Abstract
A correlative study was performed to determine if variation in streambed microbial community structure in low-order forested streams can be directly or indirectly linked to the chemical nature of the parental bedrock of the environments through which the streams flow. Total microbial and photosynthetic biomass (phospholipid phosphate [PLP] and chlorophyll a), community structure (phospholipid fatty acid analysis), and physical and chemical parameters were measured in six streams, three located in sandstone and three in limestone regions of the Bankhead National Forest in northern Alabama. Although stream water flowing through the two different bedrock types differed significantly in chemical composition, there were no significant differences in total microbial and photosynthetic biomass in the sediments. In contrast, sedimentary microbial community structure differed between the bedrock types and was significantly correlated with stream water ion concentrations. A pattern of seasonal variation in microbial community structure was also observed. Further statistical analysis indicated dissolved organic matter (DOM) quality, which was previously shown to be influenced by geological variation, correlated with variation in bacterial community structure. These results indicate that the geology of underlying bedrock influences benthic microbial communities directly via changes in water chemistry and also indirectly via stream water DOM quality.
INTRODUCTION
Microorganisms are one of the most important groups of living organisms. Their small size, ubiquitous distribution, metabolic diversity, and genetic plasticity cast microorganisms in the role of recycling agents for the biosphere, making life possible for more complex organisms (47). To better understand the ecology of microbial communities, it is important not only to describe the community composition but also to identify biological and/or environmental factors that regulate their diversity (34, 52).
Baas-Becking's dictum that “everything is everywhere, but the environment selects” has inspired many studies and debates since its inception and is considered the precursor to the niche concept adopted by macroecologists (10). Microbes were thought to be cosmopolitan due to large population sizes and short generation times, resulting in high dispersal rates (18, 22). Many studies in the past were limited by methodological tools to elucidate bacterial species, as most bacteria found in the natural environment have yet to be cultured and functional roles and diversity were largely unknown (56, 64). With the recent advances in non-culture-dependent techniques, there has been an increase of studies that have shown evidence of varying distributions, abundance, and diversity of microorganisms in the natural environment across spatial and environmental scales, including freshwater lakes and rivers (60, 65, 70), streams (28), soils (8, 21, 51, 57), and marine ecosystems (33, 54). Yet, it is not clear whether limits in distribution or strong environmental selection lead to the observed patterns. Our study focused on bedrock, a most basic component of the environment, and its influence on microbial community structure in stream sediments, as stream ecosystems provide critical links in global hydrological and biogeochemical cycles and are now viewed as transformative rather than transportive ecosystems (2).
The geochemical setting influences several key environmental determinants and determines the availability of resources that can be physiologically exploited by microorganisms. It is these interactions between microorganisms and their resources that most likely contribute to metabolic diversity (43). Several studies have identified environmental factors that structure sedimentary microbial communities in streams, including light, flow, water temperature, organic carbon concentrations, anthropogenic pollution, hydrology, pH, sediment grain size, inorganic nutrients, and dissolved oxygen (1, 6, 9, 19, 30, 55, 59, 63), but the structuring role of geological formation is unknown.
The linkage between geology and water chemistry in streams is due to minerals released by the weathering of parent bedrock. The input of dissolved chemicals will directly affect pH, alkalinity, and ion concentrations and can potentially change the proportion of essential nutrients available to organisms (44, 67). Studies addressing the relationship between sedimentary microbial communities and water chemistry have focused on pH, nutrients, or anthropogenic influences (3, 4, 20, 46, 53). It is evident that differences in water chemistry influence microbial community structure, but to date natural variation in cation/anion concentrations remains to be investigated. This study's primary objective was to determine if patterns in sedimentary microbial community structure in forested low-order streams is influenced by geological differences in stream channel bedrock.
Mosher et al. (49) showed that dissolved organic matter (DOM) quality was correlated to geological variation in the same streams used for this study. Since previous studies have shown bacterial uptake and growth efficiency are dependent of the source of DOM (37, 39) and DOM quality can influence bacterial community composition (36), this study investigated a possible indirect linkage of geology and microbial community structure via DOM quality. To accomplish this second objective, factor scores from a principal component analysis describing the variation in DOM quality in the six streams (49) were included in the environmental descriptions used in this study.
MATERIALS AND METHODS
Site description.
The study area was in the William B. Bankhead National Forest, in northern Alabama directly south of the Tennessee Valley Divide. All streams are second- or third-order tributaries to the Sipsey Fork of the Black Warrior River system in the Mobile River drainage basin. All streams have similar elevations, discharge, and sediment grain size and are located within 21 km of each other in the north central portion of the forest. The creek beds consist of exposed bedrock, cobble, and unconsolidated sediment.
The forest is located in the Appalachian Plateau physiographic province, and the primary bedrock within the region is Pennsylvanian-age Pottsville formation comprised of sandstone/shale with thin, discontinuous layers of coal (11). Some valleys and streambeds in this region have eroded through the Pottsville bedrock to older Mississippian-age bedrock, specifically Parkwood sandstone and Bangor limestone formations. Two of the streams (Beech and Brushy Creeks: 34°17′49.85″N, 87°18′44.03″W and 34°17′58.8″N, 87°16′29.41″W; see reference 49 for geologic map of study site) flow over exposed Parkwood formation bedrock, characterized by shale/sandstone and clayey coal. Three streams (Thompson, Flannagin, and Borden Creeks: 34°20′26.89″N, 87°28′15.82″W; 34°20′19.79″N, 87°23′18.31″W; 34°19′42.53″N, 87°22′30.15″W, respectively) have eroded through both the Pottsville and Parkwood bedrock and flow over exposed Bangor limestone. Only one stream in this study (Hubbard Creek: 34°18′30.22″N, 87°30′7.83″W) flows solely over the overlying Pottsville sandstone formation. The streams that have eroded through the upper layer(s) still contain Pottsville formation sandstone in the upland portion of their watershed (45). The catchments are heavily forested, primarily by mixed hardwoods interspersed with pine stands.
Sampling.
Sampling was conducted on a quarterly basis for 1.25 years under base flow conditions, at least 2 weeks following the last recorded precipitation in the area. Sediment from each stream was sampled in triplicate with a push core. The top 1 cm of sediment of each core was homogenized, and subsamples were taken for each measurement. Stream water was collected in duplicate, and physical parameters were measured in situ with portable meters. All samples were placed on ice and processed immediately upon return to the laboratory; some samples were preserved (frozen [sediments for lipid analysis] and/or acidified [water for DOC analysis]) and stored prior to analysis (16).
Physical parameters.
