Abstract
Responses to NaCl stress were investigated in phototrophically grown Alphaproteobacterium Rhodobacter sphaeroides by transcriptome profiling, mutational analysis, and measurements of compatible solutes and membrane phospholipids. After exposure to salt stress, genes encoding two putative glycine betaine uptake systems, proVWX and betS, were highly upregulated. Mutational analysis revealed that BetS, not ProVWX, was the primary transporter of this compatible solute. Upon the addition of salt, exogenous glycine betaine was taken up rapidly, and maximal intracellular levels were reached within minutes. In contrast, synthesis of another important compatible solute in R. sphaeroides, trehalose, increased slowly following salt stress, reaching maximal levels only after several hours. This accumulation pattern was consistent with the more gradual increase in salt-induced transcription of the trehalose biosynthesis operon otsBA. Several genes encoding putative transcription factors were highly induced by salt stress. Multiple copies of one of these factors, crpO (RSP1275), whose product is a member of the cyclic AMP receptor protein/fumarate and nitrate reduction regulator (CRP/FNR) family, improved NaCl tolerance. When crpO was provided in multicopy, expression of genes for synthesis or transport of compatible solutes was unaltered, but the membrane phospholipid composition became biased toward that found in salt-stressed cells. Collectively, this study characterized transcriptional responses to salt stress, correlated changes in transcription with compatible solute accumulation rates, identified the main glycine betaine transporter and trehalose synthase, characterized salt-induced changes in phospholipid composition, and uncovered a transcription factor associated with changes in phospholipids. These findings set the stage for deciphering the salt stress-responsive regulatory network in R. sphaeroides.
INTRODUCTION
Microorganisms adapt to fluctuations in medium osmolarity via two common strategies. One involves reestablishing osmotic balance by accumulation, through import from the medium or through synthesis, of intracellular osmoprotectants known as compatible solutes. The second common strategy involves altering membrane composition to better cope with the changed turgor pressure. Compatible solutes used by various microorganisms include, among others, the amino acids glutamate and proline, the amino acid derivatives glycine betaine ([GB] N,N,N-trimethyl glycine) and proline betaine, and the sugar trehalose (α-d-glucopyranosyl-α-d-glucopyranoside) (6, 14). Alterations to the membrane include both changes in fatty acid saturation and phospholipid composition (reviewed in reference 34).
We are interested in salt stress responses in the remarkably metabolically versatile facultatively phototrophic Alphaproteobacterium Rhodobacter sphaeroides 2.4.1. That this freshwater bacterium actually requires sodium for growth was an interesting characteristic established over 40 years ago (38). R. sphaeroides 2.4.1 can adapt to concentrations of NaCl up to approximately 0.4 M, but higher concentrations prevent cell growth (1).
Another freshwater strain closely related to strain 2.4.1, R. sphaeroides f. sp. denitrificans, synthesizes trehalose as a compatible solute in response to salt stress. The roles of trehalose biosynthesis and degradation in adaptation of this bacterium to salt stress have been described (24). R. sphaeroides f. sp. denitrificans also accumulates potassium cations to counter increased sodium levels brought about by salt stress. However, the organism apparently does not respond by synthesizing GB or amino acids to an appreciable degree (43). With respect to R. sphaeroides 2.4.1, Abee et al. reported that GB, when present in the medium, can be taken up for osmoprotection, whereas exogenous proline is ineffective (1). Also, Catucci et al. reported that the relative cardiolipin content of R. sphaeroides 2.4.1 membranes increases in response to osmotic stresses (7).
The aims of the present study were (i) to obtain a more comprehensive view of salt stress responses in R. sphaeroides 2.4.1, (ii) to investigate the dynamics of engagement of known salt stress systems following exposure to NaCl, and (iii) to identify and begin characterizing potential regulators of NaCl stress. For transcriptome profiling, the R. sphaeroides 2.4.1 whole-genome GeneChip described by Pappas et al. (28) was used. The experimental setup involved subjecting phototrophically grown cells to a significant increase in NaCl concentration, i.e., from 85 mM Na+ in the growth medium to a final concentration of 265 mM Na+ (described in the text as an addition of 1.5% NaCl). The response was examined at two time points. The first time point, 7 min after the NaCl addition, was chosen to capture the earliest transcriptional responses, whereas the later time point, 45 min, was chosen to identify longer-term adaptation mechanisms. This range of time points has proven informative in our transcriptome analyses of responses to other environmental stressors in R. sphaeroides 2.4.1 (5, 47).
Following transcriptome profiling, the transcriptional data were correlated with transport and synthesis of known compatible solutes. Mutational analysis was then used to investigate the relative importance of several genes predicted to be involved in compatible solute metabolism. Changes in membrane lipid composition in response to salt stress were also investigated, and a transcription factor designated CrpO that controls these changes was identified.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Growth was measured by determining the change in optical densities (ODs; measured as the OD at 600 nm [OD600] or by Klett meter) versus time. For microarray experiments, R. sphaeroides 2.4.1 was grown in Sistrom's minimal medium A containing succinate as a carbon source (38). The 60-ml cultures were grown in 100-ml glass tubes with continuous vigorous sparging with a gas mixture containing 95% N2 and 5% CO2. The tubes were placed in a transparent water bath (30°C) and exposed to white light at 10 W m−2. Salt stress was induced by the addition to the mid-exponential-growth-phase cultures of an anoxic NaCl solution amounting to an addition of 1.5% NaCl, which increased the Na+ concentration in the growth medium from 85 to 265 mM. Cultures were collected at 0, 7, and 45 min following addition of NaCl.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| R. sphaeroides | ||
| 2.4.1 | Wild type | W. Sistrom |
| RWAE | 2.4.1 ΔproXW::Ω(Smr/Spr) | This study |
| RBCCT | 2.4.1 betS::pRBCCT | This study |
| E. coli | ||
| JM109 λpir | JM109 lysogenized with λpir | 25, 44 |
| S17-1 λpir | Tpr SmrhsdR pro recA λpir RP4-2-Tc::Mu-Km::Tn7 | 37 |
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | 18 |
| DH5αphe | DH5α phe::Tn10d; Cmr | 11 |
| HB101 | F−Δ(gpt-proA)62 leuB6 supE44 ara-14 galK2 lacY1; used in triparental matings | 9 |
| XL1-Blue | [F′::Tn10 (Tcr) proAB lacIqΔ(lacZ)M15] recA1 endA1 gyrA96 thi-1 supE44 relA1 lac | Agilent Technol., Inc. |
| Plasmids | ||
| pJP5603 | Kmr; R6K-based suicide vector | 29 |
| pRWAE001 | 948-bp PCR product containing RSP3058 (proW) upstream sequences in pJP5603 | This study |
| pRWAE002 | 872-bp PCR product containing RSP3059 (proX) downstream sequences in pRWAE001 | This study |
| pRWAE003 | Ω(Smr/Spr) inserted into the SmaI site of pRWAE002b | This study |
| pHP45 | Source of Ω(Smr/Spr) cassette | 33 |
| pRBCCT | 675-bp PCR product containing RSP3080 (betS) sequences in pJP5603b | This study |
| pBSIISK+ | ColE1; Apr | Agilent Technol., Inc. |
| pUI1087 | pBSIISK+ with modified polylinker; Apr | 46 |
| pCrpO | PCR-amplified crpO DNA inserted into MscI-EcoRV-restricted pUI1087; Aprc | This study |
| pBBR1MCS-2 | Broad-host-range plasmid, Mob+ Knr | 21 |
| pBBR-CrpO | HindIII-XbaI fragment from pCrpO inserted into pBBR1MCS-2c | This study |
| pCrpO′ | Truncated crpO′ following oligonucleotide-directed mutagenesis, in pUI1087c | This study |
| pBBR-CrpO′ | HindIII-StuI fragment from pCrpO′ inserted into HindIII-EclI136II-restricted pBBR1MCS-2c | This study |
| pRK2013 | IncP1 ColE1 Tra+ of RK2 Knr; used as a helper plasmid in triparental matings | 9, 12 |
Km, kanamycin; Sm, streptomycin; Sp, spectinomycin; Ap, ampicillin; Tc, tetracycline.
