Abstract
The majority of marine dissolved organic carbon (DOC) is resistant to biological degradation and thus can remain in the water column for thousands of years, constituting carbon sequestration in the ocean. To date the origin of such recalcitrant DOC (RDOC) is unclear. A recently proposed conceptual framework, the microbial carbon pump (MCP), emphasizes the microbial transformation of organic carbon from labile to recalcitrant states. The MCP is concerned with both microbial uptakes and outputs of DOC compounds, covering a wide range from gene to ecosystem levels. In this minireview, the ATP binding cassette (ABC) transporter is used as an example for the microbial processing of DOC at the genetic level. The compositions of the ABC transporter genes of the two major marine bacterial clades Roseobacter and SAR11 demonstrate that they have distinct patterns in DOC utilization: Roseobacter strains have the advantage of taking up carbohydrate DOC, while SAR11 bacteria prefer nitrogen-containing DOC. At the ecosystem level, bacterially derived RDOC based on d-amino acid biomarkers is reported to be responsible for about a quarter of the total marine RDOC pool. Under future global warming scenarios, partitioning of primary production into DOC could be enhanced, and thus the MCP could play an even more important role in carbon sequestration by the ocean. Joint efforts to study the MCP from multiple disciplines are required to obtain a better understanding of ocean carbon cycle and its coupling with global change.
INTRODUCTION
The ocean acts as a “sink” of atmospheric CO2, thus mitigating global warming (44, 48). The recognized biological mechanism for this sink is the “biological pump,” which is based on the photosynthetic fixation of CO2 and subsequent carbon export driven mainly by sinking of particulate organic carbon (POC). However, the buried POC in the ocean sediment is <0.1% of primary production formed in the surface ocean, while the rest of the organic carbon is mainly respired back to CO2 (14, 28). Nevertheless, the ocean possesses a huge dissolved organic carbon (DOC) pool, which accounts for about 95% of the total remaining organic carbon (22, 39). The majority of this DOC pool is recalcitrant to biological degradation and can persist in the water column for thousands of years, constituting carbon sequestration in the ocean (4, 22, 25, 39, 40). However, the origin of this recalcitrant DOC pool is presently unclear.
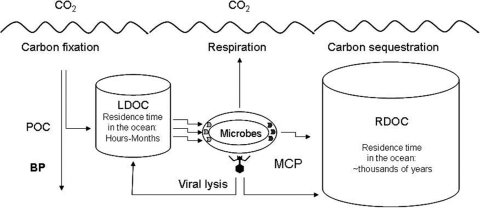
Recently a new concept was proposed to address this matter, the “microbial carbon pump” (MCP), which refers to the microbial processes that transform labile DOC (LDOC) into recalcitrant DOC (RDOC) (25). The MCP is a conceptual framework to explicitly integrate environmental, trophic, physiological, molecular, and genomic processes relevant to the in situ microbial activities that regulate RDOC production and dynamics (25, 26) (Fig. 1).
Fig. 1.
Diagram showing the microbial carbon pump (MCP) in the context of carbon cycling in the ocean. LDOC, labile dissolved organic carbon; RDOC, recalcitrant dissolved organic carbon; POC, particulate organic carbon; BP, biological pump, the known carbon-sequestration mechanism which is based on POC sinking from the surface to depths and even the bottom of the ocean. The MCP, a newly proposed mechanism of carbon sequestration, is based on microbial formation of RDOC, which is resistant to biological degradation and persistent in the water column for thousands of years. The major MCP processes includes microbial exudation of RDOC compounds and viral lysis of the host cells that releases partially RDOC compounds. The ellipse shape indicates a microbial cell with importers (left) and exporters (right).
Diverse microbes and DOC compounds in the ocean.
The transformation of DOC is carried out by heterotrophic microbes, and the pathways and rates of DOC transformation determine the fate and the amount of carbon converted ultimately to CO2 or RDOC (34, 35, 45). The DOC pool consists of thousands of organic carbon compounds with different biological turnover rates, biological availabilities, and biogeochemical features (9). While microbial processes modify the composition of the DOC pool, the availability of DOC compounds to microbes shapes microbial diversity and community structure (19). For instance, the Roseobacter clade and the SAR11 clade are dominant bacterial groups in relatively eutrophic and oligotrophic waters, respectively. Each clade has different strategies for carbon utilization (17, 18, 33) and thus different responses to and impacts on the DOC pool in the ocean.
Getting started with ABC transporters on gene watch for DOC metabolism.