Total suspended solids (TSS) were determined by filtering well-mixed stream water samples through dried, preweighed glass fiber filters (GF/F) and drying them at 103°C to a constant mass (14). Specific surface area of combusted sediments was analyzed by the absorption isotherm of nitrogen under vacuum (7). Average weekly discharge in the streams was estimated by a rating curve constructed from instantaneous measurements of discharge and the placement of barologgers in each stream (Levelogger 3001, Solinst Canada Ltd., Georgetown, ON, Canada). The rating curve was constructed from monthly (n = 15) measurements of stream velocity (Marsh McBirney, Frederick, MD), depth measurements, and width. The average weekly gauge height obtained from the barologgers was applied to the rating curve, and discharge for the week preceding the date of sampling was estimated. Canopy cover was estimated using a hand-held convex spherical crown densitometer (Forestry Suppliers, Jackson, MS).
Chemical parameters.
Stream water samples were taken using high-density polyethylene (HDPE) bottles rinsed with a sulfuric acid-nochromix mixture and washed with phosphate-free soap. The first 20 ml of sample was filtered over a combusted GF/F and acidified with two drops of 2 N HCl, and dissolved organic carbon (DOC) concentration was analyzed by flash combustion in a Shimadzu total organic carbon analyzer TOC-500. The remainder of the stream water was filtered through the same GF/F and analyzed for NO3-N, NO2-N, NH4-N, and PO4-P concentrations in a Lachat QuickChem 8000.
Water samples for anions (Cl− and SO42−) and cations (Al3+, Ba2+, Fe2+ and Fe3+ [hereafter Fe], Na+, Ca2+, Mg2+, Mn2+ and Mn3+ and Mn4+ [hereafter Mn], Si2+ and Si4+ [hereafter Si], and K+) were collected in HDPE bottles previously washed with ultrapure water (>18.0 MΩ) and filtered over a combusted GF/F filter. Anion samples were stored at −20°C, and cation samples were acidified (2% [vol/vol]) with optimum-grade nitric acid until analysis. Samples for anion concentrations were analyzed using ion chromatography, and the cation concentration samples were subjected to inductively coupled plasma spectrometry.
Acid-neutralizing capacity (ANC) of the stream water was estimated by the Gran titration method; pH was determined after incremental (0.025-ml) additions of 0.1 N HCl to 60 ml stream water while stirring until full protonation had occurred. The Gran function was calculated using the following equation (68): F1 = (Vorig + V)[H+], where V is the volume and [H+] is the hydrogen ion concentration.
The equivalence point was found by plotting F1 versus acid volume. Dissolved inorganic carbon (DIC) was calculated from the ANC by using the following equation (68): DIC = ([Alk] − [OH−] + [H+])/(α1 + 2α2), where Alk is alkalinity and α1 and α2 are the decimal percentages of carbonate and bicarbonate ions, respectively, in the total DIC.
Conductivity (Fisher Scientific, Waltham, MA), pH (Oakton Instruments, Vernon Hills, IL), dissolved oxygen (DO), and temperature (model 95; YSI, Yellow Springs, OH) were measured in situ using hand-held meters.
Biological parameters.
Microbial biomass and microbial community structure were characterized by phospholipid phosphate (PLP) and phospholipid fatty acid (PLFA) analyses. Cellular lipids were extracted from the sediments by a modified (dichloromethane-methanol-water) Bligh-Dyer lipid extraction (5, 26). Total microbial biomass was determined by the oxidation of lipids and quantification of the resulting orthophosphate colorimetrically (26). Fractionation of phospholipids from the total lipids and esterification into fatty acid methyl esters (FAMEs) using alkylation allowed identification of functional groups comprising the microbial community structure. Purified FAMEs were separated and quantified using a gas chromatograph (23). FAMEs were identified by coelution with known standards and mass spectral analysis. A fraction of total lipids, protected from light, was used to determine chlorophyll a colorimetrically. Dried samples were suspended in 90% aqueous acetone, and absorbance was measured at 663, 645, and 630 nm. Chlorophyll a abundance was calculated using the following equation (62): chlorophyll a = 11.85(OD663) − 1.54(OD645) − 0.08(OD630), where OD663, for example, represents the optical density at 663 nm.
Percent contributions of eukaryotes and prokaryotes to the microbial community biomass were determined using the approach described by Findlay and Dobbs (24), and it was assumed that 50% of the PLFAs from eukaryotes were polyenoic (25, 69) and eukaryotic biomass was the sum of the polyenoic PLFAs × 2. Prokaryotic biomass was calculated as the difference between the total microbial biomass and eukaryotic biomass. Eukaryotic biomass in terms of C was calculated from PLP concentration × the percent eukaryotic biomass (expressed as a decimal fraction) × 0.02 g C μmol P−1 (24). Prokaryotic biomass in terms of C was calculated as the PLP concentration × the percent prokaryotic biomass (expressed as a decimal fraction) × 0.01 g C μmol P−1 (24).
Statistical analyses.
Fully nested analysis of variance (ANOVA) with Tukey's honestly significant difference (P < 0.05) was performed on each physical, chemical, and biological parameter to determine significant differences of the parameters by substrate type (limestone versus sandstone) and sampling date (Minitab 14.13). Pearson correlation coefficient was used to determine colinearity among the environmental parameters (SPSS 14.0).
Constrained ordination techniques were utilized to identify patterns of variation in microbial community structure among streams and correlations between microbial community structure and environmental descriptors. Detrended correspondence analysis (DCA), an indirect gradient analysis based on segment length, was performed to determine the modality of the PLFA data and environmental predictor variables. The analysis resulted in short (<1.0) segment lengths, indicating the data set was linear and suitable for indirect gradient analysis; therefore, redundancy analysis (RDA) was applied (Canoco 4.5). In the RDA, the response variables were individual PLFAs (transformed using ln[weight percentPLFA + 1]), and the predictor variables were measured environmental parameters. Forward selection of the predictor variables followed by Monte Carlo permutation tests were used to prevent artificial inflation of variation due to autocorrelation in the constrained ordination model (42). A second RDA, limited to data collected during the March 2006 sampling, describing the influence of DOM quality on sedimentary microbial community structure was performed and included factor scores (factors 1 and 2) from a principal component analysis that examined variation in DOM quality of the six streams. DOM was sampled concurrently with the March 2006 sampling, and the analysis of the influence of bedrock geology on dissolved organic matter quality has been published elsewhere (49).
RESULTS
Physical, chemical, and biological factors.
The water chemistry of the streams reflected the nature of the bedrock over which the streams flowed. Limestone streams had significantly higher values for ANC, pH, conductivity, and concentrations of Ca2+, DIC, Mg2+, Na+, and SO42− than those of the sandstone streams (Table 1). One of the limestone streams, Thompson Creek, had significantly lower values for these parameters than the other two limestone streams, but the values were significantly higher than those observed in the sandstone streams. Water samples taken from sandstone streams had significantly higher concentrations of NO3-N and Fe than the limestone streams. The differences found in these chemical parameters, with the exception of NO3-N, were in accordance with previous published studies examining differences in ion concentrations in relation to geology. No significant differences were found between the remainder of the chemical parameters (DO and concentrations of Ba2+, Cl−, DOC, K+, Mn, NH4-N, and Si) or any of the physical parameters (percent canopy cover, discharge, temperature, and TSS).
