including 24.7% (48/194) of human and 33.3% (24/72) of food isolates. Among nonmotile isolates, 15 carried the T551 → G mutation and, significantly, 13 were recovered from food, including 7 from eggs, but only 2 were from human sources. These results suggest that the presence of paralyzed flagella may impair the ability of S. Enteritidis to cause disease in the human host but does not prevent its ability to colonize chickens and infect eggs.
INTRODUCTION
Salmonella enterica serovar Enteritidis represents a major cause of food-borne human gastroenteritis worldwide, with a significant impact on public health. Infection usually occurs by ingestion of contaminated water or food, particularly eggs and other poultry-derived products (14). In humans, the disease is characterized by acute intestinal inflammation and diarrhea, which is normally self-limiting. In chickens, S. Enteritidis can asymptomatically colonize the gastrointestinal tract and deeper organs of the animal and can be transmitted to the forming eggs through the transovarian route and to laid eggs through the eggshell (14, 18, 21).
Although rarely isolated in Uruguay before 1994, S. Enteritidis was the etiological agent of an epidemic of human gastroenteritis that occurred in the country between 1995 and 2004, peaking in 2001-2002 (5). Starting in 2009, S. Enteritidis reemerged, overtaking Salmonella enterica serovar Typhimurium as the primary cause of salmonellosis in Uruguay (data from the National Salmonella Center, NSC, Institute of Hygiene, Uruguay).
Similarly, FoodNet surveillance for 2009 in 10 U.S. states reported that salmonellae were the most common causes of food-borne infections, and among them, S. Enteritidis was the serovar most commonly isolated (http://jama.ama-assn.org/cgi/content/full/303/21/2130).
Motility and flagella are considered to be important virulence factors contributing to gastrointestinal disease caused by Salmonella. Previous studies in streptomycin-pretreated mice have shown that during infection, S. Typhimurium flagella and motility contribute to early cecal inflammation (37, 38). A flagellated but nonmotile mutant of S. Enteritidis showed reduced attachment to rat ileal explants compared to that of the corresponding wild-type strain in the presence of an intact mucus layer, suggesting that active flagella are important for S. Enteritidis to penetrate this layer (31). In vitro, it has been found that aflagellate S. Typhimurium mutants can attach to, but are defective in entering, cultured intestinal epithelial cells (32). Similarly, aflagellate mutants or mutants of S. Enteritidis with paralyzed flagella showed reduced kinetics of invasion of cultured intestinal epithelial cells compared to findings for the wild-type strain, even if the adhesion levels were similar (41). Moreover, flagella are the main stimuli for triggering of host proinflammatory responses through activation of Toll-like receptor 5 (TLR5) and Ipaf (26, 30). Although these data indicate important roles for flagella and motility in Salmonella virulence, most of the studies so far rely on work done with artificially constructed mutant strains, but little is known about variations in flagellar function among field isolates and their effects on Salmonella-host interactions.
In Salmonella, about 50 genes are needed for flagellar assembly and function, but among these, only a few are directly involved in torque generation (24). MotA is one of the two proteins composing the stator responsible for torque generation in the proton-driven flagellar motor. Together with MotB, it forms a complex embedded in the cytoplasmic membrane, which is composed of four copies of MotA and two copies of MotB, which is essential for the proton translocation process that fuels the flagellar motion (3, 22, 28, 36).
Here we report the identification of a point mutation in motA in 15 S. Enteritidis isolates from a comprehensive collection of clinical and environmental samples. This mutation results in bacteria that harbor paralyzed flagella and are impaired for invasion of Caco-2 cells. All but two of the isolates identified as carrying this mutation were isolated from nonhuman sources.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
A set of 266 S. Enteritidis strains isolated before, during, and after the epidemics of this serovar in Uruguay (covering 1988 to 2006) was obtained from the NSC collection. These included 194 human clinical isolates and 72 from animals or food as previously reported (4). Specific bacterial strains used in this study are listed in Table 1. LB broth and LB agar, supplemented with antibiotics when required (ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml), were used for routine cultures of Escherichia coli or S. enterica at 37°C in an orbital shaking incubator (200 rpm). E. coli DH5α was used as a host for cloning.
Table 1.
Bacterial strains and plasmid used in this study
| Strain or plasmid | Description | Source and/or reference |
|---|---|---|
| S. Enteritidis strains | ||
| 251/01 | Chicken egg isolate [motA(T551G)] | 4 |
| 8/02 | Human gastroenteritis isolate | 4 |
| LVR18 | 251/01, pLVR10 (Apr) | This work |
| LVR19 | 251/01, pBAD22 (Apr) | This work |
| LVR20 | 251/01 derivative, motA::cat (Cmr) | This work |
| LVR21 | 8/02 derivative, motA::cat (Cmr) | This work |
| LVR22 | motA+ derivative of LVR20 (Cms) | This work |
| LVR23 | motA+ derivative of LVR21 (Cms) | This work |
| PT4 P125109 | Wild-type, sequenced strain | Wellcome Trust Sanger Institute (39) |
| S. Typhimurium SL5338 | galE r− m+ | 9 |
| E. coli strains | ||
| DH5α | F−endA1 hsdR17 supE44 thi1 recA1 gyrA relA1 Δ(lacZYA-argF) U169 (φ80lacZΔM15) | 16 |
| SY327λpir | Δ(lac pro) argE(Am) rif 20 nalA recA56 (λpir) | 27 |
| Plasmids | ||
| pBAD22 | Expression vector with PBAD and AraC control (Apr) | 15 |
| pKD3 | Template plasmid, pANTSγ derivative (Apr Cmr) | 10 |
| pKD46 | Red recombinase expression plasmid, pINT-ts derivative (Apr) | 10 |
| pLVR10 | motA inserted between sites EcoRI and HindIII of pBAD22 | This work |
All Salmonella isolates were confirmed biochemically and serologically at the NSC and were stored in replicates at −80°C in Luria-Bertani (LB) broth containing 25% glycerol. They were resuscitated by gently scraping the surface of the frozen content with a sterile loop and streaked on LB agar plates. After overnight incubation, isolated colonies were inoculated in LB broth for further analysis.