ATP binding cassette (ABC) transporter genes are present in all species from the lowliest microbes to humans, and their products can be recognized as either importers or exporters that mediate the movement of DOC compounds into or out of cells (23, 29, 46). ABC importers have been found exclusively in prokaryotic organisms, whereas exporters are produced ubiquitously by all the domains of life (29, 46). An ABC transporter consists of three molecular components: the ATP-binding protein, the transporter domain, and the substrate-binding protein (23, 24, 46). Bacterial ABC transporters are commonly observed proteins in marine environments (12, 43, 57). Combined metagenomic and metatranscriptomic data of marine microbial communities have revealed a great number of homologous genes involved in ABC transporters related to carbon and nutrient metabolism (12, 37, 42, 57). In oceanic waters such as the Sargasso Sea, ABC transporter proteins from SAR11 are reported to be important components of the metaproteome (54). In coastal waters, most DOC-related transporters produced by the bacterial community are recognized to be components of ABC transporters (43).
All of these findings indicate that ABC transporter-mediated microbial processes are very important for the cycling of carbon and nutrients in marine environments (36). We do not discuss the possible role of secondary transport systems but assume, for the sake of clarity, that bacterial ABC importers and exporters can be used as indicators of the capabilities of a bacterium or bacterial group or community to take up and generate corresponding DOC compounds respectively. On one hand, bacterial ABC importers are essential for the uptake of amino acids, sugars, and trace metals (24, 38, 46). On the other hand, many exporters are involved either in eliminating waste products, toxins, or muramic acids (23, 52) or in excreting cellular components which function outside the plasma membrane, including cell wall polysaccharides, lipoproteins, lipopolysaccharides (Table 1, transporters 9 to 11), antibiotic products, and proteins (23, 53). Many of these products are also recognized as bacterial RDOC (1, 2, 3, 27, 32, 40). Each ABC transporter is relatively specific for its own particular DOC substrates. So far, ABC transporters have been characterized with specificity for inorganic ions, sugar, amino acids, polypeptides, lipids, and complex polysaccharides (23, 50, 53). Thus, DOC utilization and generation of a bacterium or bacterial group or even a microbial community can be characterized by their ABC transporter information, such as diversity and substrate specificity.
Table 1.
Distribution of ABC transporters in some marine bacterial genomes from Roseobacter clade and SAR11 clade
| Subcluster | Clade | Strain(s) | Operon contentc |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Importer |
Exporter |
||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | |||
| Roseobacter clade | |||||||||||||
| Alpha-3 | 1 | Phaeobacter gallaeciensis 2.10 | + | + | O | + | + | + | + | + | + | ||
| 1 | Roseobacter sp. MED193 | + | + | O | + | + | |||||||
| 1 | Roseobacter sp. SK209-2-6 | O | + | + | + | O | |||||||
| 1 | Ruegeria pomeroyi DSS-3 | O | + | O | + | + | |||||||
| 1 | Ruegeria sp. R11 | O | + | + | O | + | + | + | O | + | + | ||
| 1 | Ruegeria sp. TM1040 | O | O | + | |||||||||
| 1 | Silicibacter lacuscaerulensis ITI-1157 | + | + | + | + | + | O | + | + | ||||
| 1 | Silicibacter sp. TrichCH4B | + | + | + | O | + | + | + | + | + | + | ||
| 2 | Roseobacter litoralis Och 149 | + | + | O | + | + | + | + | + | ||||
| 2 | Roseobacter denitrificans OCh 114 | + | + | + | + | + | + | + | |||||
| 2 | Roseobacter sp. GAI101 | + | + | + | O | O | O | + | O | + | + | ||
| 2 | Sulfitobacter sp. EE-36 | + | + | + | + | + | + | ||||||
| 2 | Sulfitobacter sp. NAS-14.1 | + | + | + | + | + | + | + | |||||
| 2 | Rhodobacterales bacterium HTCC2083 | O | + | + | + | O | + | + | |||||
| 3 | Oceanicola batsensis HTCC2597 | + | + | + | + | + | + | + | + | ||||
| 3 | Pelagibaca bermudensis HTCC2601 | + | + | + | + | + | + | + | O | + | + | ||
| 3 | Roseibium sp. TrichSKD4 | O | + | O | + | + | + | + | + | ||||
| 3 | Roseobacter sp. AzwK-3b | O | + | O | + | + | |||||||
| 3 | Roseovarius nubinhibens ISM | O | + | O | + | + | |||||||
| 3 | Roseovarius sp. 217 | O | + | + | + | O | |||||||
| 3 | Roseovarius sp. TM1035 | O | + | + | O | + | + | ||||||
| 3 | Sagittula stellata E-37 | + | + | + | + | + | + | O | + | + | |||
| 4 | Ketogulonicigenium vulgare Y25 | + | + | + | + | + | + | + | |||||
| 4 | Loktanella vestfoldensis SKA53 | + | + | + | + | + | O | + | O | ||||
| 4 | Oceanicola granulosus HTCC2516 | + | + | + | + | + | + | + | + | + | + | ||
| 4 | Octadecabacter antarcticus 238 | + | + | O | O | + | O | + | O | ||||
| 4 | Octadecabacter antarcticus 307 | + | + | + | + | + | + | + | + | O | + | O | |
| 4 | Roseobacter sp. CCS2 | + | + | + | + | + | + | O | + | + | |||
| 5 | Dinoroseobacter shibae DFL12 | + | + | + | + | + | + | + | + | + | + | ||
| 5 | Jannaschia sp. CCS1 | + | + | + | + | + | + | + | + | + | + | ||
| 5 | Thalassiobium sp. R2A62 | + | + | O | + | + | + | O | + | O | |||
| Erythro-Citro clade | |||||||||||||
| Alpha-4a | Erythrobacter and Citromicrobium | + | + | + | |||||||||
| SRA11b | Pelagibacter ubique | ++ | + | ||||||||||
Erythrobacter litoralis HTCC2594, Erythrobacter sp. SD-21, Erythrobacter sp. NAP1, Citromicrobium bathyomarinum JL354, and Citromicrobium sp. JLT1363.
