Abstract
Parasites from the Cryptosporidium genus are the most common cause of waterborne disease around the world. Successful management and prevention of this emerging disease requires knowledge of the diversity of species causing human disease and their zoonotic sources. This study employed a spatiotemporal approach to investigate sporadic human cryptosporidiosis in New South Wales, Australia, between January 2008 and December 2010. Analysis of 261 human fecal samples showed that sporadic human cryptosporidiosis is caused by four species; C. hominis, C. parvum, C. andersoni, and C. fayeri. Sequence analysis of the gp60 gene identified 5 subtype families and 31 subtypes. Cryptosporidium hominis IbA10G2 and C. parvum IIaA18G3R1 were the most frequent causes of human cryptosporidiosis in New South Wales, with 59% and 16% of infections, respectively, attributed to them. The results showed that infections were most prevalent in 0- to 4-year-olds. No gender bias or regional segregation was observed between the distribution of C. hominis and C. parvum infections. To determine the role of cattle in sporadic human infections in New South Wales, 205 cattle fecal samples were analyzed. Four Cryptosporidium species were identified, C. hominis, C. parvum, C. bovis, and C. ryanae. C. parvum subtype IIaA18G3R1 was the most common cause of cryptosporidiosis in cattle, with 47% of infections attributed to it. C. hominis subtype IbA10G2 was also identified in cattle isolates.
INTRODUCTION
Cryptosporidium species are capable of initiating gastrointestinal disease in over 200 vertebrate species from various taxonomic groups, including fish, birds, mammals, and reptiles (5). The emergence of human cryptosporidiosis in the mid-1980s coincided with the human immunodeficiency virus (HIV) era. Initially, it was considered a disease limited to the immunocompromised, and due to the strong link with HIV, it was used as an initial diagnosis of the virus (20). However, over the last 20 years, cryptosporidia have emerged as significant human pathogens with a global distribution, capable of causing illness in both immune-compromised and immunocompetent individuals. Cryptosporidiosis is a primary concern for water and health authorities, in addition to the livestock industry, which suffers significant economic losses from diseased animals.
The genetic heterogeneity exhibited within the Cryptosporidium genus has been highlighted by molecular analyses, which are essential for the differentiation of Cryptosporidium species. To date, 22 species and more than 40 genotypes of Cryptosporidium have been described (5, 17). DNA analysis of Cryptosporidium parasites from humans has shown the anthroponotic C. hominis and the zoonotic C. parvum to be the most common causes of human cryptosporidial infections, with 90% of reported cases attributed to them (11). However, due to the sensitivity of molecular analyses, in conjunction with the growing number of human samples analyzed, eight additional species have been identified as a public health threat, including C. meleagridis, C. felis, C. canis, C. suis, C. muris, C. fayeri, C. ubiquitum, and C. cuniculus (3, 17, 21). The contribution of these species to human disease varies globally and is often associated with seasonality, demographics, immune status, and contact with reservoir hosts.
Further intraspecies variation has been observed in Cryptosporidium isolates through sequence analysis of the hypervariable gp60 gene, which further classifies species to the subtype family and subtype levels. Sequence variation observed in the gp60 gene has identified 6 C. hominis subtype families (designated with the Roman numeral I) and 11 C. parvum subtype families (Roman numeral II) (24). Six subtype families have also been identified in C. meleagridis (Roman numeral III) and C. fayeri (Roman numeral IV) (15, 16). Within a microsatellite region of the gp60 gene, variation in the number and form of serine codons further characterizes Cryptosporidium to the subtype level (18). Molecular analysis of the gp60 gene has facilitated the identification of transmission pathways and zoonotic disease contamination sources and highlighted the importance of certain genetic variants to human health. For example, analysis of the gp60 gene has shown that cattle are an important zoonotic source for human disease. In Australia, C. parvum IIaA18G3R1 is the dominant subtype infecting cattle. The identification of IIaA18G3R1 in humans was first reported in Australia, and it has now become the dominant C. parvum subtype causing Australian sporadic cryptosporidiosis (9, 14, 22). Cattle zoonotic sources have also been shown in Portugal, where C. parvum IIaA15G2R1 is the most common subtype in cattle and humans (1). Subtype classification has also highlighted the public health risk posed by particular Cryptosporidium subtypes. The C. hominis IbA10G2 subtype is a globally distributed subtype and is the most common cause of waterborne outbreaks and sporadic human cryptosporidiosis (8, 24).
Cryptosporidiosis has been a notifiable disease in New South Wales (NSW) since 1996 (12). Notifications data have shown a significant rise in the incidence of human cryptosporidiosis in New South Wales. In 2003, the overall incidence of human cryptosporidiosis was 2.7/100,000 population, in 2006 it had risen to 10.5/100,000, and in 2009, the incidence further increased, to 19.8/100,000 (www.health.nsw.gov.au). Previous studies conducted on sporadic human cryptosporidiosis in New South Wales show that C. hominis and C. parvum contribute equally to disease, with C. hominis IbA10G2 and C. parvum IIaA18G3R1 identified as the most common subtypes (22). Here, we perform a longitudinal investigation into sporadic human cryptosporidiosis in New South Wales and examine transmission pathways and demographic groups most at risk of disease. This knowledge is essential for the management of cryptosporidiosis in Australia and for a global understanding of the disease impacts of this parasite.
MATERIALS AND METHODS
Sample sources, parasite enumeration, and DNA extraction.