See Fig. S1 in the supplemental material.
See Fig. S2 in the supplemental material.
For compatible solute analyses, R. sphaeroides was grown in tightly capped bottles in the same medium as described earlier (24). Conditions used to obtain cells subjected to lipid analysis are provided below. Multicopy analysis of NaCl tolerance of R. sphaeroides was performed by monitoring growth of cultures inoculated with equivalent numbers of cells (OD600 of 0.17) in filled screw-cap tubes containing kanamycin (final concentration, 50 μg ml−1) for plasmid maintenance and 0% (final concentration, 85 mM), 1.5% (final concentration, 265 mM), or 3.0% (final concentration, 445 mM) additional NaCl.
Genetic manipulations.
Standard recombinant DNA techniques were used (36). Plasmids were transferred from Escherichia coli into R. sphaeroides cells by conjugation as described earlier (29, 37), and recombinants were selected using appropriate antibiotics. The correctness of the mutant strains was verified by Southern blot analysis (36).
The R. sphaeroides betS (RSP3080) gene was inactivated in strain RBCCT by insertion of the Kmr suicide vector pRBCCT (Table 1; see also Fig. S1 in the supplemental material), derived from pJP5603 (29). The Δ(proW proX) double mutant strain RWAE was constructed by replacing an internal fragment of the genes with an Ω(Smr/Spr) cassette (33) using plasmid pRWAE3 (Table 1; see also Fig. S1). The primer sequences and schematics of the plasmids used in the construction of these mutants are provided in Fig. S1.
Plasmid pCrpO (Table 1) was constructed by amplifying the crpO sequences from genomic DNA using PCR and ligating the product into pUI1087 (46). A DNA fragment carrying crpO was then isolated from pCrpO and inserted into plasmid pBBR1MCS-2 (21) to create pBBR-CrpO (Table 1; see also Fig. S2 in the supplemental material). To construct pBBR-CrpO′ (Table 1; see also Fig. S2), a QuikChange II Mutagenesis Kit (Agilent Technologies, Inc.) and mutagenic oligonucleotides were used to delete the 3′ portion of the crpO gene in plasmid pCrpO while also creating a new StuI restriction site. The truncated gene was moved into pBBR1MCS-2, which added DNA in frame with CrpO′ coding for five amino acids (LQFAL) followed by a nonsense codon. Further cloning details, oligonucleotide sequences, and plasmid maps are provided in Fig. S2.
RNA extraction, DNA microarrays, and quantitative real-time reverse transcription-PCR (RT-PCR).
RNA extraction was performed as described earlier (28). Briefly, rifampin (final concentration, 200 μg ml−1) was added to R. sphaeroides cultures to halt transcription initiation, and cells were immediately collected into centrifugation bottles containing shaved ice, followed by brief centrifugation. The pellets were then resuspended in RNeasy Midikit (Qiagen) buffer. Cells were disrupted using a Mini-BeadBeater (Biospec Products), and RNA was extracted using the RNeasy Midikit. The absence of genomic DNA contamination was verified by quantitative real-time PCR using the SYBR green method and iCycler iQ Real Time PCR Detection System (Bio-Rad), as described earlier (28).
High-density oligonucleotide R. sphaeroides GeneChips (Affymetrix, Inc.) corresponding to the whole 4.6-Mb genome were used for transcriptome profiling. The construction and performance analysis of the R. sphaeroides GeneChip were described earlier (28). The quality and quantity of RNA and cDNA at all steps were monitored using capillary gel electrophoresis (BioAnalyzer, Agilent Technologies, Inc.).
DNA microarray data analysis.
Transcriptome profiles from three independently grown cultures exposed to NaCl for 7 or 45 min, as well as five controls at 0 min, were recorded and examined. GeneChip data analysis was carried out using the R statistical software environment (http://www.r-project.org) enhanced with specialized microarray-related software packages from the Bioconductor project (http://www.bioconductor.org). The expression values were obtained using robust multiarray analysis with quantile normalization (16) using the Affy package (15) from Bioconductor. A parametric empirical Bayes approach was applied as described by Kendziorski et al. (19), using the EBarrays Bioconductor package (45). The Pearson correlation between replicate arrays was high (>0.97), which suggested a high level of interreplicate reproducibility. As a criterion for differential gene expression between the growth conditions, we used a combination of a false discovery rate (FDR) cutoff of 0.05 and a median expression ratio (fold change) cutoff of 2 in either direction (i.e., ≤0.5 or ≥2). For the purposes of the current analysis, the genes with changes in expression levels that did not satisfy these criteria were considered not changed (NC).
The expression ratios are shown in the text in parentheses preceded by the locus (RSP) numbers or their corresponding genes. When expression of several genes is discussed, the lower and upper limits of expression changes are given, e.g., a range of 2.0- to 5.0-fold increases.
Trehalose and GB measurements.
