Abstract
Salmonella enterica serovar Typhi (S. Typhi) is the etiological agent of the systemic disease typhoid fever. Transmission occurs via ingestion of contaminated food or water. S. Typhi is specific to humans, and no animal or environmental reservoirs are known. As the free-living amoeba Acanthamoeba castellanii is an environmental host for many pathogenic bacteria, this study investigates interactions between S. Typhi and A. castellanii by using cocultures. Growth of both organisms was estimated by cell count, viable count, flow cytometry, and fluorescence microscopy. Results indicate that S. Typhi can survive at least 3 weeks when grown with A. castellanii, as opposed to less than 10 days when grown as singly cultured bacteria under the same conditions. Interestingly, growth rates of amoebae after 14 days were similar in cocultures or when amoebae were singly cultured, suggesting that S. Typhi is not cytotoxic to A. castellanii. Bacteria surviving in coculture were not intracellular and did not require a physical contact with amoebae for their survival. These results suggest that S. Typhi may have a selective advantage when it is associated with A. castellanii and that amoebae may contribute to S. Typhi persistence in the environment.
INTRODUCTION
Salmonella enterica serovar Typhi (S. Typhi) is the etiologic agent of typhoid fever, a global health problem resulting in more than 200,000 deaths annually in developing countries where poor sanitary conditions are still common (10). S. Typhi causes a systemic infection, survives, and multiplies within macrophages and persists in the human body mainly by colonizing the gallbladder (33). S. Typhi is specific to humans, and no animal or environmental reservoirs are known (41). Nevertheless 3 to 5% of the population infected with S. Typhi will become chronic carriers. Thus, the propagation of S. Typhi is mainly by oral-fecal contamination via ingestion of contaminated food or water containing bacteria excreted from the feces of typhoid patients or human carriers. S. Typhi must then be able to survive in the environment prior to transmission to another host. To achieve this goal, many intracellular pathogens use protozoa as environmental reservoirs. It has been suggested that virulence genes of intracellular bacteria may have evolved from genes used to overcome or survive predation by protozoa. Furthermore, the interaction with free-living protozoa is also associated with increased survival and persistence in the environment (26) and with increased virulence (8, 31, 35). Thus, there is increasing evidence that complex relationships exist between environmental free-living protozoa and intracellular pathogens such as Legionella, Chlamydia, Mycobacterium, and Shigella (5, 24, 32, 39). Salmonella enterica serovar Typhimurium was shown to replicate and survive in Acanthamoeba spp.; however, it was cytotoxic and killed the protist (15, 17, 40).
Acanthamoeba castellanii is a free-living amphizoic protozoan that is ubiquitous in nature, including natural aquatic environments (29) and drinking water (6). The life cycle of A. castellanii includes two stages: trophozoites and cysts. Trophozoites prey on bacteria, whereas cysts are metabolically inactive (25). Moreover, free-living amoebae such as A. castellanii are known to graze surfaces such as lakebeds and feed on bacteria, which in turn might have developed ways to survive within amoebae or to kill them (18). Thus far, more than 20 species of pathogenic bacteria are reportedly associated with acanthamoebae (42).
It was previously shown that Acanthamoeba interacts with bacteria in a manner that is analogous to the interaction between macrophages and bacteria (4, 9). As S. Typhi is an intracellular pathogen that survives in macrophages and has no known reservoir, we investigated whether S. Typhi can readily survive in the environment in the presence of protozoa. The present study was undertaken to characterize the interaction between S. Typhi and A. castellanii.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
S. enterica serovar Typhi wild-type strain ISP1820 (23) and its isogenic strains DEF010 (16) (expressing green fluorescent protein [GFP] constitutively) and ΔphoP (χ8521) (16) were used as well as wild-type S. Typhi strain Ty2 (11). Strains were routinely grown in Luria-Bertani (LB) broth at 37°C unless indicated otherwise.
Growth of A. castellanii.