Pelagibacter ubique HTCC1062, Pelagibacter ubique HTCC1002, Pelagibacter sp. HTCC7211, Pelagibacter sp. IMCC9063, and alphaproteobacterium HIMB114.
Transporter classification was taken from the work of Higgins, Saier, and Schneider (23, 49, 50). Transporters were identified by KAAS (the KEGG automatic annotation server). Transporter no. 1, ribose (rbsABCD); no. 2, d-xylose (xylFHG); no. 3, fructose (frcABC); no. 4, rhamnose (rhaPQST); no. 5, glycerol-3-phosphate (ugpABCE); no. 6, sorbitol/mannitol (smoEFGK); no. 7, α-glucoside (aglEFGK); no. 8, amino acids; no. 9, capsular polysaccharide (kpsEMT); no. 10, lipoprotein (lolCDE); no. 11, lipopolysaccharide (lptBFG). Gene designations are given in parentheses. Operons containing all expected genes are characterized by ”+”; operons that apparently lack genes are characterized by ”O.” GenBank accession numbers are as follows: Phaeobacter gallaeciensis 2.10, NZ_ABIE00000000; Roseobacter sp. MED193, NZ_AANB00000000; Roseobacter sp. SK209-2-6, NZ_AAYC00000000; Ruegeria pomeroyi DSS-3, NC_006569; Ruegeria sp. R11, NZ_ABXM00000000; Ruegeria sp. TM1040, NC_008044; Silicibacter lacuscaerulensis ITI-1157, NZ_ACNX00000000; Silicibacter sp. TrichCH4B, NZ_ACNZ00000000; Roseobacter litoralis Och 149, NZ_ABIG00000000; Roseobacter denitrificans OCh 114, NC_008209; Roseobacter sp. GAI101, NZ_ABXS00000000; Sulfitobacter sp. EE-36, NZ_AALV00000000; Sulfitobacter sp. NAS-14.1, NZ_AALZ00000000; Rhodobacterales bacterium HTCC2083, NZ_ABXE00000000; Oceanicola batsensis HTCC2597, NZ_AAMO00000000; Pelagibaca bermudensis HTCC2601, NZ_AATQ00000000; Roseibium sp. TrichSKD4, NZ_AEFL00000000; Roseobacter sp. AzwK-3b, NZ_ABCR00000000; Roseovarius nubinhibens ISM, NZ_AALY00000000; Roseovarius sp. 217, NZ_AAMV00000000; Roseovarius sp. TM1035, NZ_ABCL00000000; Sagittula stellata E-37, NZ_AAYA00000000; Ketogulonicigenium vulgare Y25, NC_014625; Loktanella vestfoldensis SKA53, NZ_AAMS00000000; Oceanicola granulosus HTCC2516, NZ_AAOT00000000; Octadecabacter antarcticus 238, NZ_ABSK00000000; Octadecabacter antarcticus 307, NZ_ABSH00000000; Roseobacter sp. CCS2, NZ_AAYB00000000; Dinoroseobacter shibae DFL12, NC_009952; Jannaschia sp. CCS1, NC_007802; Thalassiobium sp. R2A62, NZ_ACOA00000000; Maritimibacter alkaliphilus HTCC2654, NZ_AAMT00000000; Citromicrobium bathyomarinum JL354, NZ_ADAE00000000; Citromicrobium sp. JLT1363, AEUE00000000; Erythrobacter litoralis HTCC2594, NC_007722; Erythrobacter sp. SD-21, NZ_ABCG00000000; Erythrobacter sp.NAP1, NZ_AAMW00000000; Pelagibacter ubique HTCC1062, NC_007205; Pelagibacter ubique HTCC1002, NZ_AAPV00000000; Pelagibacter sp. HTCC7211, NZ_ABVS00000000; Pelagibacter sp. IMCC9063, NC_015380; and Alpha proteobacterium HIMB114, NZ_ADAC00000000.