Two hundred five fecal samples were collected from beef and dairy cattle throughout New South Wales. In total, calves from seven dairy farms, one in Camden (n = 75) and six in Wagga Wagga (farm 1, n = 14; farm 2, n = 10; farm 3, n = 19; farm 4, n = 10; farm 5, n = 7; farm 6, n = 10; total n = 70), were sampled. In addition, one beef farm in Richmond (n = 60) was also investigated. Typical for beef cattle, adults and juveniles were housed in a mixed pen; because of this, the age of cattle was not determined. Sampling occurred in the spring of 2010 with the exception of the Camden farm, which was also sampled in 2008. DNA extraction from cattle fecal samples was performed using a Bioline isolate fecal DNA kit (Bioline, Sydney, Australia), following the manufacturer's instructions.
Four hundred forty-seven human fecal samples that were positive for Cryptosporidium were obtained from hospitals and pathology companies in New South Wales, Australia, between January 2008 and December 2010. Samples collected between January and April 2009 were attributed to a waterborne outbreak in New South Wales and were excluded from this sporadic cryptosporidiosis study. Oocysts were purified from feces using a sucrose flotation gradient (19), and DNA was extracted from purified oocysts using PrepGem (Zygem Corporation Ltd., Hamilton, New Zealand) (7). Oocysts were fluorescently stained with the Cryptosporidium-specific antibody CRY104 labeled with fluorescein isothiocyanate (FITC; Biotech Frontiers, Sydney, Australia) and enumerated by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences, Sydney, Australia) (2).
Cryptosporidium species identification.
Cryptosporidium species were identified using a previously described PCR-restriction fragment length polymorphism (RFLP) protocol targeting an 18S rRNA gene fragment (25). The primary and secondary reaction mixtures contained 6 mM MgCl2, 200 μM deoxynucleoside triphosphates (dNTPs), 200 nM each primer, and 1 U of Red Hot Taq DNA polymerase (Thermo Scientific, Australia). Two microliters of the DNA template was used in the primary reaction mixture, and 1 μl of the primary PCR product was used as the template in the secondary reaction mixture. The reaction conditions comprised an initial denaturation of 94°C for 3 min, followed by 35 cycles of 94°C for 45 s, 56°C for 45 s, and 72°C for 1 min, with a final extension step of 72°C for 7 min. All PCRs performed included a negative control containing PCR water and a positive control containing C. parvum DNA. Reactions were run on Eppendorf Mastercycler personal instruments (Eppendorf, North Ryde, Australia). PCR products were resolved by electrophoresis using a 2% (wt/vol) agarose gel containing Sybr safe (Invitrogen, Mulgrave, Australia) and visualized under UV light.
Secondary products of the correct size (∼830 bp) were subjected to RFLP analysis. Restriction analysis of Cryptosporidium 18S rRNA gene amplicons from cattle was performed using a previously described protocol with the restriction enzymes SspI and MboII (New England BioLabs) (6). Restriction analysis of 18S rRNA gene amplicons from human samples was performed using a previously described protocol with the restriction enzyme VspI (25). Restriction fragments were separated on 3.5% (wt/vol) agarose gels containing Sybr safe at 100 V, and patterns were visualized under UV light.
gp60 amplification.
Cryptosporidium subtype family and subtype identification were determined using a previously described nested PCR targeting the gp60 gene (22). The primary and secondary reaction mixtures contained 4 mM MgCl2, 200 nM dNTPs, 200 nM each forward and reverse primer, and 1 U of Red Hot Taq polymerase. The primary PCR mixtures contained 2 μl of DNA template, and the secondary reaction mixtures used 1 μl of the primary PCR product as the DNA template. The reaction conditions comprised an initial denaturation at 94°C for 3 min followed by 35 cycles of 94°C for 45 s, 58°C for 45 s, and 72°C for 1 min 30 s, with a final extension at 72°C for 7 min. The PCR products were separated by 2% (wt/vol) agarose gel containing Sybr safe and were visualized under UV light. Products containing the correct size fragment (∼1,000 bp) were purified using a QIAquick PCR purification kit (Qiagen, Melbourne, Australia), following the manufacturer's instructions, and sequenced using an ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, California) with a BigDye Terminator kit (Applied Biosystems).
Nucleotide sequences were analyzed using Geneious version 4.8.2 (Biomatters Ltd., Auckland, New Zealand). Isolates were assigned a subtype according to the nomenclature system as described previously (18).
Parasite enumeration and cloning of C. hominis IbA10G2 from cattle.
Oocysts from three cattle fecal samples which contained C. hominis IbA10G2 were purified using a sucrose flotation gradient method (19) and enumerated by flow cytometry using a FACSCalibur flow cytometer (BD Biosciences) after fluorescent staining with CRY104-FITC (Biotech Frontiers) (2). Purified gp60 PCR products were cloned using the TOPO-TA vector cloning system (Invitrogen, Australia), and plasmid DNA was recovered using the Qiagen plasmid kit (Qiagen, Melbourne, Australia). Sequencing was performed using an ABI 3130xl genetic analyzer (Applied Biosystems, Foster City, California) with a BigDye Terminator kit (Applied Biosystems).
Patient information and spatial analysis.
Patient data (age, gender, and residential postal code) for each sample were obtained from NSW Health. Spatial analyses were achieved using Esri ArcGIS version 10.0 (http://esriaustralia.com.au/esri/default.html) in conjunction with New South Wales digital postal boundary postcodes 2006 (http://www.abs.gov.au/AUSSTATS/abs@.nsf/DetailsPage/2923.0.30.0012006). To examine the spatial distribution of sporadic cases, the numbers of patients infected with the various Cryptosporidium subtypes were mapped. Cattle farms sampled in this study were also incorporated in the spatial analysis. The temporal analyses were performed using the pathology date provided for each sample; a map was then generated for each year of the study, as well as for each of the four seasons.
Statistical analysis.
To determine if a gender or age bias occurred in sporadic cryptosporidiosis, a chi-squared analysis was performed.