Table 1.
Summary of chemical and environmental characteristics of the six study streams located in the Bankhead National Forest, ALa
| Parameter | Value for bedrock type and specific stream |
P valueb | |||||
|---|---|---|---|---|---|---|---|
| Limestone |
Sandstone |
||||||
| Borden | Flannagin | Thompson | Beech | Brushy | Hubbard | ||
| Al3+ (mg/liter) | 0.04 ± 0.03 | 0.04 ± 0.03 | 0.05 ± 0.04 | 0.08 ± 0.04 | 0.05 ± 0.02 | 0.06 ± 0.02 | NS |
| ANC (meq/liter) | 2.17 ± 0.49 | 2.31 ± 0.39 | 0.90 ± 0.34 | 0.25 ± 0.12 | 0.14 ± 0.07 | 0.12 ± 0.05 | 0.01 |
| Ba2+ (mg/liter) | 0.03 ± 0.00 | 0.03 ± 0.01 | 0.02 ± 0.00 | 0.03 ± 0.02 | 0.02 ± 0.00 | 0.02 ± 0.00 | NS |
| Ca2+ (mg/liter) | 41.24 ± 8.16 | 42.27 ± 8.76 | 14.05 ± 5.32 | 2.60 ± 0.79 | 1.50 ± 0.48 | 1.35 ± 0.11 | 0.01 |
| Canopy (%) | 61.83 ± 33.40 | 44.38 ± 41.68 | 40.27 ± 34.85 | 66.99 ± 31.25 | 96.53 ± 3.69 | 69.11 ± 26.66 | NS |
| Cl− (mg/liter) | 1.42 ± 0.28 | 1.28 ± 0.28 | 1.11 ± 0.13 | 1.33 ± 0.23 | 1.09 ± 0.15 | 1.34 ± 0.28 | NS |
| Conductance (μΩ) | 207.09 ± 46.55 | 223.39 ± 46.35 | 77.80 ± 29.77 | 31.72 ± 5.83 | 23.47 ± 5.07 | 20.75 ± 1.56 | 0.01 |
| DIC (mg/liter) | 0.53 ± 0.12 | 0.56 ± 0.09 | 0.37 ± 0.31 | 0.07 ± 0.03 | 0.04 ± 0.02 | 0.03 ± 0.01 | 0.01 |
| Discharge (m3/s) | 0.25 ± 0.26 | 0.14 ± 0.13 | 0.22 ± 0.20 | 0.17 ± 0.13 | 0.14 ± 0.14 | 0.23 ± 0.13 | NS |
| DO (mg/liter) | 10.88 ± 3.99 | 11.65 ± 4.34 | 9.94 ± 3.62 | 10.52 ± 3.90 | 10.54 ± 4.42 | 10.40 ± 3.36 | NS |
| DOC (mg/liter) | 1.29 ± 0.56 | 1.17 ± 0.37 | 1.64 ± 1.02 | 1.33 ± 0.14 | 1.39 ± 0.50 | 1.08 ± 0.18 | NS |
| Fe (mg/liter) | 0.05 ± 0.02 | 0.05 ± 0.02 | 0.05 ± 0.03 | 0.50 ± 0.36 | 0.38 ± 0.31 | 0.18 ± 0.09 | 0.05 |
| K+ (mg/liter) | 0.86 ± 0.25 | 0.71 ± 0.25 | 0.85 ± 0.16 | 0.80 ± 0.21 | 0.71 ± 0.19 | 0.83 ± 0.20 | NS |
| Mg2+ (mg/liter) | 14.50 | 13.50 | 6.00 | 20.00 | 23.00 | 2.50 | NA |
| Mn (mg/liter) | 2.16 ± 0.42 | 2.45 ± 0.56 | 1.44 ± 0.31 | 1.26 ± 0.20 | 1.07 ± 0.26 | 0.80 ± 0.07 | 0.01 |
| Na+ (mg/liter) | 0.23 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.00 | 0.06 ± 0.03 | 0.01 ± 0.01 | 0.01 ± 0.01 | NS |
| NH4-N (μg/liter) | 1.36 ± 0.14 | 1.20 ± 0.10 | 1.20 ± 0.16 | 1.16 ± 0.15 | 1.03 ± 0.15 | 0.89 ± 0.12 | 0.01 |
| NO3-N (μg/liter) | 2.95 ± 2.45 | 5.80 ± 6.15 | 2.52 ± 2.20 | 13.97 ± 19.41 | 3.39 ± 2.11 | 3.03 ± 2.75 | NS |
| NO2-N (μg/liter) | 17.07 ± 16.53 | 20.22 ± 17.27 | 42.94 ± 21.62 | 99.27 ± 41.86 | 33.33 ± 41.33 | 190.62 ± 71.51 | 0.05 |
| pH | 0.11 ± 0.13 | 0.19 ± 0.29 | 0.16 ± 0.27 | 0.54 ± 0.78 | 0.36 ± 0.60 | 0.31 ± 0.32 | NS |
| PO4-P (μg/liter) | 8.09 ± 0.16 | 8.17 ± 0.66 | 7.73 ± 0.13 | 7.20 ± 0.19 | 7.17 ± 0.13 | 7.24 ± 0.24 | 0.01 |
| Positionc (km) | 0.42 ± 0.65 | 0.36 ± 0.56 | 0.92 ± 1.04 | 0.45 ± 0.49 | 0.26 ± 0.36 | 0.43 ± 0.42 | NS |
| Si (mg/liter) | 2.66 ± 0.27 | 2.62 ± 0.24 | 2.87 ± 0.25 | 3.00 ± 0.42 | 3.09 ± 0.46 | 2.65 ± 0.34 | NS |
| SO42− (mg/liter) | 3.94 ± 0.81 | 5.50 ± 1.44 | 4.30 ± 0.14 | 2.96 ± 0.45 | 3.44 ± 0.31 | 2.52 ± 0.71 | 0.01 |
| Surface area (m3 g of sediment−1) | 1.79 ± 3.28 | 1.86 ± 0.81 | 2.88 ± 0.61 | 1.07 ± 0.77 | 0.31 ± 0.35 | 1.64 ± 2.21 | NS |
| Temp (°C) | 15.05 ± 6.27 | 14.54 ± 6.48 | 16.73 ± 5.79 | 15.59 ± 6.72 | 15.08 ± 7.16 | 14.79 ± 6.09 | NS |
| TSS (mg/liter) | 1.59 ± 1.10 | 3.62 ± 3.07 | 1.37 ± 0.82 | 10.80 ± 9.97 | 3.23 ± 3.14 | 1.58 ± 0.42 | NS |
Adapted from reference 49.
Significance of differences between substrate type (limestone versus sandstone) as determined by fully nested ANOVA. Abbreviations: NA, statistics not applied; NS, no significant difference.
Distance from western park boundary.