In order to determine intracellular levels of trehalose and GB, R. sphaeroides cells were harvested by centrifugation. For trehalose measurements, cell pellets were resuspended in 80% (vol/vol) aqueous solution of acetonitrile (5 ml per g of wet mass) and then ultrasonically homogenized. The suspension was heated for 5 min at 100°C followed by centrifugation at 18,500 × g for 5 min. Subsequently, the supernatant was filtered through a Minisart-RC 4 filter (Sartorius, Germany), and the filtrate was subjected to high-performance liquid chromatography (HPLC) analysis on a Shodex sugar SZ5532 column (6 mm by 150 mm) at a flow rate of 1 ml of 80% acetonitrile min−1. Trehalose concentrations were determined by using refractive index (RI) monitor RI-930 (JASCO, Japan), and a standard curve was generated by using standard solutions of trehalose ranging from 0.1 to 10 mM.
For GB measurements, cell pellets were resuspended in 70% ethanol (10 ml per g of wet mass) and ultrasonically homogenized. The mixture was heated for 5 min at 100°C and centrifuged at 18,500 × g for 5 min. The supernatant was evaporated to dryness, and the residue was solubilized in 0.5 volume of distilled water. Esterification of GB was performed as described earlier (25). Briefly, p-bromophenacyl bromide (20 mg ml−1 in acetonitrile) and reaction buffer (100 mM KHCO3 · KH2PO4 · acetonitrile at 1:1:4, vol/vol) were added to the solution, and the entire contents was then heated at 80°C for 90 min. The heat-treated solution was evaporated to dryness. Subsequently, 300 μl of 100 mM NaH2PO4, pH 3.0, was added to the pellet, which was mixed and then centrifuged at 18,500 × g for 5 min. The supernatant was applied to a CAPI-3300 (Otsuka Electronics, Japan) capillary electrophoresis system, and esterified GB was detected at 254 nm. Concentrations were determined based on a standard curve generated using GB solutions of 0.01 to 10 mM.
Extraction of phospholipids.
Resting cells subjected to lipid analysis were prepared essentially as described by De Leo et al. (8) using a mid-exponential-phase phototrophic R. sphaeroides culture (grown under the same conditions as in the microarray experiments), except that the culture was divided equally before the cells were pelleted. Potassium phosphate (0.1 M; pH 7.4) was used to wash the cells from the residual medium, and one pellet of washed cells was resuspended in 50 ml of the same buffer with chloramphenicol at a final concentration of 0.3 mg ml−1 and the other in buffer alone. Salt was added to the “resting” cell suspensions to achieve either the same concentration present in culture medium (85 mM) or to a final concentration of 0.8 M in order to replicate the experimental conditions described by De Leo et al. (8). The resuspended cells were transferred to 250-ml Erlenmeyer flasks, which were then incubated with gentle rotation at 30°C overnight.
For all other phospholipid analyses, after a 45-min incubation following the addition of 3% NaCl to phototrophic cultures of R. sphaeroides, chloramphenicol was added to a final concentration of 0.3 mg ml−1 to halt further protein synthesis.
In all cases, the cells were pelleted in 50-ml conical tubes by centrifugation. Cell pellets were then resuspended in 0.1 M potassium phosphate buffer, pH 7.4, and lipid extraction was performed using standard protocols (4, 8). Following evaporation of the solvents used in extraction, the dry weights of the extracts were determined. The lipids were then dissolved in chloroform-methanol (1:1, vol/vol) to a final concentration of approximately 20 μg μl−1. All phospholipid samples were stored at −20°C.
TLC analysis.
The extracted lipid samples, together with a standard solution of 10 μg μl−1 each of cardiolipin (CL), phosphatidylcholine (PC), and phosphatidylethanolamine (PE) (Sigma-Aldrich) in chloroform-methanol (1:1, vol/vol), were applied to thin-layer chromatography (TLC) silica gel plates (layer thickness, 0.2 mm; Sigma-Aldrich) that had been washed twice with chloroform-methanol (1:1, vol/vol) and then activated by placing the plates in a dry oven for 30 min at 120°C. The development conditions that were used were previously described (7, 8). Visualization of the phospholipids was achieved by exposing the charred TLC plates to UV light. Digital images were captured using a DC290 zoom digital camera (Eastman Kodak Company). Phospholipid ratios were determined using ImageJ software (http://rsb.info.nih.gov/ij).
Chemical determination of polar phospholipid concentrations.
The concentrations of polar phospholipids were determined using a colorimetric method based on the formation of a complex between phospholipids and ammonium ferrothiocyanate as described previously (39). Absorption spectroscopy was performed using a Hitachi U-2010 UV/visible light (Vis) spectrophotometer. A standard curve was generated using a PC solution (39; http://www.cyberlipid.org).
Microarray data accession number.
The expression data obtained here were deposited in the Gene Expression Omnibus (GEO) database of NCBI (www.ncbi.nlm.nih.gov/projects/geo/), platform GPL162, series GSE8082.
RESULTS AND DISCUSSION
Scope of transcriptome changes in response to salt stress.
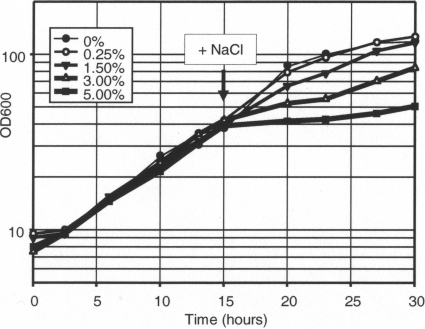
The growth of R. sphaeroides 2.4.1 under anoxic phototrophic conditions is inhibited by high-salt concentrations (Fig. 1). In the presence of 1.5% added NaCl, the generation time is approximately 1.8-fold longer than that in the standard medium. Growth in the presence of 3% added NaCl was inhibited further and was generally less reproducible (data not shown), suggesting that the population accumulated spontaneous, salt-tolerant mutants. Therefore, for transcriptome analysis, we exposed phototrophic cultures to an additional 1.5% NaCl; however, for other types of analyses we often used both 1.5% and 3% additional NaCl.
Fig. 1.
Effect of added NaCl on anoxic phototrophic growth of R. sphaeroides 2.4.1. The arrow indicates the time point at which the NaCl was added. Representative growth curves are shown.
Whole-genome transcription profiling revealed that, compared to the untreated culture (0 min), expression of approximately 900 genes changed at least under one time point following exposure to additional 1.5% NaCl, according to the criteria described in Materials and Methods. The numbers of up- and downregulated genes were approximately the same, i.e., approximately 450 genes. All of the genes that passed our selection criteria are listed in Table S1 in the supplemental material.