Nucleotide sequence accession numbers.
Cryptosporidium gp60 nucleotide sequences generated from cattle were submitted to GenBank under accession numbers JF727776 to JF727779. gp60 nucleotide sequences from human Cryptosporidium infections were submitted to GenBank under accession numbers JF727783 to JF727809. Cloned C. hominis gp60 sequences from cattle were submitted to GenBank under accession numbers JF727780 to JF727782.
RESULTS
Cryptosporidium species and subtypes in cattle.
Cattle fecal samples exhibiting positive PCR products for the target 18S rRNA gene fragment were obtained for 62/205 (30%) samples. RFLP analysis showed that of those, 45/62 (73%) cattle were infected with C. parvum, 11/62 (18%) with C. bovis, 3/62 (5%) with C. hominis, and 1/62 (2%) with C. ryanae. Mixed infections with C. parvum/C. bovis and C. ryanae/C. bovis were both reported in a single cattle sample (Table 1). Cryptosporidium parvum was the most common species detected on all farms except Wagga Wagga farms 5 and 6, where the C. bovis incidence was higher. Cryptosporidium bovis and C. ryanae were not detected on the Camden farm in 2008 but were present in 2010. Cryptosporidium hominis was identified on two farms, Richmond (n = 2) and Wagga Wagga farm 6 (n = 1). To determine whether cattle positive for C. hominis were shedding oocysts, oocyst loads were determined by epifluorescence microscopy (EFM), which showed that the Richmond cattle contained parasite loads of 300 and 148 oocysts/g of feces and the Wagga Wagga farm 6 cow was shedding 50 oocysts/g of feces.
Table 1.
Summary of the Cryptosporidium species and subtypes identified in cattle in New South Wales, Australia
| Cryptosporidium sp. | gp60 subtype | No. of isolates in samples from indicated farm |
Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Richmond, beef (n = 60) | Camden |
Wagga Wagga farm no.: |
|||||||||
| 2008, dairy (n = 11) | 2010, dairy (n = 64) | 1, dairy (n = 14) | 2, dairy (n = 10) | 3, dairy (n = 19) | 4, dairy (n = 10) | 5, dairy (n = 7) | 6, dairy (n = 10) | ||||
| C. parvum | IIaA16G3R1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| IIaA17G4R1 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | |
| IIaA18G3R1 | 0 | 6 | 9 | 2 | 4 | 8 | 0 | 0 | 0 | 29 | |
| IIaA20G3R1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | |
| Not identified | 5 | 0 | 0 | 0 | 1 | 1 | 3 | 0 | 0 | 10 | |
| C. parvum subtotal | 4 | 5 | 8 | 12 | 2 | 5 | 9 | 4 | 0 | 0 | 45 |
| C. hominis | IbA10G2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
| C. bovis | Not applicablea | 0 | 0 | 6 | 0 | 1 | 0 | 1 | 1 | 2 | 11 |
| C. ryanae | Not applicable | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| C. parvum/C. bovis | IIaA20G3R1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| C. ryanae/C. bovis | Not applicable | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Total | 5 | 7 | 8 | 20 | 2 | 6 | 9 | 6 | 1 | 3 | 62 |
gp60 subtype identification is not performed on all Cryptosporidium species.
Amplification of the gp60 gene was successful for 39/49 (80%) C. parvum and C. hominis samples typed at the 18S rRNA gene (Table 1). Two subtype families were detected in the cattle samples, C. parvum IIa (36/39) and C. hominis Ib (3/39). These two families comprised five subtypes, with the most common subtype, IIaA18G3R1, identified in 29/39 (74%) isolates. Cryptosporidium hominis gp60 products were cloned to confirm this finding. Sequence analysis of C. hominis identified subtype IbA10G2 in all 3 isolates.
Cryptosporidium species and subtypes and parasite enumeration in humans.
PCR products that were positive for the 18S rRNA gene were obtained for 261/447 (58%) fecal samples. RFLP analysis showed the presence of four species, C. hominis (172/261, 66%), C. parvum (87/261, 33%), C. andersoni (1/261, 0.5%), and C. fayeri (1/261, 0.5%) (21).
Amplification of the gp60 gene was successful for 245/261 (94%) isolates successfully typed at the 18S rRNA gene (Table 2). Three C. hominis subtype families were identified, Ia (4/245), Ib (156/245), and If (4/245). Analysis of the microsatellite serine region identified 11 C. hominis subtypes. The most commonly identified subtype, identified in 147/245 (60%) isolates, was C. hominis IbA10G2. Within C. parvum isolates, two subtype families were identified: IIa (79/245) and IId (1/245). Twenty C. parvum subtypes were detected, the most common being IIaA18G3R1, which was present in 37/245 (15%) isolates. Typing of the C. fayeri isolate at the gp60 locus identified subtype IVaA10G3T1R1 (21).
Table 2.