Characterizing DOC utilization in major bacterial groups by ABC transporters.
The Roseobacter clade is one of the major marine groups, accounting for up to 20% and 15% of bacterial communities in coastal waters and the mixed layer of oceanic waters, respectively (52, 58). Members of the Roseobacter clade are present in diverse marine habitats, and their metabolic activity plays a crucial role in the global carbon cycle (58). Their isolated bacterial strains are very closely related to the environmental clones (33, 58), providing fair representatives for genome sequencing. Based on these sequences, diverse transporters for acquiring carbon have been reported (33). The present analyses reveals seven carbohydrate-related ABC importers widely existing in the Roseobacter clade, covering monosaccharides (Table 1, transporters 1 to 4), oligopolysaccharides, and polyols (Table 1, transporters 5 to 7), suggesting that this “eutrophic” clade is more capable of utilizing carbohydrate-related DOC compounds than closely related alpha-4 subcluster bacteria (Table 1). In coastal waters, carbohydrate-related ABC transporter transcript sequences belonging to the Roseobacter clade account for more than 40% of all the DOC-related transporter sequences (43).
The SAR11 clade is another important marine alphaproteobacterial group that is found in all marine environments, accounting for ∼25% of all heterotrophic bacterial abundance (8, 17, 18). The SAR11 clade is comprised of so-called “oligotrophic bacteria” in terms of dominant habitats, genome size (and composition), and cell size. In contrast to the Roseobacter clade, in the SAR11 clade expressed transporters for amino acids or nitrogenous compounds (37%) are much more common than those for carbohydrates (6%) (17, 18, 43). Consistent with genomic results, few carbohydrate-related ABC transporters are found in SAR11 bacterial genomes (17) (Table 1). The carbohydrate utilization patterns of cultivated strains also indicate that SAR11 cannot utilize common carbohydrates, such as fucose, rhamnose, mannose, galactose, ribose, and arabinose, or the disaccharides maltose and trehalose (51). The proportion of genes encoding amino acid transporters in SAR11 is higher than that in other alphaproteobacterial genomes (17). The various ABC transporters in SAR11 are mostly nutrient uptake related, and their high affinity for specialized substrates makes SAR11 capable of taking up carbon and nutrients at very low levels, enabling SAR11 to prevail in many marine environments but especially in the oligotrophic oceans (17, 31).
Bacterially originating potential RDOC compounds.
Marine bacteria can be both major consumers and producers of DOC in the ocean (6, 25, 40). It has been experimentally demonstrated that heterotrophic bacteria can take up LDOC even at very low concentrations (nanomolar) and generate RDOC very rapidly (3, 20, 40). The known bacterium-derived RDOC compounds include cell wall components, such as peptidoglycan, lipopolysaccharides, lipoprotein (Table 1, transporters 9 to 11), aromatic or olefinic components, and d-enantiomer amino acids (1, 2, 3, 27, 32, 40). Marine bacterium cell wall materials can persist throughout the water column (32), and structural polymers can display long-term geochemical stability (11). Bacteria export polysaccharides (Table 1, transporters 9 to 11) to their outer membrane as a component of the cell wall via ABC transporters, and some of them are released into the environment (10, 23, 55). The basic structures of these polysaccharides are known but with various species specificities or stage specificities (21). Complex polymers are usually recalcitrant, at least to the bacteria releasing them (47). Some of these polymers are semilabile DOC or situation-specific RDOC; e.g., metagenomic data suggest a greater number of genes putatively involved in polysaccharide degradation in deep-sea microbial populations than those found in the surface populations (12). This indicates that DOC that is resistant to microbial degradation at one depth horizon may serve as a substrate for deeper populations of heterotrophic microbes (7, 12). Binding of d-amino acids synthesized by bacteria to polysaccharides could also make these polymers recalcitrant. The interwoven polysaccharide matrix of peptidoglycan, coupled with the unusual peptide substituents and structural variability, creates heteropolymers resistant to common hydrolytic enzymes in the ocean (32, 47).
The MCP at the ecosystem level.
As illustrated above, heterotrophic microbes generate RDOC while taking up LDOC. The resulting RDOC may be species specific or group specific and might be recalcitrant to some microbes but could be available to some others. Successive microbial processes would make the carbon increasingly recalcitrant in the environment (25, 26). In fact, the above-mentioned bacterial excretion of RDOC is only one of the pathways of the MCP. Another important mechanism releasing the bacterial organic carbon is viral lysis, which redistributes organic carbon across a continuum of dissolved to particulate organic materials (59). While the majority of these lysis products can be rapidly assimilated again by heterotrophs, some of the lysis products are resistant to further microbial use, such as some cell wall materials mentioned above, and thus remain in the environment as RDOC (Fig. 1). Although the proportion of RDOC in total viral lysis production is small, the accumulative production of RDOC via viruses would be significant given that about half of the bacterial production flows through the viral shunt (5, 56, 60). Bacterially derived RDOC based on d-amino-acid biomarkers has been reported to be responsible for about a quarter (155 gigatons of carbon) of total marine RDOC (1).