The large number of affected genes suggests that both salt-specific and nonspecific stress responses were triggered. For example, among the nonspecific responses were the upregulation at 7 min of the SOS response gene lexA (RSP1997; 2.3-fold) and the downregulation at 45 min of the σ70 gene (RSP0395; 0.43-fold) together with a number of genes encoding ribosomal proteins (RSP0021-RSP0022, RSP1048, RSP1700, RSP1709-RSP1710, RSP1716 to RSP1718 [RSP1716-RSP1718], RSP1720-RSP1733, and RSP2860; 0.30- to 0.48-fold), the latter trend being consistent with the decreased growth rate of the culture in the presence of additional 1.5% (Fig. 1). However, the prototypical osmotic stress responses could also be discerned by changes in expression of genes responsible for compatible solute synthesis and transport (Table 2). These changes are described below.
Table 2.
Expression of the most relevant salt-stress response systems in R. sphaeroides 2.4.1
| Function and locus | Annotated producta | Fold change at:b |
|
|---|---|---|---|
| 7 min | 45 min | ||
| Compatible solute synthesis | |||
| RSP0948-RSP0949 | OtsB, trehalose phosphatase | 6.5 | 11.8 |
| OtsA, trehalose 6-phosphate synthase | 2.8 | 16.2 | |
| RSP2446 | TreS, trehalose hydrolase | NC | 3.5 |
| RSP2450, RSP2452 | TreZ, maltooligosyltrehalose trehalohydrolase | NC | NC |
| TreY, maltooligosyltrehalose synthase | NC | NC | |
| Compatible solute transport | |||
| RSP3057-RSP3059 | ProX, ABC-type glycine betaine/l-proline transporter, periplasmic substrate binding protein | NC | 17.4 |
| ProW, transporter permease protein | 3.7 | 56.4 | |
| ProV, transporter inner membrane ATPase protein | 8.9 | 53.5 | |
| RSP3998-RSP4000 | ProV1, ABC-type glycine betaine/l-proline transporter, inner membrane ATPase protein | 3.2 | NC |
| ProW1, transporter permease protein | NC | NC | |
| ProX1, transporter periplasmic substrate binding protein | NC | NC | |
| RSP2179-RSP2181 | ProX2W2V2, ABC-type glycine betaine/l-proline transporter | NC | NC |
| RSP3080 | BetS, BCCT family choline/carnitine/betaine transporter | NC | 4.0 |
| Cation (potassium) uptake | |||
| RSP1265-RSP1269 | KdpABC, K+-transporter | NC | NC |
| RSP1853-RSP1854, RSP2841-RSP2842 | TrkH2H3, TrkA, TrkH1, Trk-type K+ transporter | NC | NC |
| Additional candidate salt stress tolerance genes | |||
| RSP0697 | UspA, universal stress response protein A | NC | 2.5 |
| RSP2380 | CatC, catalase | NC | 2.9 |
| RSP2395 | BCCP, cytochrome c peroxidase | 5.6 | 5.7 |
| RSP0910-RSP0912 | DctP, TRAP dicarboxylate family transporter substrate-binding protein | NC | 0.06 |
| DctQ, transporter subunit | NC | 0.04 | |
| DctM, transporter subunit | NC | 0.06 | |
| RSP0634-RSP0636 | DctP-type subunit, TRAP dicarboxylate family transporter | NC | 12.1 |
| DctQ-type transporter subunit | NC | 17.7 | |
| DctM-type transporter subunit | NC | 6.7 | |
As annotated at NCBI (http://www.ncbi.nlm.nih.gov).
NC, no change.
Compatible solute transport.
GB is known to be an important compatible solute for R. sphaeroides (1). Three sets of genes (apparently organized in operons) are annotated as encoding components of proline or GB ABC-type transporters. The proXWV genes are likely to be important for coping with the NaCl stress since at 45 min they were among the most highly upregulated genes (RSP3057-RSP3059; 17.4- to 56.4-fold) (Table 2). The genes predicted to encode the other two proline/GB ABC-type transporters, RSP3998-RSP4000 and RSP2179-RSP2181, were much less responsive or nonresponsive (Table 2). A betS gene (RSP3080) encoding a putative betaine/carnitine/choline transporter, which can potentially be involved in GB uptake, is present in the R. sphaeroides 2.4.1 genome, and its mRNA level was increased in the presence of added salt (4.0-fold) (Table 2).
In order to evaluate the significance of the observed transcriptional responses of the potential GB transporter genes and to decipher their roles in NaCl stress tolerance, the dynamics of GB uptake were examined. Intracellular GB accumulation was monitored in cultures that were grown in the presence of 1 mM GB and exposed to 3% NaCl. Already at 7 min postexposure to NaCl, intracellular GB levels had increased by approximately 36-fold from an average value of 1.6 ± 1.2 preexposure to an average value of 56.9 ± 3.1 μmol of GB (g of wet weight)−1 postexposure, based on three biological replicates. Such rapid and robust GB uptake suggests that the GB transporter must be present in R. sphaeroides prior to NaCl stress. These results are consistent with the report by Abee et al. (1) that GB uptake was unhindered by the addition of the protein synthesis inhibitor chloramphenicol. Since in the absence of added NaCl betS transcripts were present at much higher absolute levels than the absolute levels of proVWX transcripts, we hypothesized that GB transport relies primarily upon BetS.
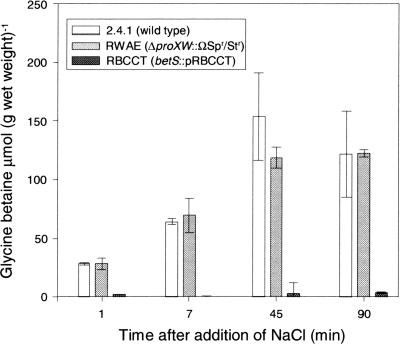
To further evaluate the relative contributions of proVWX and betS to GB transport, null mutations in proVWX and betS were constructed (Table 1, strains RWAE and RBCCT, respectively), and GB accumulation rates in these strains following NaCl stress were measured. While GB levels in cells lacking the ProVWX transporter did not differ from those of the wild type, cells lacking BetS were impaired in GB accumulation. For example, at 90 min the GB concentrations in the wild-type strain 2.4.1 and mutant RWAE were 119.6 and 121.9 μmol/g of wet weight, respectively, whereas the GB concentration in mutant RBCCT was 4.4 μmol/g of wet weight (Fig. 2).
Fig. 2.