S. Enteritidis PT4 P125109 (NCTC 13349, here referred to as PT4 [39]), was obtained from The Wellcome Trust Sanger Institute (Hinxton, United Kingdom) and used as a reference in phenotypic assays. Strain S. Typhimurium SL5338 (r− m+) was used as an intermediate host for plasmid constructs to be transferred from E. coli to Salmonella.
S. Enteritidis strains LVR18 and LVR19 are isolate 251/01 transformed with plasmids pLVR10 and pBAD22, respectively (see below).
S. Enteritidis strains LVR20 and LVR21 are motA::cat derivatives of isolates 251/01 and 8/02, respectively, and were constructed using the Lambda Red method (10). Briefly, plasmid pKD3 was used as a template for PCR amplification of a DNA fragment containing the chloramphenicol resistance gene cassette, flanked by regions of homology to motA, using the primers motAP1b and motAP2b (see Table S1 in the supplemental material). Both primers have a 5′ 47- to 50-mer region exhibiting perfect homology with the motAB locus of the S. Enteritidis PT4 chromosome and a 3′ region homologous to priming site 1 or priming site 2 of pKD3, respectively. Three hundred nanograms of the resulting 1.1-kb PCR fragment was gel purified and electroporated into PT4 carrying pKD46 for insertion into its chromosome. Loss of the helper plasmid in the obtained chloramphenicol-resistant colonies was verified by growth on LB plates at 37°C, and sensitivity to ampicillin was checked by replica plating on LB plates containing ampicillin. Then, the motA::cat gene was introduced into the chromosome of isolate 251/01 or 8/02 by P22 transduction and selection of Cmr and nonmotile transductants, resulting in strains LVR20 and LVR21, respectively. All chromosomal insertions were verified by PCR using primers C1 and C2 (see Table S1 in the supplemental material).
Strains LVR22 and LVR23 were obtained by P22 transduction of wild-type motA from PT4 into strains LVR20 and LVR21, respectively, and selection of motile and Cms transductants. This was achieved by spot plating the transduction mix onto soft agar LB plates (containing 0.3% agar) for 6 h at 37°C and picking bacteria with an inoculation loop about 2 cm away from the plating spot. The loopful of bacteria was streaked out onto LB plates, and after overnight incubation at 37°C, isolated colonies were replica plated on LB plates with or without chloramphenicol to verify sensitivity. Plating the transduction mix on LB plates without the recipient strain resulted in no colonies after overnight incubation at 37°C. The absence of cat insertion into motA was verified by PCR using primers motA1 and motB2.
All transductants were streaked out on Evans blue uridine agar to confirm the absence of contaminating phages.
Plasmid constructs and DNA manipulation.
Genetic techniques were performed using standard laboratory methods. Plasmid and chromosomal DNA purifications were done according to protocols recommended by the supplier (Qiagen, Germany). DNA was digested with restriction endonucleases or ligated with T4 DNA ligase under standard conditions as per the manufacturer's instructions (Fermentas, Invitrogen). Preparation of electrocompetent E. coli and S. enterica cells and DNA transformation were performed as previously described (12).
Plasmids and primers used in this study are listed in Table 1 and in Table S1 in the supplemental material, respectively. pLVR10 contains the motA gene cloned into pBAD22 under the control of the arabinose-inducible PBAD promoter and was constructed as follows. motA was PCR amplified from purified genomic PT4 DNA using the primers motA5′and motA3′ and a 10:1 mixture of Taq-Pfu DNA polymerases (Fermentas). The resulting amplicon was digested with EcoRI and HindIII and ligated into pBAD22 previously cut with the same restriction enzymes. The plasmid construct was verified by sequencing of the cloned insert.
DNA sequencing.
For sequencing of flagellar genes, genomic DNA was extracted from the bacterial strains using the DNeasy blood and tissue kit (Qiagen). Specific genes or regions were PCR amplified using Proof Start DNA polymerase (Qiagen) and the primers listed in Table S1 in the supplemental material. fliC (including regulatory regions) was amplified using the primers fliC1/fliC4, and the resulting 1.71-kb PCR product was sequenced with the primers fliC1, -2, -3, and -4, GFor, and GRev. For fliG sequencing, its coding region was amplified using the primers fliG1/fliG3 and sequenced with fliG1, -2, and -3. fliM and fliN were amplified using primers fliM1/fliN2, and the resulting 1.55-kb PCR product was sequenced with fliM1, -2, -3 and fliN1 and -2. For sequencing of motA and motB, the primers motA1/motB2 were used to PCR amplify a 1.96-kb fragment, which was sequenced using the primers motA1, -2, and -3 and motB1 and -2. Before sequencing, all PCR products were purified using a QIAquick PCR purification kit (Qiagen).
Motility tests.
Motility tests were performed as described by Yim et al. (43). Briefly, 2 μl of overnight cultures grown in LB broth were spotted onto the surface of an LB plate containing 0.3% agar (and arabinose when indicated) and incubated for 6 h at 37°C. Those isolates showing no halo of growth from the inoculation spot after 6 h of incubation (indistinguishable from a nonflagellated strain) were considered nonmotile. Values are expressed as a percentage of the diameter of growth (in mm) obtained for PT4. The assays were repeated three times, and the results were confirmed by phase-contrast microscope visualization of mid-log-phase bacterial cultures grown in LB broth.
Bacterial cell fractionation and protein analysis.
For evaluation of bacterial surface protein profiles, heat extracts (HE), which are composed predominantly of flagella and other surface proteins, were obtained as described by Nicholas (29), with modifications. Briefly, mid-log-phase bacteria were harvested by centrifugation, resuspended in phosphate-buffered saline (PBS), and incubated for 1 h at 65°C. The samples were then centrifuged for 25 min at 1,000 × g, and supernatants were quantified by Bradford assays (7). Equivalent amounts of protein (20 μg) were loaded into 12% SDS-PAGE gels and visualized with Coomassie blue R250 staining. Protein extracts from all strains were prepared at the same time and by the same procedure.
Secreted proteins were obtained from the supernatant of mid-log-phase bacterial cultures, filtered through 0.22-μm filters, and subsequently precipitated with trichloroacetic acid (TCA) 25% (final concentration). For preparation of total protein extracts, mid-log-phase bacterial cultures were centrifuged and resuspended in PBS, sonicated, and centrifuged again to remove unbroken cells. The supernatants (cleared lysates) were quantified by Bradford assays.