Total microbial and autotrophic biomass.
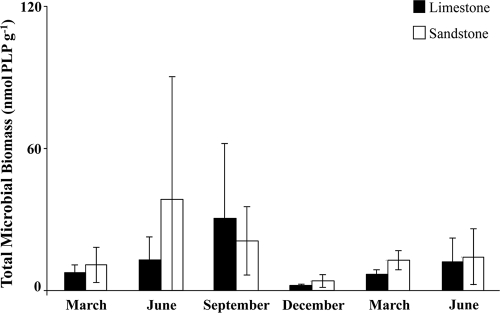
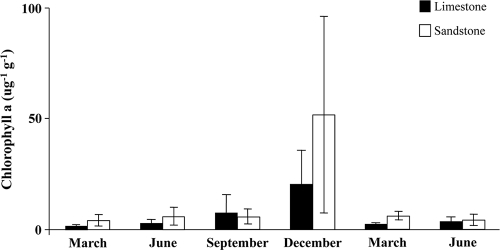
There were no significant differences between substrate types or among sampling dates for total microbial biomass (Fig. 1). Total microbial biomass values ranged from 1.5 to 100 ng PLP g of sediment−1 and the limestone streams averaged 17.5 ng PLP g of sediment−1, while the sandstone streams averaged 21.34 ng PLP g of sediment−1. There were also no significant differences for chlorophyll a concentrations between the substrate types or among sampling dates (Fig. 2). The values for chlorophyll a ranged from 1 to 101 μg g of sediment−1, with a noticeable increase in concentrations for the December sampling date in both substrates. The average concentration of chlorophyll a in limestone streams was 6.23 μg g of sediment−1 and 12.1 μg g of sediment−1 in sandstone streams. There was a positive correlation between chlorophyll a concentration and total microbial biomass (r = 0.673), and none of the environmental variables correlated with total microbial biomass or chlorophyll a concentration.
Fig. 1.
Total microbial biomass (measured by phospholipid phosphate) of streambed sediments by sampling date from six streams (three limestone, three sandstone) located in the Bankhead National Forest. Error bars represent 1 standard deviation.
Fig. 2.
Chlorophyll a concentrations of streambed sediments by sampling date from six streams (three limestone, three sandstone) located in the Bankhead National Forest. Error bars represent 1 standard deviation.
Microbial community structure.
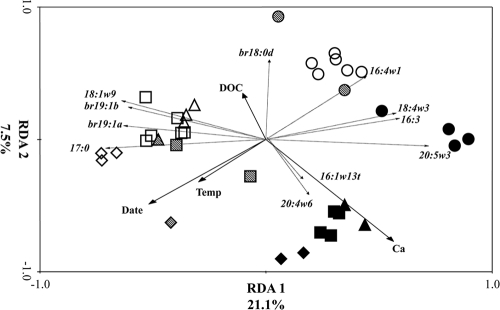
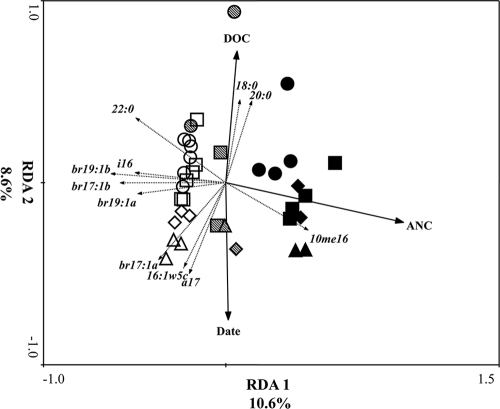
Microbial community structure displayed two distinct overlying patterns as indicated by the triplot of samples, environmental variables and individual fatty acid markers (Fig. 3). Starting at the top left of the triplot and moving toward the bottom right, samples were aligned by increasing stream water ion concentrations, which were significantly associated with Ca2+ and DOC concentrations (Table 2). Starting at the top right of the triplot and moving toward the bottom left, the samples were aligned by increasing water temperature and were significantly associated with water temperature and sampling date (F = 8.71; P = 0.002). RDA canonical axes 1 and 2 described 28.6% of the variation in microbial community structure.
Fig. 3.
Triplot of the redundancy analysis of PLFA profiles of stream bed sediments of six streams (three limestone, three sandstone) located in the Bankhead National Forest, with forward selection of predictor variables followed by Monte Carlo permutations. Solid arrows represent predictor variables significantly associated with variation in microbial community structure. Dashed arrows represent individual fatty acid markers (r > |0.65|) significantly associated with the variation among samples. The lengths of the arrows are correlated with the degree of relation between the response variable (exact values are shown in Table 2). The arrows point in the direction of the maximum change for the associated variable. Legend: closed symbols, limestone streams; open symbols, sandstone streams; stippled symbols, intermediate limestone (Thompson Creek); circles, March; squares, June; diamonds, September; triangles, December.
Table 2.
Correlation matrices (r) of environmental variables from the constrained ordination analysis (RDA) performed on PLFA profiles and environmental variables from the six streams in the Bankhead National Forest
| Factor |
r value |
|
|---|---|---|
| RDA 1 | RDA 2 | |
| Ca2+ concn | 0.4177 | −0.5782 |
| Sampling date | −0.3838 | −0.3666 |
| DOC concn | −0.0761 | 0.2609 |
| Temp | −0.2204 | −0.2404 |
The samples grouped into three clusters in the triplot. Samples taken in March were clustered together and contained two subgroups. Samples from the sandstone streams and the lowest-conductivity limestone stream (Thompson Creek) formed one subgroup within the cluster, and the other subgroup contained Borden and Flannagin Creeks. The March samples were significantly associated with fatty acids found in autotrophic eukaryotes (16:4ω1, 18:4ω3, 16:3, and 20:5ω3). The second cluster consisted of non-March sandstone streams and was described by bacterial fatty acid markers (18:1ω9, br19:1a, br19:1b, and 17:0). The third cluster was comprised of non-March Borden and Flannagin Creeks (the two highest-conductivity limestone streams) and was described by autotrophic (16:1ω13t) and heterotrophic (20:4ω6) eukaryotic markers. Thompson Creek results plotted either with the sandstone streams or between the non-March clusters.
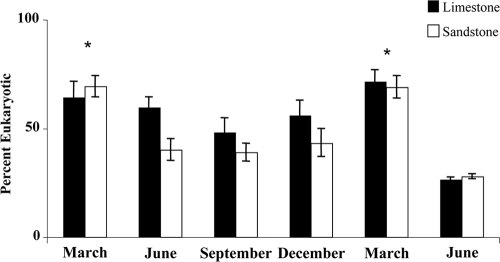
Notably, the significant fatty acid markers describing the variation among the clusters were differences in eukaryotic and bacterial fatty acids. Samples having positive RDA axis 1 scores were characterized by a significant presence of eukaryotic fatty acids, while the samples having negative RDA axis 1 scores were characterized by bacterial fatty acids. A significantly higher percentage of eukaryotes were found in the samples taken in March (P = 0.002) (Fig. 4). Removal of eukaryotic fatty acids from the response variable matrix allowed examination of bacterial community response and showed a clear clustering of samples along the RDA axis 1, with samples from limestone streams having positive scores and those from sandstone streams with negative scores (F = 7.28; P = 0.002). Again, samples taken from Thompson Creek were either grouped with the sandstone streams or in between the sandstone and other limestone streams (Fig. 5).