Dynamics of intracellular GB accumulation in salt-stressed R. sphaeroides wild-type 2.4.1 and mutants RWAE (proXWV) and RBCCT (betS). Strains were grown under anoxic phototrophic conditions in medium containing 1 mM GB. Salt stress was imposed by the addition of 3% NaCl at time zero to exponentially growing cells (OD600 of 0.4). Values are the averages of three biological replicates, and the error bars represent the standard deviations.
These data suggest that, unlike the situation in enteric bacteria, where a ProVWX homolog is the main GB transporter (reviewed in reference 32), in R. sphaeroides BetS fulfills this role. Because the proVWX gene products were highly upregulated in response to salt stress and yet play no apparent role in GB uptake, we suggest that the ProVWX transporter is involved in the uptake of an as yet unknown compatible solute, which was not tested in our experiments.
Compatible solute synthesis.
Trehalose is the main compatible solute synthesized in response to osmotic stress in the close relative of R. sphaeroides 2.4.1, R. sphaeroides f. sp. denitrificans IL106 (43). The microarray data revealed that the otsBA genes (RSP0948-RSP0949) encoding the main trehalose biosynthesis operon in strain 2.4.1 (24) responded rapidly to added NaCl; i.e., the otsBA mRNA levels had increased at 7 min (2.8- to 6.5-fold) (Table 2) and remained among the most highly upregulated genes at 45 min (11.8- to 16.2-fold). In contrast, the baseline expression of the second operon predicted to be involved in trehalose synthesis, the treYZ genes (RSP2450-RSP2452), was relatively low, and these genes were not induced in response to NaCl stress (Table 2). The treS gene (RSP2446), whose product is involved in trehalose turnover (24), was upregulated at a later (45 min) but not earlier (7 min) time point (Table 2).
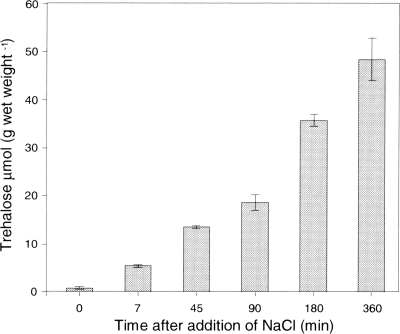
While the transcription response of the otsBA genes to added NaCl was rapid, the baseline expression of the genes was relatively low. This suggested that trehalose synthesis might require a significant amount of time before reaching osmoprotective levels. To investigate the dynamics of trehalose synthesis, intracellular trehalose concentrations in cells exposed to NaCl stress were measured (Fig. 3). At 7 min following the addition of 1.5% NaCl, the intracellular trehalose concentration had increased approximately 7-fold (5.3 versus 0.6 μmol of trehalose g of wet weight−1), but it also continued to rise for many hours, reaching a concentration that was approximately 80-fold higher than that prior to induction (47.7 versus 0.6 μmol of trehalose g of wet weight−1).
Fig. 3.
Dynamics of intracellular trehalose accumulation in salt-stressed R. sphaeroides 2.4.1 cells grown under anoxic phototrophic conditions. Salt stress was imposed by the addition of 1.5% NaCl at time zero to exponentially growing cells (OD600 of 0.4). Values are the averages of three biological replicates, and the error bars represent the standard deviations.
These results confirm that trehalose is important as a long-term osmoprotectant in R. sphaeroides 2.4.1 and reveal a pivotal role of induced otsBA expression in trehalose synthesis. They are also consistent with the hypothesis that OtsBA (and not TreYZ) play the primary role in trehalose synthesis in the R. sphaeroides species (24).
Membrane lipid alterations.
In several bacteria, osmotic stressors have been shown to evoke changes in the degree of saturation in membrane fatty acids and to increase the proportion of anionic phospholipids (reviewed in reference 34). An increase in the relative amount of CL has been documented in R. sphaeroides (7).
To identify the reasons underlying changes in membrane lipid composition, the expression of lipid biosynthesis genes was closely examined. Genes whose products are involved in CL production, i.e., pgsA (RSP1073) encoding phosphatidylglycerol (PG) 3-phosphate synthase (10), pgpA (RSP2834) encoding PG 3-phosphate phosphatase, and cls (RSP0473) encoding a CL synthase (40), were not responsive to the addition of NaCl. In fact, the transcript levels of only one gene involved in fatty acid biosynthesis, acpP (RSP2463), changed (decreased 0.45-fold) in response to added salt (see Table S1 in the supplemental material). While the present knowledge of fatty acid and lipid metabolism in R. sphaeroides is incomplete (40) and while some genes involved in these processes may not yet have been identified, the transcriptome analysis suggests that the membrane lipid changes are achieved primarily by posttranscriptional regulation.
To test this conclusion, we performed an experiment with resting cells that had been used previously by De Leo et al. (8) to demonstrate that CL was induced by NaCl in R. sphaeroides. We modified the experimental setup in order to compare the NaCl-induced changes in the phospholipid profile when protein synthesis was either permitted or halted (by chloramphenicol). PC was used as an internal reference in measuring relative abundances of CL and PG since its concentration does not change in response to osmotic stress (7). As already reported by De Leo et al. (8), upon NaCl stress both the CL/PC and PG/PC ratios increased under the conditions of permitted protein synthesis. However, when protein synthesis was halted, the CL/PC ratio still increased while the PG/PC ratio decreased (Table 3). Since CL is formed from two molecules of PG, the data suggest that NaCl stress increases the activity of CL synthase, whereas new protein synthesis is required to replenish PG. Overall, these experiments confirm the primarily posttranscriptional regulation of NaCl-induced changes in membrane lipid composition.
Table 3.
NaCl-induced changes in phospholipid composition in phototrophic R. sphaeroides 2.4.1 resting cells
| Incubation condition(s)a | Phospholipid ratiob |
|
|---|---|---|
| CL/PC | PG/PC | |
| 0.85 mM NaCl | 0.57 | 0.59 |
| 0.8 M NaCl | 0.65 | 0.79 |
| 0.8 M NaCl, chloramphenicolc | 0.85 | 0.30 |
Phospholipids were measured in R. sphaeroides 2.4.1 cells resting from phototrophic growth.
CL, cardiolipin; PC, phosphatidylcholine; PG, phosphatidylglycerol.
Final concentration, 0.3 mg ml−1.
Other features of the salt response transcriptome.
In addition to the components of the salt stress response described above, both potassium and its counter ion glutamate have been reported to play roles in bacterial tolerance to salt stress (42). R. sphaeroides f. sp. denitrificans accumulates K+ in proportion to the levels of exogenous NaCl (43). Therefore, the expression data of genes for K+ uptake and for synthesis and transport of glutamate were examined. They indicated that mRNA levels of neither the kdp-type (RSP1265-RSP1269) nor the trk-type (RSP2041, RSP1853-RSP1854, and RSP2842) K+ uptake genes were affected by NaCl stress (Table 2). While it is possible that R. sphaeroides 2.4.1 differs from R. sphaeroides f. sp. denitrificans with respect to the role of K+, it is more likely that in both species K+ accumulation is controlled posttranscriptionally.