For Western blot analysis, 10 μg of HE extracts and secreted proteins or 80 μg of total extracts were loaded onto a 12% SDS-PAGE gel and analyzed by Western blotting using rabbit anti-Flic (Hm) antiserum (Salmonella H antiserum m; Difco).
Flagellar staining.
For detection of flagella in live cells, we performed a previously described method using Alexa Fluor 594 carboxylic acid succinimidyl ester (Molecular Probes) (40). Briefly, overnight cultures of bacteria grown in LB broth at 37°C and 200 rpm. were diluted 1/100 in fresh medium and grown in the same conditions to mid-log phase (optical density at 600 nm [OD600] = 0.4 to 0.6). Then, the protocol was followed exactly as described previously (40). The samples were visualized on a Leica DM6000B fluorescence microscope using the Cy3.5 filter, and the images were acquired and manipulated using Leica AF6000 software. A minimum of 10 fields was recorded for each isolate.
Cell lines, medium, and growth conditions.
The human colon carcinoma Caco-2 cell line was obtained from the American Type Culture Collection. The cells were maintained in minimal essential medium with Earle's Salts (high glucose, 4.5 g/liter), supplemented with 4 mM l-glutamine and 20% fetal calf serum at 37°C in 5% CO2, at up to 80% confluence.
To polarize Caco-2 cells, 5 × 104 cells were seeded in Costar transwells (6.5-mm diameter, 4 μM pore, polycarbonate) in 24-well plates and grown for 19 days, changing the culture medium every other day, until transepithelial electrical resistance (TEER) stabilization (TEER > 200 Ω · cm2). Impermeability of the monolayer, indicative of the differentiation level of the cells, was verified by a diffusion test using fluorescent beads (Fluospheres carboxylate-modified microspheres, 0.2 μm, yellow/green fluorescent [505/515]; Molecular Probes).
Cell invasion assays.
Caco-2 invasion assays were performed as previously described (43). Briefly, log-phase-grown bacteria were added to the cells at an MOI of ∼30:1, the plates were centrifuged for 5 min at 200 × g, and invasion was allowed to proceed for 1 h. Then, the medium was changed to gentamicin-containing medium (100 μg/ml), and after 1.5 h the cells were washed and lysed with 0.1% Triton X-100 for bacterial release and counting. Data are expressed as the percentage of the initial inoculum. Each isolate was tested in duplicate in two independent experiments.
Quantitative real-time PCR.
Polarized Caco-2 cells were infected apically at an MOI of ∼30:1 and processed as for the invasion assays, but after 1.5 h of incubation with culture medium supplemented with 100 μg/ml of gentamicin, the antibiotic concentration was changed to 10 μg/ml and the cells were incubated for an additional 1.5 h. Then, at 3 h postinfection, the cells were gently washed 3 times with prewarmed PBS and resuspended in TRIzol (Invitrogen) for extraction of total RNA. After reverse transcription with random hexamers and real-time PCR using specific primers, threshold cycle (CT) values were normalized with values of 18S RNA and referred to values of uninfected cells. Total RNA extraction, reverse transcription, and quantitative real-time PCR (qRT-PCR) were carried out as previously reported (43). Mean results of four independent experiments are shown.
PCR screening for motA(T551G) mutation.
From the 266 field isolates subjected to motility tests, those totally devoid of motility were studied further using a specific PCR designed to detect the presence of the motA(T551G) mutation. Isolates with reduced motility compared to the reference were not included in the analysis. In brief, genomic DNA was extracted by resuspending one bacterial colony in 200 μl of sterile milliQ water and boiling the suspension for 5 min. After that, the extract was centrifuged for 5 min at 10,000 ×g, and 0.5 μl of the supernatant used as a template in a PCR using the primer motA2 as a forward primer and motAWT2 as a reverse primer (see Table S1 in the supplemental material). The cycling conditions were as follows: 3 min at 94°C and 30 cycles of 45 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Those strains that gave no product carried a nucleotide in position 551 different from that of the wild type and were further analyzed by PCR with the primers motA2 and motAmut2. In this case the cycling conditions were as described above but the annealing temperature was raised to 66°C to increase specificity. Obtaining a product of 113 bp in size indicated the presence of the motA(T551G) mutation. Those DNAs positive for the mutation were sequenced with primer motA3 for verification of the T551 → G change.
Statistical analysis.
For analysis of differences in motility, invasiveness for Caco-2 cells, and the transcriptional response to the infection, we used the Mann-Whitney U test (GraphPad Prism 4.0 software program), considering a P value of <0.05 (two-tailed) to be statistically significant. For analysis of the frequency of strains positive for the motA(T551G) mutation, we used Fisher's exact test (GraphPad Prism software), considering alpha to be <0.05.
RESULTS
Motility analysis of S. Enteritidis isolates.
Previously, we characterized a collection of 29 Uruguayan S. Enteritidis isolates from human, animal, and environmental sources (43). Ten isolates were impaired for motility, and this correlated with diminished invasion of Caco-2 cells in centrifuge-assisted assays. In this work, we performed a wider screening of motility in 266 natural isolates of S. Enteritidis, including 194 from humans and 72 from other sources, and found that 72 of them (48 from humans and 24 from nonhuman origins) were devoid of motility. Thus, 24.7% of human isolates were nonmotile, compared to 33.3% of the nonhuman isolates. However, this difference was not statistically significant (P = 0.16 in Fisher's exact test).
Characterization of the nonmotile phenotype of isolate 251/01.
One of the nonmotile isolates (strain 251/01), obtained from a nationwide microbiological survey of chicken eggs (5), was selected for further comparative studies with one motile isolate (strain 8/02), obtained from a case of human gastroenteritis. Both strains were isolated during the peak of the S. Enteritidis epidemic in Uruguay and had previously revealed identical genetic profiles with all of the typing methods utilized (4), yet they differed significantly in their ability to invade epithelial cells, survive in egg albumen, and colonize chicken organs (43). The in vitro growth rates of the two strains in rich medium were similar between them and to that of the PT4 reference strain (data not shown).