Summary of oocyst numbers in feces and the Cryptosporidium species and subtypes causing sporadic human illness from January 2008 to December 2010 in New South Wales, Australia
| Cryptosporidium species | gp60 subtype | No. of cases | Oocysts/g feces |
|||
|---|---|---|---|---|---|---|
| ≤102 | 103 to 104 | 105 to 106 | ≥107 | |||
| C. hominis | IaA11R1 | 1 | 0 | 1 | 0 | 0 |
| IaA13R1 | 1 | 0 | 1 | 0 | 0 | |
| IaA14R1 | 1 | 0 | 0 | 1 | 0 | |
| IaA32R1 | 1 | 0 | 1 | 0 | 0 | |
| IbA6G3 | 4 | 1 | 0 | 3 | 0 | |
| IbA7G3 | 1 | 0 | 0 | 1 | 0 | |
| IbA9G3 | 4 | 0 | 2 | 2 | 0 | |
| IbA10G2 | 147 | 19 | 53 | 71 | 4 | |
| IfA14G1 | 1 | 0 | 0 | 1 | 0 | |
| IfA19G1 | 1 | 0 | 1 | 0 | 0 | |
| IfA20G1 | 2 | 0 | 1 | 1 | 0 | |
| Subtotal | 11 | 164 | 20 | 60 | 80 | 4 |
| C. parvum | IIaA10G3 | 2 | 0 | 0 | 2 | 0 |
| IIaA14G3 | 1 | 0 | 0 | 1 | 0 | |
| IIaA14G3R1 | 1 | 0 | 0 | 1 | 0 | |
| IIaA15G1R1 | 1 | 0 | 1 | 0 | 0 | |
| IIaA15G2R1 | 1 | 0 | 1 | 0 | 0 | |
| IIaA15G4R1 | 1 | 1 | 0 | 0 | 0 | |
| IIaA16G3R1 | 7 | 0 | 3 | 4 | 0 | |
| IIaA17G2R1 | 3 | 0 | 2 | 1 | 0 | |
| IIaA17G3R1 | 1 | 0 | 0 | 1 | 0 | |
| IIaA17G4R1 | 3 | 0 | 1 | 2 | 0 | |
| IIaA18G3R1 | 37 | 0 | 15 | 20 | 2 | |
| IIaA19G2R1 | 3 | 0 | 2 | 1 | 0 | |
| IIaA19G3R1 | 5 | 0 | 2 | 3 | 0 | |
| IIaA19G4R1 | 3 | 1 | 1 | 1 | 0 | |
| IIaA20G3R1 | 4 | 2 | 1 | 0 | 1 | |
| IIaA20G5R1 | 2 | 0 | 1 | 1 | 0 | |
| IIaA21G3R1 | 1 | 0 | 1 | 0 | 0 | |
| IIaA22G3R1 | 2 | 0 | 1 | 1 | 0 | |
| IIaA23G3R1 | 1 | 0 | 0 | 0 | 1 | |
| IIdA24G1 | 1 | 0 | 0 | 1 | 0 | |
| Subtotal | 20 | 80 | 4 | 32 | 40 | 4 |
| C. fayeri | IVaA10G3T1R1 | 1 | 0 | 0 | 1 | 0 |
| Subtotal | 1 | 1 | 0 | 0 | 1 | 0 |
| Total | 32 | 245 | 24 | 92 | 121 | 8 |
The parasite counts (oocysts/g), determined by flow cytometry, ranged between ≤102 and ≥107 oocysts/g in both C. parvum- and C. hominis-infected patients (Table 2). The majority of C. hominis IbA10G2 and C. parvum IIaA18G3R1 cases were shedding oocysts at numbers between 105 and 106 oocysts/g.
Age and gender of patients infected with Cryptosporidium.
Information on patient age and gender was obtained for 216/261 (83%) samples. The results showed that the genders were affected approximately equally (chi square = 1.5, df = 1, P value = 0.247), with 117/216 (54%) patients being female and 99/216 (46%) male (Table 3). Cryptosporidiosis was bimodal, with the 0- to 9-year and 25- to 39-year age categories showing the highest incidence. Overall, the 0- to 4-year age group was the most commonly affected group for both genders, with 42/117 (36%) of the female (chi square = 168.27, df = 1, P ≪ 0.01) and 51/99 (52%) of the male (chi square = 297.07, df = 1, P ≪ 0.01) infections. Combined, the 0- to 4-year age group had 93/216 (43%) of all Cryptosporidium infections, and both females (78/117, 67%) and males (67/99, 68%) were most likely to be infected with C. hominis; this observation was seen in all age groups except for the 20- to 24-, 45- to 49-, and 50- to 54-year groups, where the numbers were low.
Table 3.
Age and gender distribution of sporadic Cryptosporidium infections January 2008 to December 2010, NSW, Australia
| Cryptosporidium sp. | Gender of patient | No. of infections in age group (yr) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0–4 | 5–9 | 10–14 | 15–19 | 20–24 | 25–29 | 30–34 | 35–39 | 40–44 | 45–49 | 50–54 | 55–59 | 60–64 | 65+ | Total | ||
| C. hominis | Female | 30 | 7 | 2 | 5 | 1 | 7 | 8 | 6 | 3 | 0 | 0 | 3 | 2 | 4 | 78 |
| Male | 33 | 9 | 6 | 0 | 1 | 1 | 4 | 5 | 3 | 1 | 0 | 0 | 1 | 3 | 67 | |
| Subtotal | 63 | 16 | 8 | 5 | 2 | 8 | 12 | 11 | 6 | 1 | 0 | 3 | 3 | 7 | 145 | |
| C. parvum | Female | 12 | 4 | 5 | 1 | 1 | 3 | 4 | 5 | 0 | 1 | 0 | 0 | 0 | 2 | 38 |
| Male | 18 | 3 | 2 | 0 | 2 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 32 | |
| Subtotal | 30 | 7 | 7 | 1 | 3 | 6 | 5 | 6 | 1 | 1 | 0 | 0 | 0 | 3 | 70 | |
| C. fayeri | Female | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Male | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Subtotal | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | |
| Total female | 42 | 11 | 7 | 6 | 2 | 11 | 12 | 11 | 3 | 1 | 0 | 3 | 2 | 6 | 117 | |
| Total male | 51 | 12 | 8 | 0 | 3 | 4 | 5 | 6 | 4 | 1 | 0 | 0 | 1 | 4 | 99 | |
| Total (%) | 93 (43) | 23 (11) | 15 (7) | 6 (3) | 5 (2) | 15 (7) | 17 (8) | 17 (8) | 7 (3) | 2 (0.9) | 0 (0) | 3 (1) | 3 (1) | 10 (5) | 216 (100) | |
Spatial distribution of sporadic infections, species, and subtypes between January 2008 and December 2010.