With respect to glutamate uptake, Jacobs et al. (13) have described a tripartite ATP-independent periplasmic (TRAP)-type glutamate transporter that is detected only in R. sphaeroides exposed to osmotic stress (17). The identities of the genes encoding that transporter are not yet known. Curiously, a putative TRAP transporter encoded by RSP0634-RSP0636 is greatly upregulated at 45 min (6.7- to 17.7-fold) (Table 2). Furthermore, salt stress is the only condition out of many for which DNA microarray data are available when this transporter is upregulated (26). Therefore, it is possible that RSP0634-RSP0636 encode the osmotic shock-induced glutamate transporter reported by Jacobs et al. (17).
Alternatively, RSP0634-RSP0636 may play a different role, i.e., that of a succinate uptake system. The dctPQM genes (RSP0910-RSP0912), which encode a known TRAP-type transporter of succinate (13), were strongly downregulated following the addition of NaCl (0.04- to 0.06-fold at 45 min) (Table 2). Succinate is the main carbon source in the growth medium, and it is required for growth. According to Jacobs et al. (17), succinate uptake is stimulated, not inhibited, by NaCl. Together, these observations argue that an alternative succinate uptake system is engaged during NaCl stress, and it is possible that the RSP0634-RSP0636 TRAP transporter fulfills this role. These predictions originating from the NaCl stress transcriptome analysis have yet to be tested experimentally.
Other strongly upregulated genes that may contribute to increased salt stress tolerance include RSP0697 encoding a putative universal stress response protein (2-fold at 45 min) (Table 2), catalase catC (RSP2380; 2.9-fold at 45 min), and cytochrome c peroxidase ccp (RSP2395; 5.6-fold at 7 min). RSP0697 must be somewhat specific to salt stress because it was not upregulated upon exposure to oxidative stresses, unlike catC and ccp (5, 47).
NaCl-responsive transcription factor genes.
Over 80 putative transcriptional regulator genes were activated or repressed by NaCl stress under phototrophic conditions (see Table S2 in the supplemental material), suggesting that many regulatory networks were affected. Among these, five σ-factor genes were strongly upregulated (RSP0527, RSP0601, RSP1272, RSP2681, and RSP3606; >3-fold), and three (RSP0032 and RSP0068 at < 0.2-fold and RSP0395 at 0.43-fold) were strongly downregulated, including the main σ70 factor (RSP0395).
The upregulated σ-factor genes include a previously characterized oxidative stress response σ-factor gene, rpoH2 (RSP0601), which is a member of the heat shock family (3, 5, 48), and two σ54 factors, RSP0527 and RSP3606 (30, 31). One of the strongly downregulated σ-factor genes, RSP0068, also belongs to the σ54 family. Differential regulation of the members of this family reinforces the conclusions from earlier studies that the four σ54 factors of R. sphaeroides 2.4.1 have different functional roles (30, 31). The second significantly downregulated gene encoding a σ-factor, fliA (RSP0032), is involved in flagellar gene-specific transcription (reviewed in reference 2). The decreased expression of fliA is probably responsible for the reduced transcription of flagellar and motility genes in response to NaCl (see Table S1 in the supplemental material). A decrease in flagellar gene expression has been observed in diverse salt-stressed bacteria, e.g., E. coli, Shewanella oneidensis, Sinorhizobium meliloti, and Desulfovibrio vulgaris (23, 27, 35, 41), suggesting that this phenomenon is a common bacterial response to salt stress.
The two remaining upregulated σ-factor genes, RSP2681 and RSP1272, have no known functions. These were upregulated already at 7 min following NaCl addition, and the extent of their upregulation was the highest under salt stress of all other conditions tested by microarrays thus far (26). Therefore, we expect RSP2681 and RSP1272 to play important roles in salt tolerance, a prediction that remains to be tested.
The transcription factor CrpO is important for NaCl tolerance.
To begin exploring the contributions to salt tolerance of various regulatory systems, we selected four upregulated genes encoding putative transcription factors of unknown function. Among these genes, RSP1435 was the most highly upregulated (38.8-fold at 7 min and 5.6-fold at 45 min) (see Table S2 in the supplemental material). RSP1945 had a similar pattern (11.3-fold at 7 min and 6.5-fold at 45 min) to that of RSP1435, which is characterized by higher expression at 7 min and somewhat lower expression at 45 min. The mRNA levels of two other transcription factor genes, RSP0018 (2.5-fold at 7 min and 8.6-fold at 45 min) and RSP1275 (3.1-fold at 7 min and 4.6-fold at 45 min), increased more gradually.
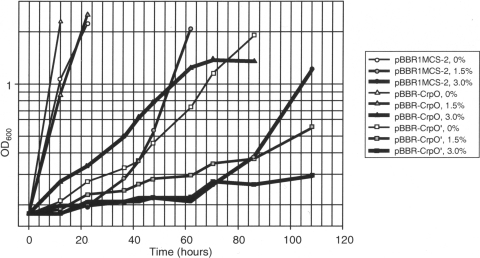
To test whether increased expression of these four putative transcription factors contributes to salt tolerance, all were cloned into a 4- to 6-copy number vector under their native promoters, and salt tolerance of strain 2.4.1 containing the expression plasmids was compared to strain 2.4.1 containing the empty vector, pBBR1MCS-2. The relative growth rate of strain 2.4.1 expressing RSP1435, RSP1945, or RSP0018 in multicopy was not improved in the presence of added salt (data not shown). However, one of four genes in multicopy, RSP1275, greatly improved salt tolerance for 2.4.1(pBBR-CrpO) (Fig. 4). The RSP1275 gene product belongs to the Crp-Fnr regulatory protein family (reviewed in reference 20). Since the effector domain of the protein encoded by RSP1275 lacks cysteine residues involved in iron-sulfur cluster formation, which is characteristic of the Fnr homologs, we designated it CrpO (where O stands for osmotic stress).
Fig. 4.
Effect of the phospholipid regulator CrpO on salt tolerance. Anoxic phototrophic cultures of R. sphaeroides 2.4.1 containing the plasmids indicated were grown in the presence of added NaCl (0%, 1.5%, or 3%). Plasmids are as follows: ●, pBBR1MCS-2 (vector); ▴, pBBR-CrpO expressing crpO; and ■, pBBR-CrpO′ expressing truncated crpO, i.e., crpO′.