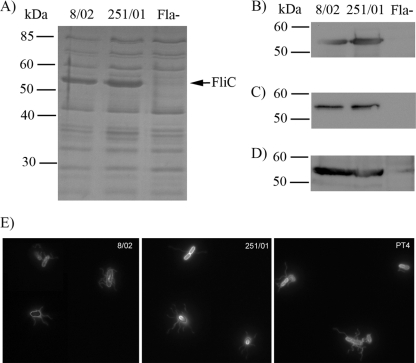
The lack of motility could be due to defects in flagellin synthesis, export, or assembly (in which case no flagellar filament would be observed at all) or in the mechanisms of flagellar motion (in which case the bacteria would harbor structurally normal but paralyzed flagella). To gain further information about this phenotype, we analyzed the protein content of heat extract (HE) fractions of the isolates, which are composed predominantly of flagellar filaments and other surface proteins. As seen in Fig. 1A, the two isolates contained similar amounts of flagellin (FliC) protein on their surfaces, suggesting no differences in FliC production or in assembly of the flagellar filament. FliC levels were further verified by Western blot analyses of HE, secreted proteins, and total protein extracts (Fig. 1B to D). Furthermore, the nucleotide sequences of the fliC gene and its promoter region were identical in both isolates and identical to that of the sequenced S. Enteritidis PT4 strain (data not shown).
Fig. 1.
(A) SDS-PAGE analysis of heat extracts from isolates 8/02 and 251/01. Location of FliC (53 kDa) in the gel is indicated to the right. Western blot analysis of heat extracts (B), secreted proteins (C), or total protein extracts (D) of isolates using antiflagellin antibody. Fla−, a Salmonella isolate lacking flagella. (E) Fluorescent labeling of flagellar filaments in live cells of strain 8/02, 251/01, or PT4.
We further investigated the presence, appearance, and movement of flagella by directly labeling the flagellar filaments with an amino-specific fluorescent dye in live cells (Fig. 1E). The flagellar filaments of isolate 251/01 were readily visualized, and the shape, length, and number were indistinguishable from those of 8/02 or PT4. However, while 8/02 and PT4 cells were actively swimming, no movement of the 251/01 cells was observed (data not shown). This result demonstrates that isolate 251/01 is flagellated but nonmotile, suggesting that the lack of motility in this strain is probably due to impaired function of motor proteins.
Sequence analysis of motor protein-coding genes.
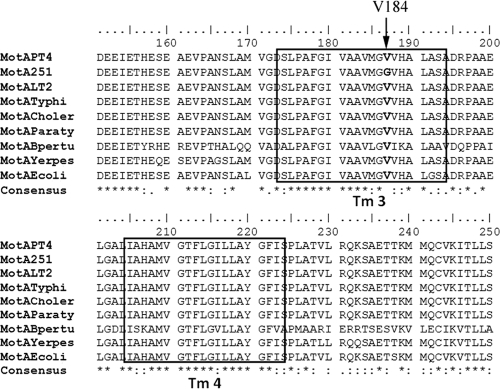
Since motA and motB encode essential components of the flagellar motor, we analyzed the sequence of both genes from isolates 8/02 and 251/01. While motB sequences were identical between the two isolates and to that of the PT4 strain, a T → G substitution was found in nucleotide 551 of motA from isolate 251/01 which changes an amino acid residue of the protein (Val184Gly). This residue is located in a highly conserved region of MotA, comprising one of the four membrane-spanning segments of the protein (Tm3), where several mutations that render the protein nonfunctional were previously described (Fig. 2) (6, 8). We also analyzed the nucleotide sequences of other motor genes (fliG, fliM, and fliN) from isolate 251/01, but no further differences were found compared to PT4. These results indicate that the motility impairment of isolate 251/01 is most likely due to a nonfunctional MotA.
Fig. 2.
Alignment of amino acid residues of MotA from several Gram-negative bacteria using ClustalW2. PT4, S. Enteritidis PT4 P125109; 251, S. Enteritidis isolate 251/01; LT2, S. Typhimurium strain LT2; Typhi, S. Typhi; Choler, S. enterica serovar Choleraesuis; Paraty, S. enterica serovar Paratyphi; Bpertu, Bordetella pertussis; Yerpes, Yersinia pestis; Ecoli, Escherichia coli. The Tm2 and Tm3 transmembrane segments are indicated with squares, and the mutated Val184 in 251/01 is shown in bold.
Genetic complementation of the motA(T551G) mutant.
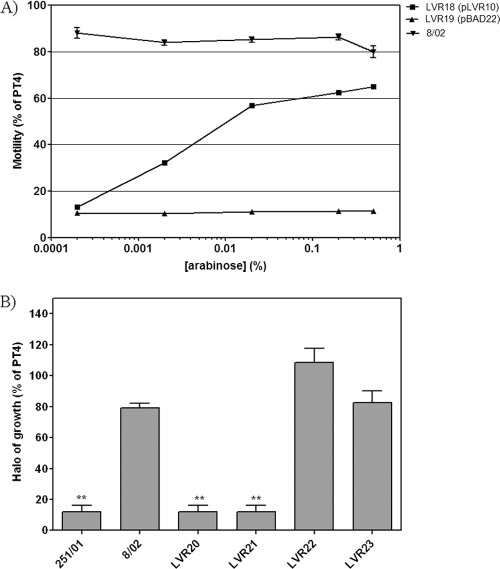
In order to confirm that the lack of motility in isolate 251/01 is due to the motA(T551G) mutation, we performed genetic complementation. Isolate 251/01 was electroporated with plasmid pLVR10, carrying wild-type motA into an arabinose-inducible expression vector, and the resulting strain, LVR18, was subjected to motility assays with increasing concentrations of inducer (Fig. 3A). LVR18 displayed motility in an arabinose-dependent fashion, whereas the 251/01 isolate transformed with the empty vector did not. This demonstrates that the absence of motility in isolate 251/01 is due exclusively to the T551 → G mutation in motA. However, the motility of LVR18 was lower than that of isolate 8/02, a naturally motile strain (Fig. 3A). This could be due to altered expression levels of motA controlled by an heterologous promoter in a high-copy-number plasmid or to a dominant-negative effect of the mutated motA gene present in the chromosome. In this regard, it was previously reported that mutations in the Tm segments of MotA frequently display dominant-negative phenotypes (6, 34). Thus, we deleted the chromosomal motA gene in isolate 251/01 by replacement with a Cmr cassette, resulting in strain LVR20. As a control, the same approach was done in parallel with isolate 8/02, resulting in strain LVR21. As expected, both strains were nonmotile (Fig. 3B). We then introduced the wild-type copy of motA from strain PT4 by P22 transduction into strains LVR20 and LVR21 and selected for motile and Cms strains. The resulting strains, named LVR22 and LVR23, respectively, showed motility levels similar to that of the original isolate, 8/02, as expected (Fig. 3B).