Fig. 4.
Percentage of total microbial biomass that was comprised of microeukaryotes, by date in the streambed sediments of six streams (three limestone, three sandstone) located in the Bankhead National Forest. Error bars represent 1 standard deviation. *, significantly greater eukaryotic contribution to total biomass for the March sampling date (fully nested ANOVA with Tukey's honestly significant difference, P < 0.05).
Fig. 5.
Triplot of redundancy analysis of bacterial PLFA profiles of streambed sediments from six streams (three limestone, three sandstone) located in the Bankhead National Forest from all sampling dates. Significant predictor variables were selected by forward selection followed by Monte Carlo permutations. Arrows and symbols are as described for Fig. 3, except correlations (r) are given in Table 3.
ANC was the environmental parameter most closely associated with the variation describing limestone streams along the RDA axis 1, and DOC concentrations and sampling date described the variation on RDA axis 2 (Table 3). Individual fatty acids describing the variation in this analysis were a mixture of branched monoenoic and saturated fatty acids.
Table 3.
Correlation matrices (r) of environmental variables from the constrained ordination analysis (RDA) performed on bacterial PLFA profiles and environmental variables from the six streams in the Bankhead National Forest
| Factor |
r value |
||
|---|---|---|---|
| RDA 1 | RDA 2 | ||
| ANC | 0.7620 | −0.1338 | |
| DOC concn | 0.0487 | 0.4454 | |
| Sampling date | 0.0110 | −0.4644 | |
Influence of DOM quality on bacterial community structure.
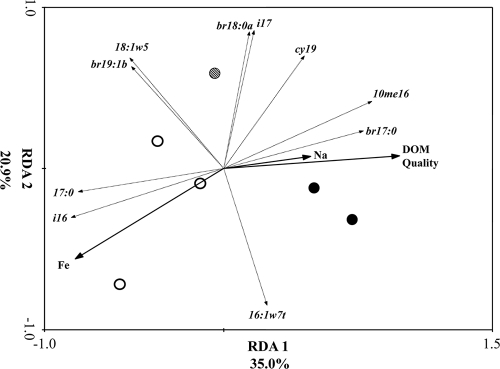
The number, nature, and relative abundance of molecules that comprise stream water DOM (here referred to as DOM quality) from the six streams, sampled March 2006, was significantly influenced by stream water chemistry and the presence of bituminous coal beds within the watershed (49). These data were added to the March 2006 description of the environment, and RDA was performed to determine if DOM quality contributed to the variation in bacterial community structure. RDA axes 1 and 2 described 55.9% of the variation (Fig. 6). Stream water ion composition (Na+ and Fe2 concentration) and DOM quality were significantly associated with the variation (Table 4) (F = 3.31; P = 0.004). The samples plotted according to stream water ion concentrations. The sandstone samples, containing lower ion concentrations, had negative RDA factor scores, and the samples from two of the limestone streams (Borden and Flannagin Creeks), containing higher stream water ion concentrations, had positive RDA factor scores. Thompson Creek, with intermediate stream water ion concentrations, had values near zero and the results were between the sandstone and the two limestone streams that had higher stream water ion concentrations. There was a mixture of individual fatty acids significantly associated with the variation, the majority being indicative of anaerobic and/or Gram-positive bacteria.
Fig. 6.
Triplot of redundancy analysis of bacterial PLFA profiles of streambed sediments from six streams (three limestone, three sandstone) located in the Bankhead National Forest from the March 2006 sampling date. Factor scores from a principal component analysis describing variation in DOM quality (49) were included in the analysis. Significant predictor variables were selected by forward selection followed by Monte Carlo permutations. Arrows and symbols are as described for Fig. 3, except correlations (r) are given in Table 3.
Table 4.
Correlation matrices (r) of environmental variables from the constrained ordination analysis (RDA) performed on bacterial PLFA profiles and environmental variables including DOM quality from the six streams in the Bankhead National Forest on March 2006
| Factor |
r value |
|
|---|---|---|
| RDA 1 | RDA 2 | |
| DOM quality | 0.928 | 0.070 |
| Fe concn | −0.783 | −0.522 |
| Na+ concn | 0.4593 | 0.072 |
DISCUSSION
Total microbial and autotrophic biomass levels.
No significant differences were observed in total microbial biomass and chlorophyll a concentrations between the two substrate types. Microbial biomass was comparable to reports from previous studies of streambed sediments (6, 12, 63), and chlorophyll a concentrations were also within ranges previously reported (31, 58). These previous studies found environmental and physical factors that correlated with microbial and autotrophic biomass levels. For instance, variation in sediment particle size (59) and DOC concentrations (6) have been found to be influential on microbial biomass, while inorganic nutrients (41) and light attenuation (17) have been associated with variation in chlorophyll a concentrations. With this knowledge, 6 streams were chosen within a single watershed that had similar physical and environmental characteristics, with the exception of bedrock geology, to eliminate potential confounding factors and to maximize the ability to detect the influence of geological formations. We conclude that there was no direct influence of geology on microbial or autotrophic biomass in streambed sediments stemming from variations in stream water ion concentrations.
Microbial community structure.
In contrast to microbial and autotrophic biomass, the variation described in streambed microbial community structure was associated with geological formations, specifically, stream water ion concentrations, DOM quality, and sampling date. Streams flowing over sandstone bedrock consistently showed lower pH and cation concentrations, while streams flowing over limestone bedrock showed higher pH and cation concentrations. Previously, Findlay et al. (28) showed differences in microbial community structure in stream sediments at the biome level; while they noted differences in stream water chemistry among biomes they did not determine proximate causes contributing to the variations in microbial community structure. The current study describes variation in stream water chemistry and streambed microbial community structure of six streams within one biome. Again, by confining this study to a single forest, many physical and environmental parameters that contribute to variations in microbial community structure were minimized, allowing the demonstration of the role of geologic bedrock in structuring microbial communities.
One parameter not controlled for in the study design was the presence of geologically influenced macroinvertebrate grazers. Reproduction and growth of pleurocerid snails (Elimia carineferia) are dependent on sufficient concentrations of stream water ions, specifically Ca2+, and therefore these organisms were only found in the limestone streams of this study (35). In a related study, an exclosure/enclosure experiment was conducted in two of the study streams, one limestone (Flannagin Creek) and one sandstone (Brushy Creek) stream, to determine if any of the variation observed in microbial community structure could be attributed to the effects of macroinvertebrate grazing. No significant differences in total microbial biomass, chlorophyll a biomass, or microbial community structure for any of the treatments were detected, thus eliminating grazing as a geology-induced effect (48).