Equivalent to the total carbon inventory of the atmospheric CO2, the RDOC pool could play a significant role in carbon cycling and climate change. The MCP concept provides a link between microscale processes and macroscale consequences (Fig. 1). Thus, MCP studies must deal with the complexity over a wide range from genes to the ecosystem, and MCP studies require joint efforts from multiple disciplines, particularly microbiology and biogeochemistry (26). This will be a great challenge, but it promises exciting breakthroughs. For instance, once genes are linked to specific RDOC compounds with known chemical structures, a black box will be opened, and questions that have long puzzled scientists can be gradually answered. For example, why do so many organic molecules persist in the ocean in the presence of abundant microbes?
From microscale to ecosystem scope, the MCP is interconnected with the simultaneously occurring biological and abiotic processes involved in carbon cycling in the ocean (26), such as the POC-based biological pump and the dissolved inorganic carbon-based solubility pump (16). In contrast to the biological pump, which emphasizes the vertical transport of carbon from the euphotic zone to the deep sea for carbon sequestration, the MCP emphasizes the formation of RDOC for carbon sequestration. RDOC can persist at any depth in the water column, including the surface ocean. Partitioning of primary production to POC and DOC and its controlling factors should be one of the focuses for integrated studies along environmental gradients. Compared with the solubility pump, which depends on solubility of CO2 in seawater and thus has negative impacts, such as ocean acidification, for marine organisms (41), the MCP does not appreciably alter the buffering capacity of seawater and has no known negative impacts on marine organisms (25).
Recently, the SOCR (Scientific Committee for Oceanic Studies) Working Group 134 on the MCP published a booklet supplemental to Science where the major MCP-related aspects are overviewed (26a). In the MCP special section here, 6 research papers are included to address some specific aspects, including bacterial carbon storage, growth and dynamics of the functional groups of heterotrophic bacteria, cyanobacteria, and archaea, interactions between viruses and their hosts, and microbial carbon products, such as lipids and colored dissolved organic matter (CDOM). The formation of polymers as cellular carbon storage is verified to be common in aerobic anoxygenic phototrophic bacteria (AAPB) when they are provided with substrates with a high C/N ratio and under light (61). Once released into the environment, polymers can be species-specific RDOC or RDOC precursors. AAPB are widely distributed in the global ocean (26b) and are shown by manipulated experiments to grow much faster than other bacteria and to play a much more important role in carbon cycling than is seen from their abundance (16a). Viruses/phages, having top-down control of the host cells, are examined with AAPB in the coastal Mediterranean waters (16a) and with Synechococcus in the Chesapeake Bay (58a); the results suggest that the virus-mediated host mortality and subsequent liberation of DOC may substantially influence the oceanic biogeochemical processes through a microbial loop as well as the MCP. Significant niche partitioning of Crenarchaeota group I between the euphotic and mesopelagic zones has been demonstrated in the East China Sea (24a). The field studies on archaeal lipids in the South China Sea shed light on potential archaeal links to RDOC and the paleoclimate (58b). The experimental studies demonstrated that CDOM is produced mainly by heterotrophic bacteria in the marine environment (48a).
Closing remarks.
Currently, the anthropogenically derived increase in atmospheric CO2 is a global challenge which will change seawater chemical characteristics and particle-size distributions, as well as microbial function and diversity (13, 15, 30). With elevated partial CO2 pressure (pCO2), both primary production and the release of DOC might increase. As a result, the POC pool may remain largely unaltered (48), whereas DOC or its derivatives, such as transparent exopolymer particles, might increase (48). Stoichiometric assays also show that more carbon than nitrogen is drawn down under elevated pCO2 levels (48), suggesting that the MCP will play a more important role under global warming and ocean acidification scenarios. However, the role of the MCP in the world's oceans, with its production and turnover of RDOC still poorly quantified, remains a challenge for further studies. A better picture of ocean carbon cycling will be achieved by addressing the central issues of the MCP, from genes to ecosystems.
ACKNOWLEDGMENTS
This work was supported by the NSFC projects (91028001 and 41076063), the SOA project (201105021), the MOST project (2007CB815904), and NSF grant OCE-0938349.
We thank the three anonymous reviewers for their useful comments and suggestions.
Biography

Dr. Nianzhi Jiao has been Cheung Kong Chair Professor at the State Key Laboratory for Marine Environmental Sciences, Xiamen University, China, since 2000. After receiving his Ph.D. from Ocean University of Qingdao in 1991, he continued his research at MIT in the United States, the University of Tokyo, and the National Institute for Environmental Studies, Japan. Dr. Jiao's research focuses on microbial ecology and carbon cycling. He is a cochair of the Scientific Committee for Ocean Research (SCOR) Working Group 134 on Microbial Carbon Pump in the Ocean.