A. castellanii (ATCC 30234), obtained from J. Barbeau (Dentistry Faculty, Université de Montréal), was used in all experiments. Amoeba cultures were routinely maintained in 75-cm2 cell culture flasks (Corning) filled with 30 ml of peptone-yeast extract-glucose medium [PYG; 2% proteose peptone, 0.1% yeast extract, 1.6 mM MgSO4 · 7H2O, 0.4 mM CaCl2, 0.1 M sodium citrate · 2H2O, 2.5 mM Na2PO4 · 7H2O, 2.5 mM KH2PO4, 0.5 mM Fe(NH4)2(SO4)2 · 12H2O, 0.1 M glucose] pH 7.0 at 30°C, unless indicated otherwise. For subculture, trophozoites were suspended by tapping the flask vigorously and using a cell scraper (Corning), centrifuged at 300 × g, washed in Page's Amoeba Saline [PAS; 1.6 mM MgSO4 · 7H2O, 0.4 mM CaCl2, 0.1% sodium citrate · 2H2O, 2.5 mM NaH2PO4, 2.5 mM K2HPO4, 0.05 mM Fe(NH4)2(SO4)2 · 12H2O], centrifuged again at 300 × g, and suspended in 10 ml of fresh PYG medium. Cell counts were done using a hemocytometer, and trypan blue staining was used to assess cell viability and to estimate concentration. A total of 2.5 × 105 amoebae were then inoculated in a new flask containing fresh PYG medium. This subculture procedure was repeated every 7 days.
Cocultures.
A. castellanii and S. Typhi were incubated together in cocultures as described previously (3) and below. Amoebae from an axenic culture were washed once with PAS and suspended in 10 ml of fresh PYG culture medium or diluted PYG medium (diluted 5-fold with PAS [1/5-PYG]). After cell counts were determined with a hemocytometer, amoebae were inoculated in sterile 75-cm2 flasks containing 30 ml of PYG medium or diluted PYG to a final concentration of 105 amoebae/ml. The bacterial count of a static overnight culture of S. Typhi was evaluated using a spectrophotometer (optical density at 600 nm [OD600] of 0.6 corresponds to 4 × 108 bacteria per ml). Bacteria were added to the flask containing the amoebae for a final concentration of 106 bacteria/ml, giving a multiplicity of infection (MOI) of 10. Control flasks included either bacteria or amoebae cultured alone and incubated under the same conditions and concentrations as for coculture experiments. Incubation was at either 18°C, 30°C, or 37°C. Cocultures were done in PYG medium or 1/5-PYG medium. After 30 min of incubation, amoebae were resuspended by vigorous tapping, and a 2-ml sample was taken from each of the three flasks. Amoeba cell counts were done on each sample. Bacterial counts were done on each sample by diluting and spreading samples on LB plates to estimate the number of CFU. Samples were taken after 0, 1, 4, 10, and 14 days and up to 21 days in some assays.
Transwell coculture experiments were carried out as described above. An insert containing a 0.4-μm-pore-size membrane (Greiner Bio-One) was placed into each well of a 24-well tissue culture dish. Amoebae were added to the bottom chamber at a concentration of 105 amoebae/ml, and S. Typhi ISP1820 was added to the top chamber in 200 μl of fresh diluted PYG medium at a concentration of 106 bacteria per ml (2 × 105 bacteria total per insert). Wells containing bacteria or amoebae alone under the same respective growth conditions and concentrations as in coculture were used as controls. Amoeba and bacterial counts were done exactly as in cocultures stated above.
Finally, a coculture was prepared as described above with the exception that gentamicin (10 μg/ml) was added at day 10 after sampling was completed and remained until the end of the coculture assay. With the exception of the gentamicin treatment, no other manipulations that differ from the coculture method stated above were added.
Flow cytometry.
Cocultures of amoebae with a fluorescent Salmonella strain (DEF010) were prepared as described above and analyzed by flow cytometry. Bacterial populations were tracked using a logarithmic setting adjusted for specific readings of bacteria and by fluorescence with a laser emitting at 488 nm (FL1-H) and collected at 530/30. At different time points, each flask was gently tapped for 30 s to suspend the amoebae that were attached to the flask surface, and 1 ml of the total suspension was taken. Flow cytometry was executed on a FACSCalibur (Becton Dickinson), and 10,000 events were collected and analyzed using CellQuest (Becton Dickinson) and FlowJo software (Tree Star Inc.). Amoeba cell populations were analyzed according to their relative granularity, using side scatter (SSC-H), and relative size, using forward-scatter (FSC-H), as trophozoites and cysts are of different sizes. The setting was linear (E 00) and did not permit bacterial quantification.
Microscopy.
Epifluorescence microscopy was done using cocultures with a GFP-expressing strain of S. Typhi (DEF010). Cocultures were executed as stated above. Microscopy was performed on a Nikon E600 fluorescence microscope. Samples were either live or fixed with 4% paraformaldehyde. Fixed samples were mounted on slides with Geltol Mounting Medium (Thermo Shandon). Samples were routinely taken from cocultures and examined the same day (fresh) or the next day (fixed).
Statistical analysis.
The statistical significance between different values was calculated by an unpaired t test with a two-tailed P value. A P value of <0.05 was considered to be statistically significant. In some cases, one-way analysis of variance (ANOVA) with a P value of <0.05 considered statistically significant was used. The particulars of the statistical analysis can be found in the figure legends.