Fig. 3.
Genetic complementation of the motA(T551G) mutant isolate. (A) Motility of isolate 251/01 carrying pLVR10 (motA) or the empty vector in soft agar plates containing increasing concentrations of arabinose (in % [wt/vol]). For comparison, isolate 8/02 was included in the analysis. (B) Motility of isolates 8/02, 251/01, and motA-inactivated and recomplemented derivatives in soft agar plates. Means ± SEM are shown. **, significant difference compared to results for PT4 (P < 0.01).
Motility restoration also restores cell invasiveness in the motA(T551G) mutant.
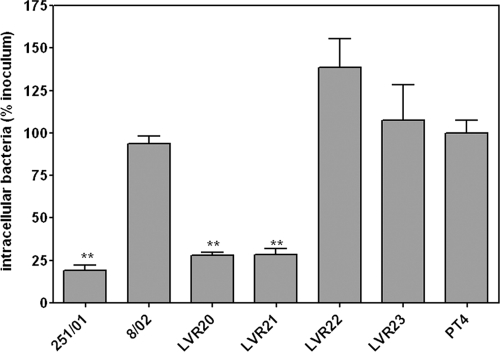
In order to evaluate if the motA mutation was responsible for the impairment in cell invasiveness previously reported for isolate 251/01 (43), we performed Caco-2 invasion assays with strains LVR22 and LVR23 and with the parental isolates and the motA-inactivated ones (Fig. 4). Both LVR22 and LVR23 were as invasive as the PT4 reference strain, whereas strains LVR20 and LVR21 (the motA::cat strains) were as hypoinvasive as the original 251/01 isolate. This indicates that the presence of a mutated motA gene was sufficient to induce the invasion-defective phenotype of isolate 251/01. It is important to note that all invasion assays were done with a mild centrifugation step after infection to synchronize bacterium-cell contact.
Fig. 4.
Nonpolarized Caco-2 invasion assays of isolates 8/02 and 251/01 and derivatives. Data are expressed as percentages of intracellular bacteria related to the initial inoculum and further normalized to PT4 (considering this value equal to 100%). Means ± SEM are shown. **, significant difference compared to results for PT4 (P < 0.01).
The motA(T551G) isolate is able to trigger proinflammatory transcriptional responses in Caco-2 cells.
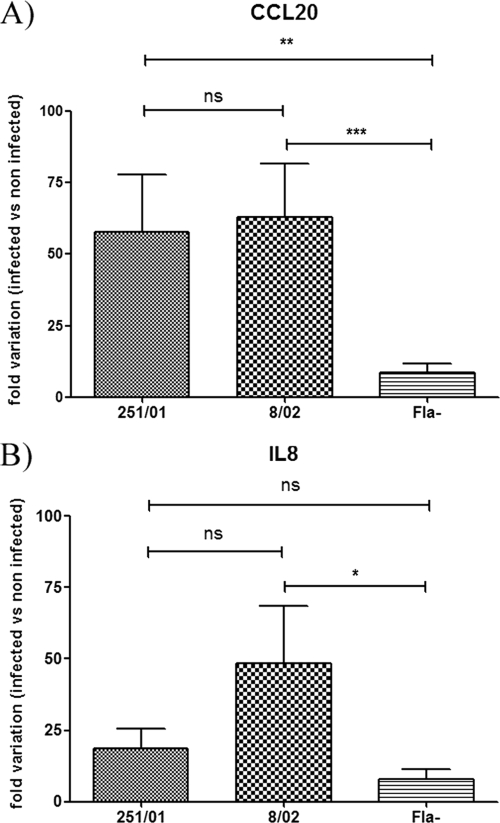
Upon interaction with enterocytes, Salmonella induces an inflammatory response, characterized by secretion of interleukin 8 (IL-8), CCL20 (MIP3A), and several proinflammatory chemokines (44), that recruits neutrophils and dendritic cells into the subepithelial compartment (35, 44). In polarized Caco-2 cells used as model epithelia, it has been demonstrated that upregulation of transcription of the CCL20 and IL8 genes in response to Salmonella infection depends on pattern recognition of flagellin by TLR5 (35). Thus, we tested if the isolate with paralyzed flagella would be able to trigger a response similar to that induced by the motile one. For this, we measured the CCL20 and IL8 mRNA levels from polarized Caco-2 cells infected with isolate 251/01 or 8/02 in comparison with those of uninfected cells (2, 44). Infection with the motA mutant induced levels of transcription of CCL20 similar to those with the motA+ isolate, while a Salmonella isolate completely lacking flagella induced significantly reduced levels of CCL20 transcript (Fig. 5A). Concerning IL8 expression, isolate 251/01 induced lower levels of transcription than isolate 8/02 and higher levels than the aflagellate strain, but these differences were not statistically significant (Fig. 5B). These results suggest that the paralyzed flagella can still drive proinflammatory responses in Caco-2 cells, although with reduced efficiency compared to those with motile flagella.
Fig. 5.
Analysis of the Caco-2 transcriptional response to infection by S. Enteritidis motile or paralyzed isolates. Polarized Caco-2 cells were infected with isolate 251/01 or 8/02, and at 3 h postinfection, the levels of mRNA transcripts for CCL20 (A) or IL8 (B) were measured by qRT-PCR. Values were normalized with values of 18S RNA and referenced to values of uninfected cells. Fla−, a Salmonella isolate lacking flagella; ns, not statistically significant (P > 0.05); *, P < 0.05; **, P < 0.01; ***, P < 0.001.
PCR screening for motA(T551G) mutation.