Footnotes
Published ahead of print on 26 August 2011.
REFERENCES
- 1. Benner R., Herndl G. J. 2011. Bacterially derived dissolved organic matter in the microbial carbon pump, p. 46–48 In Jiao N., Azam F., Sanders S. (ed.), Microbial carbon pump in the ocean. Science/AAAS, Washington, DC: doi:10.1126/science.opms.sb0001 [Google Scholar]
- 2. Benner R., Kaiser K. 2003. Abundance of amino sugars and peptidoglycan in marine particulate and dissolved organic matter. Limnol. Oceanogr. 48:118–128 [Google Scholar]
- 3. Benner R., Pakulski J. D., McCarthy M., Hedges J. I., Hatcher P. G. 1992. Bulk chemical characterization of dissolved organic matter in the ocean. Science 225:1561–1564 [DOI] [PubMed] [Google Scholar]
- 4. Brophy J. E., Carlson D. J. 1989. Production of biologically refractory dissolved organic carbon by natural seawater microbial populations. Deep Sea Res. 36:497–507 [Google Scholar]
- 5. Brussaard C. P. D., et al. 2008. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 2:575–578 [DOI] [PubMed] [Google Scholar]
- 6. Campbell L., Nolla H. A., Vaulot D. 1994. The importance of Prochlorococcus to community structure in the central North Pacific Ocean. Limnol. Oceanogr. 39:954–961 [Google Scholar]
- 7. Carlson C. A., Hansell D. A., Tamburini C. 2011. DOC persistence and its fate after export within the ocean interior, p. 57–59 In Jiao N., Azam F., Sanders S. (ed.), Microbial carbon pump in the ocean. Science/AAAS, Washington, DC: doi:10.1126/science.opms.sb0001 [Google Scholar]
- 8. Carlson C. A., et al. 2009. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 3:283–295 [DOI] [PubMed] [Google Scholar]
- 9. Cherrier J., Bauer J. E. 2004. Bacterial utilization of transient plankton-derived dissolved organic carbon and nitrogen inputs in surface ocean waters. Aquat. Microb. Ecol. 35:229–241 [Google Scholar]
- 10. Decho W. 1990. Microbial exopolymer secretions in ocean environments—their role(s) in food webs and marine processes. Oceanogr. Mar. Biol. Annu. Rev. 28:73–153 [Google Scholar]
- 11. De Leeuw J. W., Largeau C. 1993. A review of macromolecular organic compounds that comprise living organisms and their role in kerogen coal and petroleum formation, p. 23–63 In Engel M. H., Macko S. A. (ed.), Organic geochemistry. Plenum Press, New York, NY [Google Scholar]
- 12. DeLong E. F., et al. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496–503 [DOI] [PubMed] [Google Scholar]
- 13. Du H., Jiao N., Hu Y., Zeng Y. 2006. Diversity and distribution of pigmented heterotrophic bacteria in marine environments. FEMS Microbiol. Ecol. 57:92–105 [DOI] [PubMed] [Google Scholar]
- 14. Ducklow H. W., Steinberg D. K., Buesseler K. O. 2001. Upper ocean carbon export and the biological pump. Oceanography 14:50–58 [Google Scholar]
- 15. Engel A., et al. 2008. Effects of CO2 on particle size distribution and phytoplankton abundance during a mesocosm bloom experiment (PeECE II). Biogeosciences 5:509–521 [Google Scholar]
- 16. Falkowski P., et al. 2000. The global carbon cycle: a test of our knowledge of earth as a system. Science 290:291–296 [DOI] [PubMed] [Google Scholar]
- 16a. Ferrera I., Gasol J. M., Sebastián M., Hojerová E., Koblížek M. 2011. Comparison of growth rates of aerobic anoxygenic phototrophic bacteria and other bacterioplankton groups in coastal Mediterranean waters. Appl. Environ. Microbiol. 77:7451–7458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giovannoni S. J., et al. 2005. Genome streamlining in a cosmopolitan oceanic bacterium. Science 309:1242–1245 [DOI] [PubMed] [Google Scholar]
- 18. Giovannoni S. J., et al. 2005. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438:82–85 [DOI] [PubMed] [Google Scholar]
- 19. Giovannoni S. J., Stingl U. 2005. Molecular diversity and ecology of microbial plankton. Nature 437:343–348 [DOI] [PubMed] [Google Scholar]
- 20. Gruber D. F., Simjouw J., Seitzinger S. P., Taghon G. L. 2006. Dynamics and characterization of refractory dissolved organic matter produced by a pure bacterial culture in an experimental predator-prey system. Appl. Environ. Microbiol. 72:4184–4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo H., Yi W., Song J., Wang G. 2008. Current understanding on biosynthesis of microbial polysaccharides. Curr. Top. Med. Chem. 8:141–151 [DOI] [PubMed] [Google Scholar]