RESULTS
Influence of temperature.
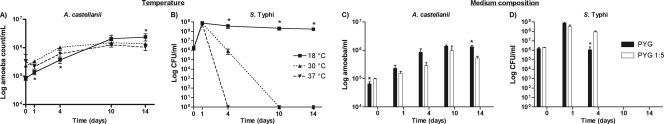
The optimal temperature for coculture experiments was evaluated because both Acanthamoeba and Salmonella can easily adapt to extreme environmental conditions, including temperatures ranging from 4°C to 45°C (14, 19, 21, 30). The optimal growth temperature for Acanthamoeba, however, is between 25 and 30°C (34), and it is between 35 and 37°C for Salmonella (22). A. castellanii and S. Typhi were grown individually in PYG medium at different temperatures (18°C, 30°C, or 37°C). For amoebae, the different temperature settings did not particularly affect global growth (Fig. 1A). From an initial 105 amoebae, an average maximum concentration of 1.6 × 106 amoebae/ml was reached after 14 days. In contrast, S. Typhi was much more sensitive to temperature changes (Fig. 1B). Cultures inoculated with 106 S. Typhi bacteria reached a maximum number after 24 h for any temperature. At 18°C, S. Typhi was able to survive significantly better than at other temperatures at every time point, but more specifically at points later than 10 days of incubation (Fig. 1B). After 14 days at 18°C, an average of 1.6 × 108 CFU/ml was obtained. Most bacteria, however, were filamentous at this temperature (data not shown). Bacteria grown at temperatures of 30°C or 37°C decreased significantly to nondetectable levels of viable bacteria after 10 and 4 days of incubation, respectively (Fig. 1B). Growth at 30°C was selected for further experiments as this temperature is optimal for the amoeba and limited bacterial survival to less than 10 days.
Fig. 1.
Growth conditions. Growth in PYG medium at 18°C, 30°C, and 37°C was evaluated for individual survival of A. castellanii (A) and S. Typhi (B). The effect of growth medium composition of pure cultures of A. castellanii (C) or S. Typhi (D) incubated at 30°C was evaluated in PYG medium or diluted PYG (1/5-PYG) medium. Values are the means ± standard errors of the means of at least three independent experiments. Asterisks indicate a significant difference (P < 0.05).
Influence of media.
Amoebae and bacteria were grown individually at 30°C in PYG medium and in diluted PYG (1/5-PYG) medium to evaluate the influence of growth medium composition. From an initial 105 amoebae, an average maximum concentration of 1.5 × 106 amoebae/ml was reached after 14 days in PYG medium, and a significantly lower number, with an average 7 × 105 amoebae/ml, was reached in 1/5-PYG medium after 14 days (P = 0.0160) (Fig. 1C). Flasks inoculated with 106 S. Typhi bacteria produced 7.5 ×108 CFU/ml after 24 h of incubation in PYG medium and 3.2 × 108 CFU/ml in 1/5-PYG medium (Fig. 1D). No viable bacteria were detected past 10 days, a situation which remained unchanged after 14 days of incubation in both media (Fig. 1D). The diluted PYG medium was selected for coculture experiments as the growth of both amoebae and S. Typhi was adequate.
Cocultures of A. castellanii and S. Typhi.
Growth of A. castellanii at 30°C in 1/5-PYG medium in the presence of S. Typhi was evaluated by amoeba cell counts (Fig. 2). The total number of amoebae from infected cultures decreased significantly in the first 24 h of incubation (P = 0.001), which coincides with exponential growth of the bacterial population (Fig. 2A). Except for this reduction and a significant lag at 4 days (P = 0.0055), amoebae in cocultures grew similarly in absence (5.3 × 105 amoebae/ml) or presence of S. Typhi (3 × 105 amoebae/ml), without any significant difference after 14 days (P = 0.19). Growth of S. Typhi in coculture with A. castellanii showed an initial increase after 24 h, similar to the growth of S. Typhi without amoebae (Fig. 2B). The bacteria were not detected after 10 days in the absence of amoebae (Fig. 2B). Interestingly, the bacteria were still detected at 10 days postinfection (2.3 × 105 CFU/ml) and even multiplied and survived at 14 days (3.6 × 108 CFU/ml) in the presence of A. castellanii (Fig. 2B). Then, the number of bacteria seemed to reach an equilibrium, and a mean of 2.7 × 108 CFU/ml was observed after 21 days (see Fig. 5B), and 1.5 × 108 CFU/ml was observed after 6 months of coculture with amoebae. No viable bacteria were ever recovered when cultured in the absence of amoebae at these time points.
Fig. 2.