To investigate the prevalence of the motA(T551G) mutation in the strains circulating in Uruguay, we developed a specific PCR using a forward primer complementary to nucleotides 459 to 478 of motA and two reverse primers complementary to nucleotides 551 to 572, containing either the T551 → G change or the wild-type sequence at the 3′ end, to specifically amplify the mutant or the wild-type sequence, respectively (Fig. 6). We analyzed the 72 nonmotile S. Enteritidis isolates identified in the motility screening (see above) and found in total 15 isolates carrying this mutation (including 251/01). Interestingly, 13 of these isolates were obtained from nonhuman sources, including 7 from eggs, and only 2 were from human sources (Table 2). This corresponds to 18.1% versus 1.0% of the total number of strains isolated from nonhuman or human sources, respectively (P < 0.0001 in Fisher's exact test). This difference is also significant if we consider only the nonmotile isolates from each origin (P < 0.0001). To rule out the possibility that the mutation occurred during prolonged storage of the isolates, we analyzed both replicates of each isolate stored, and the result was the same in both of them.
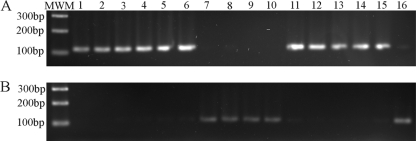
Fig. 6.
PCR screening for motA(T551G) mutation. Colony-extracted DNA was used as a template in a PCR specific for wild-type (A) or motA(T551G) (B) alleles. Amplicon size is 113 bp. (A) PCR using primers motA2/motAWT2, amplifying only wild-type motA alleles. (B) PCR using primers motA2/motAmut2, amplifying only motA(T551G) alleles. Only a fraction of isolates analyzed are shown. Sample 7, isolate 18/97; 8, 19/97; 9, 20/97; 10, 21/97; 14, PT4; 15, 8/02; 16, 251/01.
Table 2.
S. Enteritidis isolates positive for the motA(T551G) mutation in the NSC collection, isolated between 1988 and 2006
| Isolatea | Period of isolation | Sourceb |
|---|---|---|
| 18/97 | Epidemic | Food |
| 19/97 | Epidemic | Food |
| 20/97 | Epidemic | Food |
| 21/97 | Epidemic | Food |
| 11/99 | Epidemic | Human gastroenteritis |
| 48/01 | Epidemic | Food |
| 117/01 | Epidemic | Egg |
| 118/01 | Epidemic | Egg |
| 251/01 | Epidemic | Egg |
| 252/01 | Epidemic | Egg |
| 253/01 | Epidemic | Egg |
| 254/01 | Epidemic | Egg |
| 255/01 | Epidemic | Egg |
| 31/04 | Postepidemic | Food |
| 93/04 | Postepidemic | Human invasive disease |
Strain designations adhere to the following rule: number of isolate/year of isolation.
“Food” refers to any product for human consumption (e.g., cake or sandwich), with the exception of eggs.
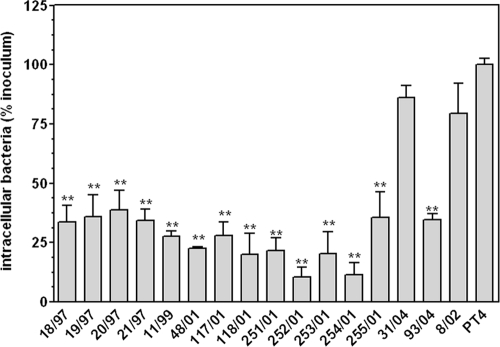
All isolates positive for the mutation in the PCR carried the T551 → G mutation, as confirmed by DNA sequencing. We tested all the motA(T551G) isolates in Caco-2 invasion assays, and all except one, the nonhuman isolate 31/04, were hypoinvasive compared to PT4 (Fig. 7).
Fig. 7.
Nonpolarized Caco-2 invasion assays of motA(T551G) isolates. Isolate 8/02, carrying wild-type motA, was included for comparison, as was PT4, used as a reference strain. Data are expressed as percentages of intracellular bacteria related to the initial inoculum and further normalized to the result for PT4 (considering this value equal to 100%). Means ± SEM are shown. **, significant difference compared to result for PT4 (P < 0.01).
DISCUSSION
Here we report the identification of a missense mutation in motA from S. Enteritidis field isolates, which is unevenly distributed among isolates according to their source. We show that the prevalence of this mutation is significantly higher in isolates from nonhuman sources than in those from clinical human sources. To our knowledge, this is the first time that a mutation in this gene has been identified in natural isolates of Salmonella. Since many of these bacteria have been stored for more than 10 years, it could be argued that the mutation may have appeared during the storage period. However, this would not explain why it is found with much more frequency among isolates derived from nonhuman sources than in those from human sources. Further, we confirmed the presence of the mutation in both replicate cryovials of each isolate, which suggests that the possibility that the mutation arose during storage is negligible. Since undirected genetic mutation arises in Salmonella in response to environmental stressors (25), this mutation could have occurred spontaneously, and its presence did not prevent the mutant bacteria from surviving in the environment or from infecting chickens. In fact, the avian-adapted Salmonella enterica Gallinarum serotype is nonmotile. The fact that the same mutation was found in strains isolated during various periods of time (Table 2) suggests that its appearance is not an unlikely event or alternatively that the same clone has persisted for long periods of time.
An isolate carrying this mutation assembled normal flagella but had totally lost motility and showed reduced invasion of Caco-2 epithelial cells in culture compared to a motile isolate. However, the ability to attach to cells was previously shown to be comparable between both isolates (43). These results are in line with those reported for nonmotile insertional mutant strains of S. Enteritidis (41) and S. Typhimurium (32) and suggest a role for motile flagella in invasion but not in adhesion to epithelial cells.
It has been previously reported that in nonflagellated S. Typhimurium, the impairment in cell invasion can be largely reversed by applying a centrifugal force upon infection (19, 20), while no increase in invasiveness after centrifugation is observed for Salmonella enterica serovar Typhi that is lacking flagella or motility (23), suggesting the existence of differences in the entry mechanisms between these serovars. For S. Enteritidis, it was demonstrated that promoting contact between bacteria and cells by application of centrifugal force greatly enhances invasion but that this is not enough to bring the levels of invasion of a nonmotile strain up to those of a motile one (11, 41, 43). Moreover, the presence of paralyzed flagella seems to be more detrimental for epithelial cell invasion than a total absence of flagella (41), which could be explained by steric hindrance caused by the paralyzed flagella (19). All these data suggest a role for functional flagella in addition to the facilitation of approximation of bacteria to the apical surface of the host cell.