- 22. Hansell D. A., Carlson C. A., Repeta D. J., Schlitzer R. 2009. Dissolved organic matter in the ocean. Oceanography 22:52–61 [Google Scholar]
- 23. Higgins C. F. 2001. ABC transporters: physiology, structure and mechanism—an overview. Res. Microbiol. 152:205–210 [DOI] [PubMed] [Google Scholar]
- 24. Hollenstein K., Dominik C. F., Kaspar P. L. 2007. Structure of an ABC transporter in complex with its binding protein. Nature 446:213–216 [DOI] [PubMed] [Google Scholar]
- 24a. Hu A., Jiao N., Zhang R., Yang Z. 2011. Niche partitioning of marine group I Crenarchaeota in the euphotic and upper mesopelagic zones of the East China Sea. Appl. Environ. Microbiol. 77:7469–7478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jiao N., et al. 2010. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8:593–599 [DOI] [PubMed] [Google Scholar]
- 26. Jiao N., Azam F. 2011. Microbial carbon pump and its significance for carbon sequestration in the ocean, p. 43–45 In Jiao N., Azam F., Sanders S. (ed.), Microbial carbon pump in the ocean. Science/AAAS, Washington DC: doi:10.1126/science.opms.sb0001 [Google Scholar]
- 26a. Jiao N., Azam F., Sanders S. (ed.). 2011. Microbial carbon pump in the ocean, p. 1–68 Science/AAAS, Washington, DC: doi:10.1126/science.opms.sb0001 [Google Scholar]
- 26b. Jiao N., et al. 2007. Distinct distribution pattern of abundance and diversity of aerobic anoxygenic phototrophic bacteria in the global ocean. Environ. Microbiol. 9:3091–3099 [DOI] [PubMed] [Google Scholar]
- 27. Kaiser K., Benner R. 2000. Determination of amino sugars in environmental samples with high salt content by high-performance anion-exchange chromatography and pulsed amperometric detection. Anal. Chem. 72:2566–2572 [DOI] [PubMed] [Google Scholar]
- 28. Karl D., et al. 2001. Biological pump working group summary. OCTET. http://www.msrc.sunysb.edu/octet/biological_pump.html
- 29. Locher K. P. 2009. Structure and mechanism of ATP-binding cassette transporters. Philos. Trans. R. Soc. B 364:239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ma Y., Jiao N., Zeng Y. 2004. Natural community structure of cyanobacteria in the South China Sea as revealed by rpoC1 gene sequence analysis. Lett. Appl. Microbiol. 39:353–358 [DOI] [PubMed] [Google Scholar]
- 31. Malmstrom R. R., Kiene R. P., Cottrell M. T., Kirchman D. L. 2004. Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl. Environ. Microbiol. 70:4129–4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCarthy M. D., Hedges J. I., Benner R. 1998. Major bacterial contribution to marine dissolved organic nitrogen. Science 281:231–234 [DOI] [PubMed] [Google Scholar]
- 33. Moran M. A., et al. 2007. Ecological genomics of marine roseobacters. Appl. Environ. Microbiol. 73:4559–4569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Moran M. A., Sheldon W. M., Sheldon J. E. 1999. Biodegradation of riverine dissolved organic carbon in five estuaries of the southeastern United States. Estuaries 22:55–64 [Google Scholar]
- 35. Moran M. A., Hodson R. E. 1994. Dissolved humic substances of vascular plant origin in a coastal marine environment. Limnol. Oceanogr. 39:762–771 [Google Scholar]
- 36. Mou X., Sun S., Rayapati P., Moran M. A. 2010. Genes for transport and metabolism of spermidine in Ruegeria pomeroyi DSS-3 and other marine bacteria. Aquat. Microb. Ecol. 58:311–321 [Google Scholar]
- 37. Mou X., Sun S., Edwards R. A., Hodson R. E., Moran M. A. 2008. Bacterial carbon processing by generalist species in the coastal ocean. Nature 451:708–711 [DOI] [PubMed] [Google Scholar]
- 38. Nikaido H., Basina M., Nguyen V., Rosenberg E. Y. 1998. Multidrug efflux pump AcrAB of Salmonella typhimurium excretes only those betalactam antibiotics containing lipophilic side chains. J. Bacteriol. 180:4686–4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ogawa H., Tanoue E. 2003. Dissolved organic matter in oceanic waters. J. Oceanogr. 59:129–147 [Google Scholar]
- 40. Ogawa H., Amagai Y., Koike I., Kaiser K., Benner R. 2001. Production of refractory dissolved organic matter by bacteria. Science 292:917–920 [DOI] [PubMed] [Google Scholar]
- 41. Orr J. C., et al. 2005. Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 437:681–686 [DOI] [PubMed] [Google Scholar]