Our attempts to construct a crpO null mutant by allele replacement were unsuccessful, suggesting that crpO may be essential. To investigate CrpO function, we constructed a dominant negative allele. Because Crp-Fnr proteins are composed of an N-terminal effector domain linked by a dimerization region to a C-terminal DNA binding domain, we expected that a truncated CrpO lacking the DNA-binding domain, CrpO′, would be able to dimerize with the wild-type protein and that CrpO-CrpO′ heterodimers would be impaired in DNA binding. In agreement with these expectations, in the presence of 1.5% added NaCl, CrpO′ inhibited phototrophic growth of the wild type; e.g., by the time 2.4.1(pBBR1MCS-2) reached stationary phase, the cell density of 2.4.1(pBBR-CrpO′) had not yet doubled (Fig. 4).
CrpO affects membrane lipids.
Measurements using transcriptional fusions revealed that CrpO in multicopy does not affect expression of genes associated with compatible solute transport and synthesis (data not shown). Therefore, we investigated the involvement of CrpO in changing membrane lipid composition. In 2.4.1(pBBR-CrpO), even in the absence of added NaCl, the CL/PC ratio was found to be similar to the ratio observed in the wild type, 2.4.1(pBBR1MCS-2), experiencing salt stress. Addition of NaCl further increased the CL/PC ratio in 2.4.1(pBBR-CrpO) (Table 4). These observations strongly suggest that CrpO primarily contributes to increased salt tolerance via changes in membrane lipids.
Table 4.
Changes in the CL/PC phospholipid ratio in R. sphaeroides 2.4.1 with multiple copies of crpO
| Strain and conditiona | CL/PC ratiob |
|---|---|
| pBBR1MCS-2 | |
| No NaCl | 0.55 |
| + NaCl | 0.75 |
| pBBR-CrpO | |
| No NaCl | 0.72 |
| + NaCl | 0.85 |
Phototrophically growing R. sphaeroides 2.4.1 containing the empty vector, pBBR1MCS-2, or pBBR-CrpO(pBBR1MCS-2::crpO).
Phospholipid ratios in cells following a 45-min incubation with 3% added NaCl versus no added NaCl. Values are averages of three biological replicates. Standard deviations did not exceed 10% of the average for all measurements.
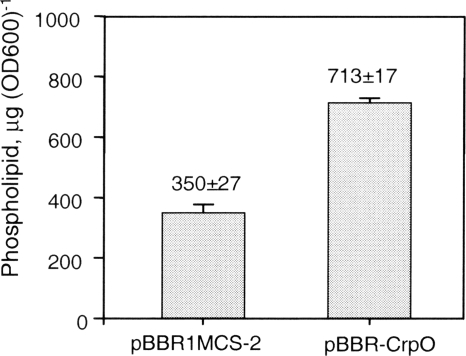
Mukhopadhyay et al. observed that polar lipid content in salt-stressed D. vulgaris was increased in response to salt stress (27). Likewise, the total phospholipid amount in 2.4.1(pBBR-CrpO) was approximately 2-fold higher than in 2.4.1(pBBR1MCS-2) (713 versus 350 μg/OD600 unit) (Fig. 5). This result suggests that CrpO may contribute to increased salt tolerance by both increasing the percentage of CL among the membrane phospholipids and elevating the overall phospholipid levels. How CrpO function mechanistically remains to be explored.
Fig. 5.
Changes in phospholipid concentrations induced by CrpO. Shown are phospholipid levels in phototrophic R. sphaeroides 2.4.1 with the empty plasmid vector pBBR1MCS-2 or plasmid pBBR-CrpO. Values are averages of a total of four data points consisting of duplicate assays of two independently grown replicates, and error bars indicate standard deviations.
In summary, this study correlated the changes in the NaCl stress-induced transcriptome with the dynamics of transport and synthesis of key compatible solutes in R. sphaeroides and identified or confirmed genes responsible for compatible solute transport and synthesis. This work also characterized NaCl-induced changes in phospholipids and identified an apparently essential transcription factor, CrpO, as an important contributor to salt tolerance that affects membrane lipids. Other known or putative transcription factors that were up- or downregulated by NaCl stress represent potential new regulators of salt tolerance whose analysis will help decipher the salt stress-responsive regulatory network in R. sphaeroides. Finally, this study may shed light on the adaptation of freshwater bacteria to a seawater environment since a recently sequenced genome of the seawater strain KD131 of R. sphaeroides has remarkably high sequence identity with the genome of strain 2.4.1 (22).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by KAKENHI (Grant-in-Aid for Scientific Research) from the Japan Society for the Promotion of Science (M.A.), by NSF 0921449 (J.Z.-R.), and by the University of Wyoming Agricultural Experimental Station (M.G.).