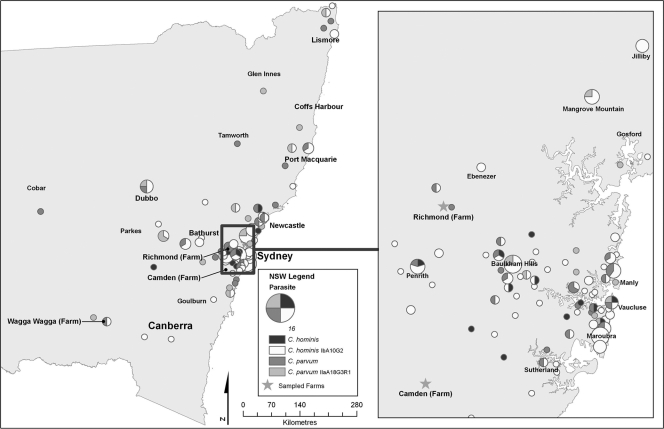
Postal codes for the locations from which isolates originated were obtained for 209/245 (85%) isolates successfully typed at the gp60 gene. Analysis of the New South Wales statewide map showed that sporadic cryptosporidiosis was confined to the east, northeast, southeast, and central regions of New South Wales (Fig. 1). The western areas of New South Wales reported no cryptosporidiosis infections and were excluded from the maps. Disease clusters were observed in the Sydney-to-Newcastle, Dubbo, Bathurst, Port Macquarie, and northeastern regions of the state. Within Sydney, infections were predominant in the eastern and the northwestern suburbs, which were localized in the Baulkham Hills region. Clustering was also observed in the Penrith, Mangrove Mountain, and Jilliby regions.
Fig. 1.
Spatial distribution by postal-code areas of the patients infected with the C. hominis IbA10G2 subtype, other C. hominis subtypes, C. parvum IIaA18G3R1, and other C. parvum subtypes in New South Wales and Sydney between January 2008 and December 2010. The size of the circle represents the number of cases.
Geographical segregation between the distributions of C. hominis and C. parvum was not observed; that is, both species were found in urban and rural areas throughout the state. However, C. parvum infections became more prominent further inland (Fig. 1). Within Sydney, C. hominis and C. parvum infections were widespread; however, C. hominis IbA10G2 was the dominant parasite causing infections in Sydney.
Spatiotemporal distribution of infections, species, and subtypes.
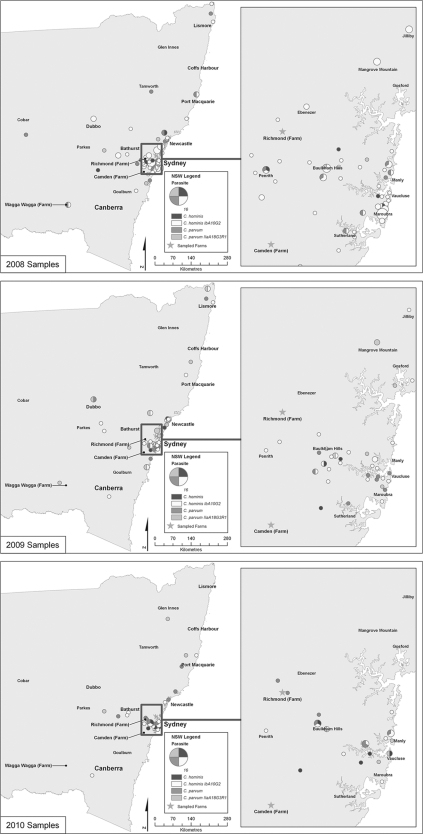
Postal codes and sample dates were obtained for 209/245 (85%) samples successfully typed at the gp60 gene. To determine the yearly distribution of sporadic cryptosporidiosis, samples were divided into their respective years, as follows: 2008, 104 samples; 2009, 67 samples; and 2010, 38 samples. The distribution of sporadic infections was widespread throughout 2008, 2009, and 2010 (Fig. 2). Clusters in urban coastal cities, such as Sydney, Newcastle, and Port Macquarie, and in the northeastern areas of the state remained constant. Cluster differences in regional areas, such as Wagga Wagga, Dubbo and Bathurst, were observed over the 3-year period. Clusters which were identified in Dubbo and Wagga Wagga in 2008 and 2009 were absent in both areas in 2010, while Bathurst clusters were observed in 2008 and 2010 but not in 2009. Infections within Sydney were also widespread. Urban areas surrounding Penrith and the eastern and northwestern suburbs maintained the highest number of infections. Large clusters were reported in the semirural areas of Mangrove Mountain and Jilliby in 2008 and 2009 but were absent in 2010.
Fig. 2.
Temporal analysis by year of the patients infected with the C. hominis IbA10G2 subtype, other C. hominis subtypes, C. parvum IIaA18G3R1, and other C. parvum subtypes in New South Wales and Sydney between January 2008 and December 2010. The size of the circle represents the number of cases.
Spatiotemporal analysis of the statewide species distribution showed that C. hominis infections were more common than C. parvum infections in 2008. Large C. hominis IbA10G2 clusters were seen in Sydney, Newcastle, Bathurst, and Dubbo and in the northeastern regions of the state (Fig. 2). Infections with C. parvum IIaA18G3R1 and other C. parvum subtypes increased in the Newcastle region and on the outskirts of Sydney. Port Macquarie and Wagga Wagga both showed clusters made up of C. hominis IbA10G2 and other C. parvum subtypes. Within Sydney, C. hominis IbA10G2 infections dominated the eastern and northwestern suburbs, Mangrove Mountain, Jilliby, and Penrith.