- 42. Poretsky R. S., et al. 2005. Analysis of microbial gene transcripts in environmental samples. Appl. Environ. Microbiol. 71:4121–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Poretsky R. S., Sun S., Mou X., Moran M. A. 2010. Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ. Microbiol. 12:616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raven J. A., Falkowski P. G. 1999. Oceanic sinks for atmospheric CO2. Plant Cell Environ. 22:741–755 [Google Scholar]
- 45. Raymond P. A., Bauer J. E. 2000. Bacterial consumption of DOC during transport through a temperate estuary. Aquat. Microb. Ecol. 22:1–12 [Google Scholar]
- 46. Rees D. C., Johnson E., Lewinson O. 2009. ABC transporters: the power to change. Nat. Rev. Mol. Cell Biol. 10:218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Repeta D. J., Quan T. M., Aluwihare L. I., Accardi A. 2002. Chemical characterization of high molecular weight dissolved organic matter in fresh and marine waters. Geochim. Cosmochim. Acta 66:955–962 [Google Scholar]
- 48. Riebesell U., et al. 2007. Enhanced biological carbon consumption in a high CO2 ocean. Nature 450:545–548 [DOI] [PubMed] [Google Scholar]
- 48a. Romera-Castillo C., Sarmento H., Álvarez-Salgado X. A., Gasol J. M., Marrasé C. 2011. Net production and consumption of fluorescent colored dissolved organic matter by natural bacterial assemblages growing on marine phytoplankton exudates. Appl. Environ. Microbiol. 77:7490–7498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Saier M. H. 2000. Families of transmembrane sugar transport proteins. Mol. Microbiol. 35:699–771 [DOI] [PubMed] [Google Scholar]
- 50. Schneider E. 2001. ABC transporters catalyzing carbohydrate uptake. Res. Microbiol. 152:303–310 [DOI] [PubMed] [Google Scholar]
- 51. Schwalbach M. S., Tripp H. J., Steindler L., Smith D. P., Giovannoni S. J. 2010. The presence of the glycolysis operon in SAR11 genomes is positively correlated with ocean productivity. Environ. Microbiol. 12:490–500 [DOI] [PubMed] [Google Scholar]
- 52. Selje N., Simon M., Brinkhoff T. 2004. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 427:445–448 [DOI] [PubMed] [Google Scholar]
- 53. Silver R. P., Prior K., Nsahlai C., Wright L. F. 2001. ABC transporters and the export of capsular polysaccharides from Gram-negative bacteria. Res. Microbiol. 152:357–364 [DOI] [PubMed] [Google Scholar]
- 54. Sowell S. M., et al. 2009. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 3:93–105 [DOI] [PubMed] [Google Scholar]
- 55. Stoderegger K. E., Herndl G. J. 1998. Production and release of bacterial capsular material and its subsequent utilization by marine bacterioplankton. Limnol. Oceanogr. 43:877–884 [Google Scholar]
- 56. Suttle C. A. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812 [DOI] [PubMed] [Google Scholar]
- 57. Venter J. C., et al. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66–74 [DOI] [PubMed] [Google Scholar]
- 58. Wagner-Döbler I., Biebl H. 2006. Environmental biology of the marine Roseobacter lineage. Annu. Rev. Microbiol. 60:255–680 [DOI] [PubMed] [Google Scholar]
- 58a. Wang K., Wommack K. E., Chen F. 2011. Abundance and distribution of Synechococcus spp. and cyanophages in the Chesapeake Bay. Appl. Environ. Microbiol. 77:7459–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58b. Wei Y., et al. 2011. Spatial variations in archaeal lipids of surface water and core-top sediments in the South China Sea and their implications for paleoclimate studies. Appl. Environ. Microbiol. 77:7479–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Weinbauer M. G., Chen F., Wilhelm S. W. 2011. Virus-mediated redistribution and partitioning of carbon in the global oceans. In Jiao N., Azam F., Sanders S. (ed.), Microbial carbon pump in the ocean, p. 54–56 Science/AAAS, Washington, DC: doi:10.1126/science.opms.sb0001 [Google Scholar]
- 60. Wilhelm S. W., Suttle C. A. 1999. Viruses and nutrient cycles in the sea. Viruses play critical roles in the structure and function of aquatic food webs. Bioscience 49:781–788 [Google Scholar]
- 61. Xiao N., Jiao N. 2011. Formation of polyhydroxyalkanoate in aerobic anoxygenic phototrophic bacteria and its relationship to carbon source and light availability. Appl. Environ. Microbiol. 77:7445–7450 [DOI] [PMC free article] [PubMed] [Google Scholar]