We were able to assign the paralyzed phenotype of isolate 251/01 to a nonfunctional mutation in motA [motA(T551G)]. The introduction of a wild-type copy of motA in the chromosome of isolate 251/01 reverted the paralyzed phenotype and rendered the bacteria fully invasive in Caco-2 cells, indicating that the absence of motility is responsible for the impairment of invasion seen in this isolate.
One crucial step in the enteropathogenesis of Salmonella is its ability to trigger a proinflammatory response from the intestinal epithelium. We found that the motA mutant isolate elicited proinflammatory transcriptional responses in polarized Caco-2 cells, indicating that the paralyzed flagella, although being associated with a significant reduction in the invasiveness of the strain, are still able to signal through TLR5 in this model epithelium. These results are consistent with those reported by Winter et al. (42), who demonstrated that an S. Typhimurium flgK mutant, which is unable to assemble flagella and is consequently nonmotile but can still secrete flagellin, is as invasive as an flgK fliC fljB mutant (a non-flagellin-expressing mutant) and is significantly less invasive than the wild type in bovine ligated ileal loops. However, the flgK mutant elicited significantly more fluid secretion and MIP3A (CCL20) gene expression than the flgK fliC fljB mutant, suggesting that invasion-independent flagellin pattern recognition contributes to diarrhea during the early phase of S. Typhimurium infection in calves (42).
Data about epidemiology and motility in Salmonella are scarce. Grossman et al. (13) reported an association between decreased motility and decreased severity of illness in clinical isolates of S. Typhi from Indonesia. More recently, it was reported that impaired motility in four poultry-associated isolates of S. Enteritidis was associated with low invasiveness in differentiated Caco-2 cells and reduced virulence in the mouse typhoid model (33). In the present work, we screened a comprehensive collection of isolates derived from human clinical, animal, and environmental sources and found that 72 out of 266 (27%) showed total absence of motility. A specific survey for the motA(T551G) mutation revealed its presence in 15 isolates obtained during the epidemic and postepidemic periods in Uruguay, with it being recovered mainly from animal sources, including eggs, rather than from human sources. Thus, it can be hypothesized that the presence of proinflammatory but paralyzed flagella impairs the ability of S. Enteritidis to cause intestinal disease in the human host but does not prevent Salmonella infections of chickens nor egg contamination. Supporting this view, it has been reported that a motAB::cat mutant of S. Enteritidis, with paralyzed flagella, was recovered from livers and spleens of chickens in numbers similar to those of the wild type (1). Further, our previous results demonstrated that two motA(T551G) S. Enteritidis isolates (251/01 and 254/01) were capable of colonizing the spleens and reproductive tracts of 3 day-old-chicks (43). However, both isolates differed extensively in their chicken invasiveness (43), demonstrating that with natural isolates the picture is more complex than that obtained with artificially constructed mutants. To the best of our knowledge, there are no reports studying the capability of Salmonella-paralyzed mutants to infect eggs, but the fact that 7 out of the 15 motA mutants were isolated from eggs supports the hypothesis that this mutation does not prevent S. Enteritidis from contaminating them.
In mice, it has been reported that motility allows S. Typhimurium to benefit from the nutrients released in the context of an inflamed gut by increasing the growth rate and improving pathogen fitness to compete with the intestinal microbiota (37). We speculate that in the human host, the motA mutants would not be able to cause disease, although they may elicit a proinflammatory response, because the lack of motility prevents them from efficient access to the localized high-energy nutrients released upon inflammation, avoiding efficient gut colonization. In addition, the diminished ability of the motA isolate to enter intestinal epithelial cells may account for a reduced proinflammatory response in the intestine and reduced pathogenesis.
The hypothesis that paralyzed S. Enteritidis mutants, though able to colonize chickens and infect eggs, would be impaired in causing disease in humans is challenging. It could be the result of the different ways the mammalian and avian hosts respond to Salmonella infection. In this regard, it has been reported that oral infection with S. Typhimurium induces upregulation of the TLR15 receptor in chicken ceca (17). Interestingly, TLR15 is an avian-specific TLR which is therefore absent in mammals, suggesting that different pathways are involved in the host response to Salmonella infection.