In 2009, C. parvum infections were more common than C. hominis infections. Statewide analyses showed an increased incidence of C. parvum IIaA18G3R1 and other C. parvum subtypes in Sydney, Newcastle, Dubbo, and Wagga Wagga and in the northeastern regions of New South Wales. The increased C. parvum infection trend continued within Sydney. Cryptosporidium parvum replaced the C. hominis IbA10G2 clusters previously seen in Mangrove Mountain and the eastern and northwestern suburbs. The incidence of C. hominis IbA10G2 remained high in Jilliby.
C. parvum and C. hominis contributed equally to sporadic disease in 2010. Bathurst remained dominated by C. hominis IbA10G2, while regions surrounding Port Macquarie exhibited both C. hominis and C. parvum infections. Isolated C. hominis and C. parvum clusters were seen throughout Sydney. However, two C. parvum subtype clusters appeared in the Richmond area.
Seasonal distribution of infections, species, and subtypes.
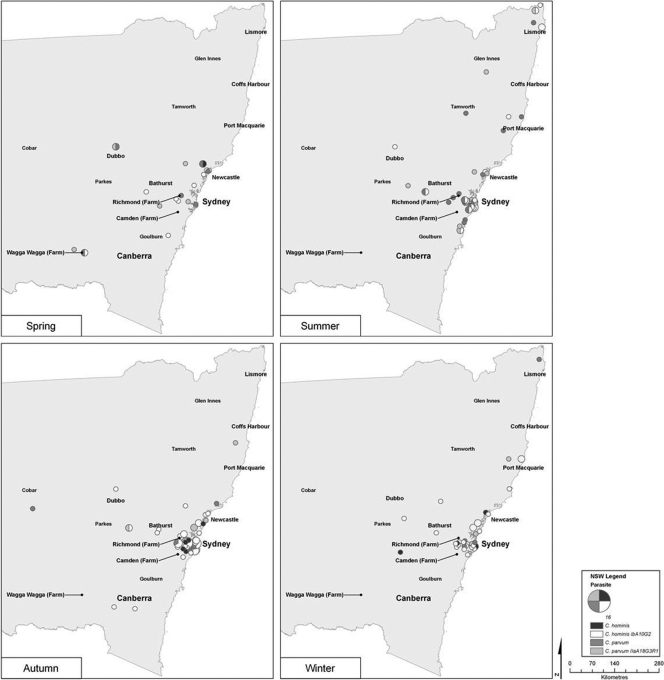
To determine the seasonal distribution of sporadic cryptosporidiosis, samples were divided into their respective seasons, as follows: spring, 22 samples: summer, 59 samples; autumn, 85 samples; and winter, 43 samples. Sporadic spring infections were dispersed throughout Sydney and the surrounding areas and were highest in Wagga Wagga and Dubbo (Fig. 3). Spring showed a higher incidence of C. parvum throughout the state, particularly in Dubbo, which was dominated by C. parvum IIaA18G3R1 and other C. parvum subtypes. This trend was repeated in Wagga Wagga and Richmond. Summer sporadic cases were concentrated along the coastal areas of the Sydney-to-Newcastle region and in the northeastern regions of the state. Both C. hominis and C. parvum were represented equally, and all C. hominis infections were attributed to subtype IbA10G2. Although previously dominated by C. parvum in spring, Dubbo summer infections were all attributed to C. hominis IbA10G2. Autumn infections increased in the western regions of Sydney and along the coast between Sydney and Newcastle. Cryptosporidium hominis IbA10G2 showed a higher incidence than C. parvum infections. Infections with other C. hominis subtypes also appeared in the western regions of Sydney. Dubbo remained dominated by C. hominis IbA10G2 throughout both summer and autumn. Winter infections showed small clusters that were confined to the Sydney region. Cryptosporidium hominis IbA10G2 was the most common parasite.
Fig. 3.
Temporal analysis by season of the patients infected with the C. hominis IbA10G2 subtype, other C. hominis subtypes, C. parvum IIaA18G3R1, and other C. parvum subtypes in New South Wales and Sydney between January 2008 and December 2010. The size of the circle represents the number of cases.
DISCUSSION
Essential to managing cryptosporidiosis and reducing risks of continued disease prevalence is knowledge of the species contributing to disease and the potential zoonotic sources. This study used a spatially based approach to investigate sporadic human cryptosporidiosis between January 2008 and December 2010.
Sixty-six percent of human infections in this study were caused by C. hominis. This observation is consistent with previous Australian studies investigating sporadic human cryptosporidiosis (4, 9, 14). No gender bias or regional segregation between the distributions of C. hominis and C. parvum infections was detected. The results indicated that both genders were most likely to be infected with C. hominis and that the disease was most prevalent in the 0- to 4-year age group.
The diversity within the gp60 of C. hominis and C. parvum isolates was extensive, with 5 subtype families and 31 subtypes within those families identified. The most common subtype was C. hominis IbA10G2, which was identified in 60% of samples. This subtype is the most common cause of sporadic cryptosporidiosis around the world, with 44.5% of total reported infections attributed to it (8). Subtype IbA10G2 was also previously identified as the most common cause of sporadic disease in Australia and was the cause of the 2009 New South Wales waterborne outbreak (9, 14, 22, 23). The second most frequently detected Cryptosporidium subtype from humans was C. parvum IIaA18G3R1, which was identified in 15% of isolates. The occurrence of this subtype in humans was first reported in Australia, and it has previously been identified as the most common C. parvum subtype causing sporadic cryptosporidiosis in New South Wales (4, 9, 10, 14, 22). Subtype IIaA18G3R1 is also the dominant type of C. parvum causing illness in humans in Ireland (26).