We are grateful to M. Suwansaard for useful discussions and to the staff of the University of Colorado Cancer Center Microarray Core Facility, where the R. sphaeroides gene chips were processed.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Abee T., Palmen R., Hellingwerf K., Konings W. 1990. Osmoregulation in Rhodobacter sphaeroides. J. Bacteriol. 172:149–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson J., Smith T., Hoover T. 2010. Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol. 18:30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anthony J., Warczak K., Donohue T. 2005. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc. Natl. Acad. Sci. U. S. A. 102:6502–6507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bligh E., Dyer W. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 5. Braatsch S., Moskvin O., Klug G., Gomelsky M. 2004. Responses of the Rhodobacter sphaeroides transcriptome to blue light under semiaerobic conditions. J. Bacteriol. 186:7726–7735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown A. 1976. Microbial water stress. Bacteriol. Rev. 40:803–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Catucci L., Depalo N., Lattanzio V., Agostiano A., Corcelli A. 2004. Neosynthesis of cardiolipin in Rhodobacter sphaeroides under osmotic stress. Biochemistry 43:15066–15072 [DOI] [PubMed] [Google Scholar]
- 8. De Leo V., et al. 2009. Cardiolipin increases in chromatophores isolated from Rhodobacter sphaeroides after osmotic stress: structural and functional roles. J. Lipid Res. 50:256–264 [DOI] [PubMed] [Google Scholar]
- 9. Ditta G., Stanfield S., Corbin D., Helinski D. 1980. Broad host range DNA cloning system for gram negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc. Natl. Acad. Sci. U. S. A. 77:7347–7351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dryden S., Dowhan W. 1996. Isolation and expression of the Rhodobacter sphaeroides gene (pgsA) encoding phosphatidylglycerophosphate synthase. J. Bacteriol. 178:1030–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eraso J. M., Kaplan S. 1994. prrA, a putative response regulator involved in oxygen regulation in photosynthesis gene expression in Rhodobacter sphaeroides. J. Bacteriol. 176:32–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Figurski D., Helinski D. 1979. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Forward J., Behrendt M., Wyborn N., Cross R., Kelly D. 1997. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse Gram-negative bacteria. J. Bacteriol. 179:5482–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Galinski E., Truper H. 1994. Microbial behavior in salt-stressed ecosystems. FEMS Microbiol. Rev. 15:95–108 [Google Scholar]
- 15. Gautier L., Cope L., Bolstad B., Irizarry R. 2004. Affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20:307–315 [DOI] [PubMed] [Google Scholar]
- 16. Irizarry R., et al. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacobs M., van der Heide T., Driessen A., Konings W. 1996. Glutamate transport in Rhodobacter sphaeroides is mediated by a novel binding protein-dependent secondary transport system. Proc. Nat. Acad. Sci. U. S. A. 93:12786–12790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jessee J. 1986. New subcloning efficiency competent cells: >1 × 106 transformants/μg. Focus 8:9–10 [Google Scholar]
- 19. Kendziorski C., Newton M., Lan H., Gould M. 2003. On parametric empirical Bayes methods for comparing multiple groups using replicated gene expression profiles. Stat. Med. 22:3899–3914 [DOI] [PubMed] [Google Scholar]
- 20. Korner H., Sofia H., Zumft W. G. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:559–592 [DOI] [PubMed] [Google Scholar]
- 21. Kovach M., et al. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 22. Lim S. K., et al. 2009. Complete genome sequence of Rhodobacter sphaeroides KD131. J. Bacteriol. 191:1118–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu Y., et al. 2005. Transcriptome analysis of Shewanella oneidensis MR-1 in response to elevated salt conditions. J. Bacteriol. 187:2501–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Makihara F., et al. 2005. Role of trehalose synthesis pathways in salt tolerance mechanism of Rhodobacter sphaeroides f. sp. denitrificans IL106. Arch. Microbiol. 184:56–65 [DOI] [PubMed] [Google Scholar]
- 25. Miller V., Mekalanos T. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires boxR. J. Bacteriol. 170:2575–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moskvin O., Bolotin D., Wang A., Ivanov P., Gomelsky M. 2011. Rhodobase, a meta-analytical tool for reconstructing gene regulatory networks in a model photosynthetic bacterium. Biosystems 103:125–131 [DOI] [PubMed] [Google Scholar]
- 27. Mukhopadhyay A., et al. 2006. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 188:4068–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pappas C., et al. 2004. Construction and validation of the Rhodobacter sphaeroides 2.4.1 DNA microarray: transcriptome flexibility at diverse growth modes. J. Bacteriol. 186:4748–4758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Penfold R., Pemberton J. 1992. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene 118:145–146 [DOI] [PubMed] [Google Scholar]
- 30. Poggio S., Osorio A., Dreyfus G., Camarena L. 2002. The four different σ54 factors of Rhodobacter sphaeroides are not functionally interchangeable. Mol. Microbiol. 46:75–85 [DOI] [PubMed] [Google Scholar]
- 31. Poggio S., Osorio A., Dreyfus G., Camarena L. 2006. Transcriptional specificity of RpoN1 and RpoN2 involves differential recognition of the promoter sequences and specific interaction with the cognate activator proteins. J. Biol. Chem. 281:27205–27215 [DOI] [PubMed] [Google Scholar]
- 32. Poolman B., Glaasker E. 1998. Regulation of compatible solute accumulation in bacteria. Mol. Microbiol. 29:397–407 [DOI] [PubMed] [Google Scholar]
- 33. Prentki P., Krisch H. M. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303–313 [DOI] [PubMed] [Google Scholar]
- 34. Romantsov T., Guan Z., Wood J. 2009. Cardiolipin and the osmotic stress responses of bacteria. Biochim. Biophys. Acta 1788:2092–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rüberg S., et al. 2003. Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: genome-wide profiling of osmoadaptive gene expression. J. Biotechnol. 106:255–268 [DOI] [PubMed] [Google Scholar]
- 36. Sambrook J., Fritsch E., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Simon R., Priefer U., Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Biotechnology 1:37–45 [Google Scholar]
- 38. Sistrom W. R. 1960. A requirement for sodium in the growth of Rhodopseudomonas sphaeroides. J. Gen. Microbiol. 22:778–785 [DOI] [PubMed] [Google Scholar]
- 39. Stewart J. 1980. Colorimeteric determination of phospholipids with ammonium ferrothiocyanate. Anal. Biochem. 104:10–14 [DOI] [PubMed] [Google Scholar]
- 40. Tamot B., Benning C. 2009. Membrane lipid biosynthesis in purple bacteria, p. 119–134 In Hunter C., Daldal F., Thurnauer M., Beatty J. (ed.), The purple phototrophic bacteria. Springer, Dordrecht, The Netherlands [Google Scholar]
- 41. Weber A., Jung K. 2002. Profiling early osmostress-dependent gene expression in Escherichia coli using DNA macroarrays. J. Bacteriol. 184:5502–5507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wood J., et al. 2001. Osmosensing and osmoregulatory compatible solute accumulation by bacteria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 130:437–460 [DOI] [PubMed] [Google Scholar]
- 43. Xu X., Abo M., Okubo A., Yamazaki S. 1998. Trehalose as osmoprotectant in Rhodobacter sphaeroides f. sp. denitrificans IL106. Biosci. Biotechnol. Biochem. 62:334–337 [DOI] [PubMed] [Google Scholar]
- 44. Yanisch-Perron C., Vieira J., Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
- 45. Yuan M., Kendziorski C. 2006. A unified approach for simultaneous gene clustering and differential expression identification. Biometrics 62:1089–1098 [DOI] [PubMed] [Google Scholar]
- 46. Zeilstra-Ryalls J. H., Kaplan S. 1995. Regulation of 5-aminolevulinic acid synthesis in Rhodobacter sphaeroides 2.4.1: the genetic basis of mutant H-5 auxotrophy. J. Bacteriol. 177:2760–2768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zeller T., Moskvin O., Li K., Klug G., Gomelsky M. 2005. Transcriptome and physiological responses to hydrogen peroxide of the facultatively phototrophic bacterium Rhodobacter sphaeroides. J. Bacteriol. 187:7232–7242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zeller T., et al. 2007. Regulation of hydrogen peroxide-dependent gene expression in Rhodobacter sphaeroides: regulatory functions of OxyR. J. Bacteriol. 189:3784–3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.