Seasonal variation in the microbial community structure was observed in both stream types. Remarkably, while distinct differences in sediment microbial communities occurred between stream types, the shifts in the microbial community structure throughout seasonal changes were similar. Eukaryotic biomarkers showed increased relative abundances in all samples taken in March. Previous studies have shown seasonal variation in microbial community structure caused by differing eukaryotic and bacterial contribution to total biomass. Most of these studies found that microeukaryotes were more abundant and responsible for describing the variation in microbial communities in colder months (marine sediments [27], reservoir sediments [61], and creosote-impacted stream sediments [40]). One study found a higher relative abundance of eukaryotes in sediments of a regulated stream in warmer months (63). In a comparison of streams within and among biomes, sampled to avoid any seasonal effects, Findlay et al. (28) also found the major component of variation in microbial community structure was caused by differing eukaryotic and bacterial contributions to total biomass. Variation occurred among biomes, among streams with a biome, and for one biome, within streams. Interestingly, the second component of variation in microbial community structure in the Findlay et al. (28) study appeared to be associated with stream water ion concentrations/geology. In our current study, the importance of eukaryotes versus bacteria was evident both seasonally and by bedrock type, with the variation in microbial community structure in the limestone streams characterized by higher relative abundances of microeukaryotic fatty acid markers, while bacterial fatty acids were significant in describing the variation in the sandstone streams. When eukaryotic biomarkers were removed from the analysis, the effects of geologically driven water chemistry parameters and seasonal variations were still significant, and DOC concentration became a significant determinant of bacterial community structure.
DOM in stream ecosystems is an essential component for many geochemical processes and is a major energy source for microbe-based food webs (66). DOM quantity and quality are important in shaping aquatic bacterial community structure and metabolism (15, 29). Judd et al. (36) cross-fed bacterial communities from stream and lake sediments with DOM from stream, lake, or soil water and found that the bacterial community shifted to reflect the source from which the DOM originated. An experiment manipulating DOM quality and quantity sources in planktonic bacterial communities from a lake and two streams reported similar results (13). While these previous studies showed evidence that the source of DOM is important for bacterial assemblages, none characterized DOM quality at the molecular level, nor did any of the studies identify factors influencing DOM quality. Mosher et al. (49) successfully accomplished both of these by using ultra-high-resolution mass spectrometry to determine DOM quality in the stream waters of Hubbard, Thompson, Flannagin, Borden, Beech, and Brushy creeks. The water in the sandstone streams was characterized by higher abundances of condensed hydrocarbons, while the limestone stream water contained more oxygenated molecules. Further, Mosher et al. (49) determined that geological variation, including the presence of coal, was the most influential factor behind the variation in DOM quality. The inclusion of the DOM quality data from Mosher et al. (49) demonstrated that DOM quality was a significant factor (along with the direct effects of bedrock type) behind the variations in bacterial community structure. This was the first study to show evidence of the influence of geological variation and DOM quality on bacterial and microbial community structures in stream sediments. In addition, this correlative experimental design clearly demonstrated the influence of geological variation on DOM quality (49); however, it was unable to differentiate the effects of variation in microbial community structure on stream water DOM and vice versa. Stream microorganisms, via DOM processing, alter DOM quality (38), and it is reasonable to expect that different microbial communities alter DOM quality to a lesser or greater extent. Nonetheless, this study provides further support for the second half of Baas-Becking's “everything is everywhere, but the environment selects” dictum. This support should be tempered by the understanding that while PLFA analysis produces quantitative data, unlike DNA-based methods (50), it is a phenotypic method (see reference 23 for a full discussion of the advantages and disadvantages of the PLFA approach). It should be noted that when PLFA and molecular methods are applied in tandem, they produce very similar analyses of microbial community structure (28, 32). These results definitively demonstrated that DOM quality and bedrock composition, via the influence on stream water chemistry, influenced microbial community structure in streambed sediments. While a complex relationship between geology, microbial community structure, and DOM quality may exist, it is clear that geologically mediated stream water ion concentrations and DOM quality selected for sedimentary microbial community structure and that bedrock type within fluvial networks should be added to the growing list of environmental determinants of microbial distribution and abundance.
ACKNOWLEDGMENTS
This research was funded, in part, by awards from the National Science Foundation (DEB-0516235; R.H.F.), Sigma Xi grants in aid of research (J.J.M.), a Henry Aldrich student research grant (Southeastern Branch of the American Society for Microbiology; J.J.M.), a University of Alabama Graduate Council Research and Creative Activity fellowship (J.J.M.), and a Department of Biological Sciences Aquatic Biology Enhancement fellowship (J.J.M.). The work was made possible through U.S. Department of Agriculture Forest Service Special Use permit BAN201601.
Eric Roden, University of Wisconsin, provided instrumentation for the surface area analysis. Numerous individuals assisted in the field portion of this study, and Rusty Ward and Richard Carroll of the Alabama Geological Survey and Tom Counts of the National Forest Service provided technical advice. The Aquatic Chemistry Laboratory, Center for Freshwater Studies, The University of Alabama, performed the analyses of inorganic nutrients and DOC concentrations. Betsy Graham performed anion and cation analyses.