In conclusion, in this study we identified 72 natural isolates of S. Enteritidis lacking motility from a comprehensive collection of 266 epidemic-spanning isolates and determined the presence of paralyzed flagella as the cause of this phenotype in 15 of them, derived mainly from nonhuman sources. The cause of the absence of motility in the remaining 57 nonmotile isolates remains to be determined.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a project grant from the Wellcome Trust (078168/Z/05/Z) and from Comisión Sectorial para la Investigación Científica (CSIC), Uruguay, I+D 2008.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Allen-Vercoe E., Sayers A. R., Woodward M. J. 1999. Virulence of Salmonella enterica serotype Enteritidis aflagellate and afimbriate mutants in a day-old chick model. Epidemiol. Infect. 122:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderle P., et al. 2005. Novel markers of the human follicle-associated epithelium identified by genomic profiling and microdissection. Gastroenterology 129:321–327 [DOI] [PubMed] [Google Scholar]
- 3. Berg H. C. 2003. The rotary motor of bacterial flagella. Annu. Rev. Biochem. 72:19–54 [DOI] [PubMed] [Google Scholar]
- 4. Betancor L., et al. 2009. Genomic and phenotypic variation in epidemic-spanning Salmonella enterica serovar Enteritidis isolates. BMC Microbiol. 9:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Betancor L., et al. 2010. Prevalence of Salmonella enterica in poultry and eggs in Uruguay during an epidemic due to Salmonella enterica serovar Enteritidis. J. Clin. Microbiol. 48:2413–2423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blair D. F., Berg H. C. 1991. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J. Mol. Biol. 221:1433–1442 [DOI] [PubMed] [Google Scholar]
- 7. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 8. Braun T. F., Al-Mawsawi L. Q., Kojima S., Blair D. F. 2004. Arrangement of core membrane segments in the MotA/MotB proton-channel complex of Escherichia coli. Biochemistry 43:35–45 [DOI] [PubMed] [Google Scholar]
- 9. Brown A., et al. 1987. An attenuated aroA Salmonella typhimurium vaccine elicits humoral and cellular immunity to cloned beta-galactosidase in mice. J. Infect. Dis. 155:86–92 [DOI] [PubMed] [Google Scholar]
- 10. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dibb-Fuller M. P., Allen-Vercoe E., Thorns C. J., Woodward M. J. 1999. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145(Pt. 5):1023–1031 [DOI] [PubMed] [Google Scholar]
- 12. Dower W. J., Miller J. F., Ragsdale C. W. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grossman D. A., et al. 1995. Flagellar serotypes of Salmonella typhi in Indonesia: relationships among motility, invasiveness, and clinical illness. J. Infect. Dis. 171:212–216 [DOI] [PubMed] [Google Scholar]
- 14. Guard-Petter J. 2001. The chicken, the egg and Salmonella enteritidis. Environ. Microbiol. 3:421–430 [DOI] [PubMed] [Google Scholar]
- 15. Guzman L. M., Belin D., Carson M. J., Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hanahan D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557–580 [DOI] [PubMed] [Google Scholar]
- 17. Higgs R., et al. 2006. Induction of a novel chicken Toll-like receptor following Salmonella enterica serovar Typhimurium infection. Infect. Immun. 74:1692–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Humphrey T. J. 1994. Contamination of egg shell and contents with Salmonella enteritidis: a review. Int. J. Food Microbiol. 21:31–40 [DOI] [PubMed] [Google Scholar]
- 19. Jones B. D., Lee C. A., Falkow S. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jones G. W., Richardson L. A., Uhlman D. 1981. The invasion of HeLa cells by Salmonella typhimurium: reversible and irreversible bacterial attachment and the role of bacterial motility. J. Gen. Microbiol. 127:351–360 [DOI] [PubMed] [Google Scholar]
- 21. Keller L. H., Benson C. E., Krotec K., Eckroade R. J. 1995. Salmonella enteritidis colonization of the reproductive tract and forming and freshly laid eggs of chickens. Infect. Immun. 63:2443–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kojima S., Blair D. F. 2004. The bacterial flagellar motor: structure and function of a complex molecular machine. Int. Rev. Cytol. 233:93–134 [DOI] [PubMed] [Google Scholar]
- 23. Liu S. L., Ezaki T., Miura H., Matsui K., Yabuuchi E. 1988. Intact motility as a Salmonella typhi invasion-related factor. Infect. Immun. 56:1967–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Macnab R. M. 1992. Genetics and biogenesis of bacterial flagella. Annu. Rev. Genet. 26:131–158 [DOI] [PubMed] [Google Scholar]
- 25. Massey R. C., Rainey P. B., Sheehan B. J., Keane O. M., Dorman C. J. 1999. Environmentally constrained mutation and adaptive evolution in Salmonella. Curr. Biol. 9:1477–1480 [DOI] [PubMed] [Google Scholar]
- 26. Miao E. A., Andersen-Nissen E., Warren S. E., Aderem A. 2007. TLR5 and Ipaf: dual sensors of bacterial flagellin in the innate immune system. Semin. Immunopathol. 29:275–288 [DOI] [PubMed] [Google Scholar]
- 27. Miller V. L., Mekalanos J. J. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Minamino T., Imada K., Namba K. 2008. Molecular motors of the bacterial flagella. Curr. Opin. Struct. Biol. 18:693–701 [DOI] [PubMed] [Google Scholar]
- 29. Nicholas R. A., Cullen G. A. 1991. Development and application of an ELISA for detecting antibodies to Salmonella enteritidis in chicken flocks. Vet. Rec. 128:74–76 [DOI] [PubMed] [Google Scholar]
- 30. Ramos H. C., Rumbo M., Sirard J. C. 2004. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12:509–517 [DOI] [PubMed] [Google Scholar]
- 31. Robertson J. M., et al. 2000. Adhesion of Salmonella enterica var Enteritidis strains lacking fimbriae and flagella to rat ileal explants cultured at the air interface or submerged in tissue culture medium. J. Med. Microbiol. 49:691–696 [DOI] [PubMed] [Google Scholar]
- 32. Schmitt C. K., et al. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619–5625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shah D. H., et al. 2011. Cell invasion of poultry-associated Salmonella Enteritidis isolates is associated with pathogenicity, motility and secretion of type-three secretion system secreted proteins. Microbiology 157:1428–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharp L. L., Zhou J., Blair D. F. 1995. Features of MotA proton channel structure revealed by tryptophan-scanning mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 92:7946–7950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sierro F., et al. 2001. Flagellin stimulation of intestinal epithelial cells triggers CCL20-mediated migration of dendritic cells. Proc. Natl. Acad. Sci. U. S. A. 98:13722–13727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sowa Y., Berry R. M. 2008. Bacterial flagellar motor. Q. Rev. Biophys. 41:103–132 [DOI] [PubMed] [Google Scholar]
- 37. Stecher B., et al. 2008. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol. 10:1166–1180 [DOI] [PubMed] [Google Scholar]
- 38. Stecher B., et al. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:4138–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomson N. R., et al. 2008. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome Res. 18:1624–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Turner L., Ryu W. S., Berg H. C. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182:2793–2801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van Asten F. J., Hendriks H. G., Koninkx J. F., van Dijk J. E. 2004. Flagella-mediated bacterial motility accelerates but is not required for Salmonella serotype Enteritidis invasion of differentiated Caco-2 cells. Int. J. Med. Microbiol. 294:395–399 [DOI] [PubMed] [Google Scholar]
- 42. Winter S. E., et al. 2009. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype Typhimurium infection. Infect. Immun. 77:1904–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yim L., et al. 2010. Differential phenotypic diversity among epidemic-spanning Salmonella enterica serovar Enteritidis isolates from humans or animals. Appl. Environ. Microbiol. 76:6812–6820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zeng H., et al. 2003. Flagellin is the major proinflammatory determinant of enteropathogenic Salmonella. J. Immunol. 171:3668–3674 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.