The epidemiology of human cryptosporidiosis is a complex interplay between humans, domestic animals, livestock, wildlife, and the environment. Knowledge of the Cryptosporidium species infecting these sources and an understanding of the factors influencing contact between these sources is essential to cryptosporidiosis management. Analysis of Cryptosporidium from cattle samples was included in this investigation to determine their zoonotic potential for contribution to human cryptosporidiosis. The results showed that all subtypes identified in cattle were frequent causes of sporadic human cryptosporidiosis in New South Wales. Of particular concern are the C. parvum IIaA18G3R1 and C. hominis IbA10G2 subtypes which were the most frequently detected subtypes in humans in New South Wales, with 47% of cattle infections attributed to C. parvum IIaA18G3R1. This observation has been previously made for Australian cattle in both Perth and Tamworth (13, 14). Interestingly, C. hominis IbA10G2 was detected on two farms, in the regional area of Wagga Wagga and in the semirural locality of Richmond, Sydney. Cryptosporidium hominis is thought to be host specific for humans, so the identification of this species in cattle was unexpected. However, the detection of a DNA sequence in a sample is not indicative that an infection is occurring. To ascertain that C. hominis infections were present in cattle, the presence of oocysts was confirmed by fluorescence microscopy, which revealed that all cattle infected with C. hominis were shedding between 101 to 102 oocysts/g, indicating low-level infections. Unfortunately, the C. andersoni infection, which was identified in one human sample by molecular analysis, could not be confirmed by flow cytometry or microscopy due to limited sample material. Increasingly, diverse species of Cryptosporidium are being recognized as having zoonotic capabilities. Knowledge of the zoonotic threats that different Cryptosporidium species pose to vertebrate hosts and how they circulate through the environment is essential to future disease management.
Expanding human populations, urbanization, and intensifying agricultural practices will influence the transmission of Cryptosporidium through the environment and will bring disease sources into closer contact. Visualization of the geographic distribution of Cryptosporidium species and infections in zoonotic sources facilitates the identification of hot spot zones and the different disease risks posed to these areas. Sporadic human cryptosporidiosis was highest in the urban coastal cities of Sydney, Newcastle, and Port Macquarie, in addition to regional inland areas of Wagga Wagga, Bathurst, and Dubbo. Both C. hominis and C. parvum were prevalent in these areas. Due to the different infection capabilities of C. parvum, it was undetermined whether infections were from anthroponotic or zoonotic disease sources. Seasonal differences were shown between the prevalence of C. hominis and C. parvum infections. Cryptosporidium hominis infections, particularly with subtype IbA10G2, dominated throughout the state and within the Sydney region in winter. Conversely, the majority of spring infections were attributed to C. parvum. This was most pronounced in the regional areas of Dubbo, Wagga Wagga, and Richmond. The increased incidence of C. parvum in these rural areas can probably be attributed to the calving season. Cattle samples analyzed in this study were sampled in spring and showed the presence of C. hominis and C. parvum subtypes frequently identified in humans. The spatial analysis of sporadic human infections in the spring showed disease clusters in Wagga Wagga and Richmond, where infections were caused by C. parvum subtypes identified in cattle in those regions. As human populations expand toward and into more-rural areas, farm management practices will become pivotal for controlling zoonotic disease transmission, especially when community-shared recreational water is in close proximity.
Spatial analyses showed 7 disease hot spot zones throughout New South Wales, 4 in the urban coastal cities of Sydney, Newcastle, Port Macquarie, and Lismore and 3 in the regional areas of Wagga Wagga, Dubbo, and Bathurst. All areas were equally impacted by C. hominis and C. parvum, except for Bathurst, which maintained C. hominis infections over the 3-year study period. From the recent finding of C. hominis infections in cattle from New South Wales, cattle in Bathurst need to be investigated to determine if they are contributing to human disease in this regional locality. Within Sydney, Mangrove Mountain was identified as a hot spot zone. This is a recreational area with numerous camping grounds, recreational water activities, agriculture, and wildlife. Incidentally, this was also the location of the human C. fayeri infection. Screening of wildlife and livestock in this area needs to be conducted to determine the zoonotic disease risks posed to both humans and animals.
Cryptosporidium is widespread in New South Wales and has complex transmission pathways involving humans, cattle, and native Australian wildlife. Urbanization, increased agriculture, and population expansion will all contribute to the continued emergence of this disease in New South Wales. From this, understanding the contact and transmission pathways that occur between different hosts, in addition to knowing the parasites present in these sources, will become an essential component for disease management. This study has provided initial data on hot spot zones and population groups most at risk of disease in New South Wales. Targeted surveillance in these regions and increased understanding of human-animal contact will enable the development and implementation of successful disease prevention measures.
ACKNOWLEDGMENTS
Funding for this research was provided through an Australian Research Council (ARC) Linkage Grant in partnership with NSW Health and was approved by the Macquarie University Human Research Ethics Committee (approval reference no. HE23FEB2007-R05007).
We thank Douglas Hanley Moir of SDS and Symbion Pathology companies in North Ryde and the Westmead, John Hunter of St. Vincent's, Blacktown, Gosford, and Davis Campbell of Wollongong and Seals Hospitals for providing the human fecal samples. Special acknowledgments to Jeremy McAnulty, Jennie Musto, and Nicola Stephens from NSW Health, Communicable Disease Branch, for their assistance in sample collection. We gratefully acknowledge Murray Austin, Craig McConnel, Dane Vandine, and Clem Whale for their assistance with cattle sampling, Kelly Lanfranca for her assistance in GIS analyses, and Stephen Hoggard for his assistance in statistical analyses.