Footnotes
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Battin T. J., Kaplan L. A., Newbold J. D., Hendricks S. P. 2003. A mixing model analysis of stream solute dynamics and the contribution of a hyporheic zone to ecosystem function. Freshw. Biol. 48:995–1014 [Google Scholar]
- 2. Battin T. J., et al. 2008. Biophysical controls on organic carbon fluxes in fluvial networks. Nat. Geosci. 1:95–100 [Google Scholar]
- 3. Bell R. T., Tranvik L. 1993. Impact of acidification and liming on the microbial ecology of lakes. Ambio 22:325–330 [Google Scholar]
- 4. Ben-David E. A., Holden P. J., Stone D. J. M., Harch B. D., Foster L. J. 2004. The use of phospholipids fatty acid analysis to measure impact of acid rock drainage on microbial communities in sediments. Microb. Ecol. 48:300–315 [DOI] [PubMed] [Google Scholar]
- 5. Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911–917 [DOI] [PubMed] [Google Scholar]
- 6. Bott T. L., Kaplan L. A. 1985. Bacterial biomass, metabolic state, and activity in stream sediments: relation to environmental variables and multiple assay comparisons. Appl. Environ. Microbiol. 50:508–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brunauer S., Emmet P. H., Teller E. 1938. Adsorption of gases in multimolecular layers. J. Am. Chem. Soc. 60:309–319 [Google Scholar]
- 8. Cho J., Tiedje J. M. 2000. Biogeography and degree of endemicity of fluorescent Pseudomonas strains in soil. Appl. Environ. Microbiol. 66:5448–5456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dale N. G. 1974. Bacteria in intertidal sediments: factors related to their distribution. Limnol. Oceanogr. 19:509–518 [Google Scholar]
- 10. de Wit R., Bouvier T. 2006. ‘Everything is everywhere, but the environment selects’; what did Baas Becking and Beijerinck really say? Environ. Microbiol. 8:755–758 [DOI] [PubMed] [Google Scholar]
- 11. Diehl S. F., Goldhaber M. B., Hatch J. R. 2004. Modes of occurrence of mercury and other trace elements in coals from the Warrior Field, Black Warrior Basin, northwestern Alabama. Int. J. Coal Geol. 59:193–208 [Google Scholar]
- 12. Dobbs F. C., Findlay R. H. 1993. Analysis of microbial lipids to determine biomass and detect the response of sedimentary microorganisms to disturbance, p. 347–358 In Kemp P. F., Sherr B. F., Sherr E. B., Cole J. J. (ed.), Handbook of methods in aquatic microbiology. Lewis Publishers, Boca Raton, FL [Google Scholar]
- 13. Docherty K. M., Young K. C., Maurice P. A., Bridgham S. D. 2006. Dissolved organic matter concentration and quality influences upon structure and function of freshwater microbial communities. Microb. Ecol. 52:378–388 [DOI] [PubMed] [Google Scholar]
- 14. Fanson M. A. 1985. Standard methods for the examination of water and wastewater, 19th ed American Public Health Association, Washington, DC [Google Scholar]
- 15. Fazi S., Amalfitano S., Pernthaler J., Puddu A. 2005. Bacterial communities associated with benthic organic matter in headwater stream microhabitats. Environ. Microbiol. 7:1633–1640 [DOI] [PubMed] [Google Scholar]
- 16. Federle T. W., White D. C. 1982. Preservation of estuarine sediments for lipid analysis of biomass and community structure of microbiota. Appl. Environ. Microbiol. 44:1166–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feminella J. W., Power M. E., Resh V. H. 1989. Periphyton responses to invertebrate grazing and riparian canopy in three northern California coastal streams. Freshw. Biol. 22:445–457 [Google Scholar]
- 18. Fenchel T., Finlay B. J. 2004. The ubiquity of small species: patterns of local and global diversity. Bioscience 54:777–784 [Google Scholar]
- 19. Feris K. P., et al. 2003. Structure and seasonal dynamics of hyporheic zone microbial communities in free-stone rivers of the western United States. Microb. Ecol. 46:200–215 [DOI] [PubMed] [Google Scholar]
- 20. Feris K. P., et al. 2004. Seasonal dynamics of shallow-hyporheic-zone microbial community structure along a heavy-metal contamination gradient. Appl. Environ. Microbiol. 70:2323–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fierer N., Jackson R. B. 2006. The diversity and biogeography of soil bacterial communities. Proc. Nat. Acad. Sci. U. S. A. 103:626–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Finlay B. J., Clarke K. J. 1999. Ubiquitous dispersal of microbial species. Nature 400:828 [Google Scholar]
- 23. Findlay R. H. 2004. Determination of microbial community structure using phospholipid fatty acid profiles, p. 983–1004 In Kowalchuk G. A., De Bruijn F. J. , Head I. M., Akkermans A. D. L., Van Elsas J. D. (ed.), Molecular microbial ecology manual, 2nd ed Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 24. Findlay R. H., Dobbs F. C. 1993. Quantitative description of microbial communities using lipid analysis, p. 271–284 In Kemp P. R., Sherr B. F., Sherr E. B., Cole J. J. (ed.), Handbook of methods in aquatic microbial ecology. Lewis, Boca Raton, FL [Google Scholar]
- 25. Findlay R. H., Fell J. W., Coleman N. K., Vestal J. R. 1986. Biochemical indicators of the role of fungi and thraustochytrids in mangrove detrital systems, p. 91–104 In Moss S. T. (ed.), Biology of marine fungi. Cambridge University Press, Cambridge, England [Google Scholar]
- 26. Findlay R. H., King G. M., Watling L. 1989. Efficacy of phospholipid analysis in determining microbial biomass in sediments. Appl. Environ. Microbiol. 55:2888–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Findlay R. H., Watling L. 1998. Seasonal variation in the structure of a marine benthic microbial community. Microb. Ecol. 36:23–30 [DOI] [PubMed] [Google Scholar]
- 28. Findlay R. H., Yeates C., Hullar M. A. J., Stahl D. A., Kaplan L. A. 2008. Biome level biogeography in streambed sediments. Appl. Environ. Microbiol. 74:3014–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Findlay S. E. G., Sinsabaugh R. L., Sobczak W. V. 2003. Metabolic and structural response of hyporheic microbial commuities to variations in supply of dissolved organic matter. Limnol. Oceanogr. 48:1608–1617 [Google Scholar]
- 30. Franken R., Storey R., Williams D. 2001. Biological, chemical and physical characteristics of downwelling and upwelling zones in the hyporheic zone of a north-temperate stream. Hydrobiologia 444:183–195 [Google Scholar]
- 31. Gao X., Olapade O. A., Leff L. G. 2005. Comparison of benthic bacterial community composition in nine streams. Aquat. Microb. Ecol. 40:51–60 [Google Scholar]
- 32. Hazen T. C., et al. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208 [DOI] [PubMed] [Google Scholar]
- 33. Hewson I., Jacobson-Meyers M. E., Fuhrman J. A. 2007. Diversity and biogeography of bacterial assemblages in surface sediments across the San Pedro Basin, southern California borderlands. Environ. Microbiol. 9:923–933 [DOI] [PubMed] [Google Scholar]
- 34. Horner-Devine M. C., Carney K. M., Bohannan B. J. M. 2004. An ecological perspective on bacterial biodiversity. Proc. R. Soc. Lond. 271:113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huryn A. D., Benke A. C., Ward G. M. 1995. Direct and indirect effects of geology on the distribution, biomass, and production of the freshwater snail Elimia. J. North Am. Benthol. Soc. 14:519–534 [Google Scholar]
- 36. Judd K. E., Crump B. C., Kling G. W. 2006. Variation in dissolved organic matter controls bacterial production and community composition. Ecology 87:2068–2079 [DOI] [PubMed] [Google Scholar]
- 37. Kaplan L. A., Bott T. L. 1983. Microbial heterotrophic utilization of dissolved organic matter in a piedmont stream. Freshw. Biol. 13:363–377 [Google Scholar]
- 38. Kaplan L. A., Wiegner T. N., Newbold J. D., Ostrom P. H., Gandhi H. 2008. Untangling the complex issue of dissolved organic carbon uptake: a stable isotope approach. Freshw. Biol. 53:855–864 [Google Scholar]
- 39. Kreutzweiser D. P., Capell S. S. 2003. Benthic microbial utilization of differential dissolved organic matter sources in a forest headwater stream. Can. J. Forest Res. 33:1444–1451 [Google Scholar]
- 40. Langworthy D. E., Stapleton R. D., Sayler G. S., Findlay R. H. 1998. Genotypic and phenotypic responses of a riverine microbial community to polycyclic aromatic hydrocarbon contamination. Appl. Environ. Microbiol. 64:3422–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Le J., Wehr J. D., Campbell L. 1994. Uncoupling of bacterioplankton and phytoplankton production in fresh waters is affected by inorganic nutrient limitations. Appl. Environ. Microbiol. 60:2089–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leps J., Smilauer P. 2003. Multivariate analysis of ecological data using CANOCO, p. 64–66, 168-182 Cambridge University Press, Cambridge, England [Google Scholar]
- 43. Madsen E. L. 1998. Epistemology of environmental microbiology. Environ. Sci. Technol. 32:429–439 [Google Scholar]
- 44. Markewitz D., Davidson E. A., Figueiredo R. D., Victoria R. L., Krusche A. V. 2001. Control of cation concentrations in stream waters by surface soil processes in an Amazonion watershed. Nature 410:802–805 [DOI] [PubMed] [Google Scholar]
- 45. Mast M. A., Turk J. T. 1999. Environmental characteristics and water quality of hydrologic benchmark network stations in the eastern United States, 1963–95 USGS circular 1173-A. U.S. Geological Survey, Reston, VA [Google Scholar]
- 46. Methe B. A., Zehr J. P. 1999. Diversity of bacterial communities in Adirondack lakes: do species assemblages reflect lake water chemistry? Hydrobiologia 401:77–96 [Google Scholar]
- 47. Morehead D. L., Sinsabaugh R. L., Linkins A. E., Reynolds J. F. 1996. Decomposition processes: modeling approaches and applications. Sci. Total Environ. 183:137–149 [Google Scholar]
- 48. Mosher J. J. 2008. Geological influences on microbial community structure and dissolved organic matter quality in forested streams. Ph.D dissertation. University of Alabama, Tuscaloosa, AL [Google Scholar]
- 49. Mosher J. J., Klein G. C., Marshall A. G., Findlay R. H. 2010. Influence of bedrock geology on dissolved organic matter quality in stream water. Org. Geochem. 41:1177–1188 [Google Scholar]
- 50. Mumy K. L., Findlay R. H. 2004. Convenient determination of DNA extraction efficiency using an external DNA recovery standard and quantitative competitive PCR. J. Microbiol. Methods 57:259–268 [DOI] [PubMed] [Google Scholar]
- 51. Noguez A. M., et al. 2005. Microbial macroecology: highly structured prokaryotic soil assemblages in a tropical deciduous forest. Global Ecol. Biogeogr. 14:241–248 [Google Scholar]
- 52. Nold S. C., Zwart G. 1998. Patterns and governing forces in aquatic microbial communities. Aquat. Ecol. 32:17–35 [Google Scholar]
- 53. Osgood M. P., Boylen C. W. 1990. Seasonal-variations in bacterial communities in Adirondack streams exhibiting pH gradients. Microb. Ecol. 20:211–230 [DOI] [PubMed] [Google Scholar]
- 54. Pommier T., et al. 2007. Global patterns of diversity and community structure in marine bacterioplankton. Mol. Ecol. 16:867–880 [DOI] [PubMed] [Google Scholar]
- 55. Pusch M., et al. 1998. The role of micro-organisms in the ecological connectivity of running waters. Freshw. Biol. 40:453–495 [Google Scholar]
- 56. Ramette A., Tiedje J. M. 2007. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microbiol. Ecol. 53:197–206 [DOI] [PubMed] [Google Scholar]
- 57. Ramette A., Tiedje J. M. 2007. Multiscale responses of microbial life to spatial distance and environmental heterogeneity in a patchy ecosystem. Proc. Nat. Acad. Sci. U. S. A. 104:2761–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Romani A. M., Butturini A., Sabater F., Sabater S. 1998. Heterotrophic metabolism in a forest stream sediment: surface versus subsurface zones. Aquat. Microb. Ecol. 16:143–151 [Google Scholar]
- 59. Schallenberg M., Kalff J. 1993. The ecology of sediment bacteria in lakes and comparisons with other aquatic ecosystems. Ecology 74:919–934 [Google Scholar]
- 60. Schauer M., Kamenik C., Hahn M. W. 2005. Ecological differentiation within a cosmopolitan group of planktonic freshwater bacteria. (SOL cluster, Saprospiraceae, Bacteroidetes). Appl. Environ. Microbiol. 71:5900–5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Smoot J. C., Findlay R. H. 2001. Spatial and seasonal variation in a freshwater reservoir sedimentary microbial community as determined by phospholipid fatty acid analysis. Microb. Ecol. 42:350–358 [DOI] [PubMed] [Google Scholar]
- 62. Smoot J. C., Langworthy D. E., Levy M., Findlay R. H. 1998. Periphyton growth on submerged artificial substrate as a predictor of phytoplankton response to nutrient enrichment. J. Microbiol. Methods 32:11–19 [Google Scholar]
- 63. Sutton S. D., Findlay R. H. 2003. Sedimentary microbial community dynamics in a regulated stream: East Fork of the Little Miami River, Ohio. Environ. Microbiol. 5:256–266 [DOI] [PubMed] [Google Scholar]
- 64. Torsvik V., Ovreas L., Thingstad T. F. 2002. Prokaryotic diversity-magnitude, dynamics, and controlling factors. Science 296:1064–1066 [DOI] [PubMed] [Google Scholar]
- 65. Urbach E., et al. 2001. Unusual bacterioplankton community structure in ultra-oligotrophic Crater Lake. Limnol. Oceanogr. 46:557–572 [Google Scholar]
- 66. Volk C. J., Volk C. B., Kaplan L. A. 1997. Chemical composition of biodegradable dissolved organic matter in streamwater. Limnol. Oceanogr. 42:39–44 [Google Scholar]
- 67. Ward A. K., Ward G. M., Harris S. C. 1991. Water quality and biological communities of the Mobile River drainage, eastern Gulf of Mexico region, p. 279–304 In Becker C. D., Neitzel D. A. (ed.), Water quality in North American river systems. Battelle Press, Columbus, OH [Google Scholar]
- 68. Wetzel R. G., Likens G. E. 2000. Limnological analysis, 3rd ed., p. 117–123 Springer-Verlag, New York, NY. [Google Scholar]
- 69. Wood B. J. B. 1988. Lipids of algae and protozoa, p. 807–867 In Ratledge C., Wilkinson S. G. (ed.), Microbial lipids 1. Harcourt Brace Jovanovich, San Diego, CA [Google Scholar]
- 70. Yannarell A. C., Triplett E. W. 2004. Within- and between-lake variability in the composition of bacterioplankton communities: investigations using multiple spatial scales. Appl. Environ. Microbiol. 70:214–223 [DOI] [PMC free article] [PubMed] [Google Scholar]