Footnotes
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Alves M., Xiao L., Antunes F., Matos O. 2006. Distribution of Cryptosporidium subtypes in humans and domestic and wild ruminants in Portugal. Parasitol. Res. 99:287–292 [DOI] [PubMed] [Google Scholar]
- 2. Bennett J. W., Gauci M. R., Le Moënic S., Schaefer F. W., III, Lindquist H. D. A. 1999. A comparison of enumeration techniques for Cryptosporidium parvum oocysts. J. Parasitol. 85:1165–1168 [PubMed] [Google Scholar]
- 3. Chalmers R. M., Davies A. P. 2010. Clinical cryptosporidiosis. Exp. Parasitol. 124:138–146 [DOI] [PubMed] [Google Scholar]
- 4. Chalmers R. M., et al. 2005. Direct comparison of selected methods for genetic categorisation of Cryptosporidium parvum and Cryptosporidium hominis species. Int. J. Parasitol. 35:397–410 [DOI] [PubMed] [Google Scholar]
- 5. Fayer R. 2010. Taxonomy and species delimitation in Cryptosporidium. Exp. Parasitol. 124:90–97 [DOI] [PubMed] [Google Scholar]
- 6. Feng Y., et al. 2007. Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet. Parasitol. 144:1–9 [DOI] [PubMed] [Google Scholar]
- 7. Ferrari B. C., Power M. L., Bergquist P. L. 2007. Closed-tube DNA extraction using a thermostable proteinase is highly sensitive, capable of single parasite detection. Biotechnol. Lett. 29:1831–1837 [DOI] [PubMed] [Google Scholar]
- 8. Jex A. R., Gasser R. B. 2010. Genetic richness and diversity in Cryptosporidium hominis and C. parvum reveals major knowledge gaps and a need for the application of “next generation” technologies. Biotechnol. Adv. 28:17–26 [DOI] [PubMed] [Google Scholar]
- 9. Jex A. R., et al. 2008. Classification of Cryptosporidium species from patients with sporadic cryptosporidiosis by use of sequence-based multilocus analysis following mutation scanning. J. Clin. Microbiol. 46:2252–2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jex A. R., et al. 2007. A practical and cost-effective mutation scanning-based approach for investigating genetic variation in Cryptosporidium. Electrophoresis 28:3875–3883 [DOI] [PubMed] [Google Scholar]
- 11. Morgan-Ryan U. M., et al. 2002. Cryptosporidium hominis n. sp. (Apicomplexa: Cryptosporidiidae) from Homo sapiens. J. Eukaryot. Microbiol. 49:433–440 [DOI] [PubMed] [Google Scholar]
- 12. New South Wales Public Health Act of 1991 New South Wales Government. http://www.legislation.nsw.gov.au/viewtop/inforce/act+10+1991+FIRST+0+N
- 13. Ng J., et al. 2008. Evidence supporting zoonotic transmission of Cryptosporidium in rural New South Wales. Exp. Parasitol. 119:192–195 [DOI] [PubMed] [Google Scholar]
- 14. O'Brien E., McInnes L., Ryan U. 2008. Cryptosporidium GP60 genotypes from humans and domesticated animals in Australia, North America and Europe. Exp. Parasitol. 118:118–121 [DOI] [PubMed] [Google Scholar]
- 15. Plutzer J., Karanis P. 2009. Genetic polymorphism in Cryptosporidium species: an update. Vet. Parasitol. 165:187–199 [DOI] [PubMed] [Google Scholar]
- 16. Power M. L., Cheung-Kwok-Sang C., Slade M., Williamson S. 2009. Cryptosporidium fayeri: diversity within the GP60 locus of isolates from different marsupial hosts. Exp. Parasitol. 121:219–223 [DOI] [PubMed] [Google Scholar]
- 17. Robinson G., et al. 2010. Re-description of Cryptosporidium cuniculus Inman and Takeuchi, 1979 (Apicomplexa: Cryptosporidiidae): morphology, biology and phylogeny. Int. J. Parasitol. 40:1539–1548 [DOI] [PubMed] [Google Scholar]
- 18. Sulaiman I. M., et al. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol. 43:2805–2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Truong Q., Ferrari B. C. 2006. Quantitative and qualitative comparisons of Cryptosporidium faecal purification procedures for the isolation of oocysts suitable for proteomic analysis. Int. J. Parasitol. 36:811–819 [DOI] [PubMed] [Google Scholar]
- 20. Tzipori S., Widmer G. 2008. A hundred-year retrospective on cryptosporidiosis. Trends Parasitol. 24:184–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Waldron L. S., Cheung-Kwok-Sang C., Power M. L. 2010. Wildlife-associated Cryptosporidium fayeri in human, Australia. Emerg. Infect. Dis. 16:2006–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waldron L. S., Ferrari B. C., Power M. L. 2009. Glycoprotein 60 diversity in C. hominis and C. parvum causing human cryptosporidiosis in NSW, Australia. Exp. Parasitol. 122:124–127 [DOI] [PubMed] [Google Scholar]
- 23. Waldron L. S., Ferrari B. C., Cheung-Kwok-Sang C., Beggs P. J., Stephens N., Power M. L. 2011. Molecular epidemiology and spatial distribution of a waterborne cryptosporidiosis outbreak in Australia. Appl. Environ. Microbiol. 77:7766–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xiao L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80–89 [DOI] [PubMed] [Google Scholar]
- 25. Xiao L., et al. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386–3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zintl A., et al. 2009. The prevalence of Cryptosporidium species and subtypes in human faecal samples in Ireland. Epidemiol. Infect. 137:270–277 [DOI] [PubMed] [Google Scholar]