Amoebae-bacteria interaction. Coculture of A. castellanii and S. Typhi at 30°C in 1/5-PYG medium. (A) Growth of A. castellanii with (coculture) and without (alone) S. Typhi. (B) Growth of S. Typhi with and without A. castellanii. Values are the means ± standard errors of the means of at least three independent experiments. Asterisks indicate a significant difference (P < 0.05).
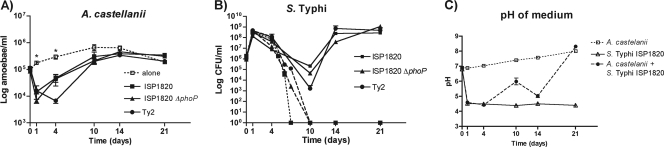
Fig. 5.
Coculture of various S. Typhi strains with A. castellanii for 21 days and pH of the cultures. (A) Growth of A. castellanii alone and in the presence of S. Typhi strain ISP1820, its isogenic phoP mutant (ISP1820 ΔphoP; involved in intracellular survival), and S. Typhi strain Ty2. (B) Growth of S. Typhi strain ISP1820 (squares), its isogenic mutant ISP1820 ΔphoP (triangles), and S. Typhi strain Ty2 (circles) with (solid lines) and without (dashed lines) A. castellanii. (C) pH value of culture medium containing only amoebae (square), only bacteria (triangle), or both microorganisms (circle). Values are the means ± standard errors of the means of at least two independent experiments. Asterisks indicate a significant difference (P < 0.05).
Characterization of cell populations by cytometry.
Coculture with S. Typhi strain DEF010 constitutively expressing the green fluorescent protein (GFP) with A. castellanii was analyzed by flow cytometry. The cell populations were evaluated by fluorescence using a setting that allowed bacterial detection. Fluorescence analysis of the culture containing only bacteria showed that 40% of the population was highly fluorescent at the beginning of incubation (Fig. 3A). The fluorescence level reached 89%, the highest, after 24 h and then significantly decreased. After 6 days, no signs of fluorescence remained; only low-intensity background fluorescence was present. The fluorescence signal continued to fade until the end of the coculture at 14 days (Fig. 3A), which correlated with the absence of CFU by plating of the bacteria under these conditions. Fluorescence analysis of amoeba cultures revealed an auto-fluorescence level that initially corresponded to 50% of the population (Fig. 3). After 14 days, only 11% of amoebae showed a similar level of auto-fluorescence (Fig. 3). Fluorescence analysis of cocultures revealed that 63% of the amoebae-bacteria population was highly fluorescent at the beginning of infection, corresponding to the highest initial level of fluorescence. Then, the percentage of fluorescence reached a maximum of 93% after 24 h of infection and decreased between 24 h and 6 days in a similar way in both the coculture and control culture containing only bacteria. There was, however, a difference in fluorescence at 14 days postinfection between the coculture and bacterial control culture: a fluorescent signal was visible in the coculture and was produced by 25% of the population. This fluorescent population was not present in the S. Typhi control culture or in the A. castellanii culture. Plating of these coculture samples at 14 days confirmed the presence of viable S. Typhi in the coculture, while no viable bacteria were detected in the control culture.
Fig. 3.
Association and localization of bacteria during interaction with amoeba. (A) Fluorescence-activated cell sorting analysis showing fluorescence of cell populations from pure culture of strain DEF010 (GFP positive) (bacteria; top), cocultivation of bacteria and amoebae (middle), or pure culture of amoebae (bottom) over a 14-day period. The vertical axis represents cell number, and the horizontal axis represents log fluorescence intensity. The percentage of fluorescent cells is indicated on the right of the histogram, and the mean intensity of fluorescence is indicated in the top right corner in italics. One representative experiment is shown. (B) Epifluorescence microscopy of samples taken from coculture flasks at various time points. Arrows show bacteria; V, vacuole; C, cyst; T, trophozoite. Original magnification, ×1,000. Scale bar, 10 um.
Microscopy.
Samples from cocultures using a GFP-expressing S. Typhi were observed by fluorescence microscopy. As observed by cytometry, uninfected amoebae and cysts have a certain level of auto-fluorescence. Many bacteria were found associated with amoebae following infection (Fig. 3B). Uniformly fluorescent vacuoles were observed during the first 24 h of interaction. Adherent bacteria were clearly visible around trophozoites at 24 h postinfection. Occasionally, bacteria were also observed associated with cysts. Bacteria were found free or associated with amoebae at day 14. After that period, many bacteria were found free in culture medium (data not shown).
Bacterial localization and interaction with amoebae during cocultures.
A gentamicin protection assay was used to kill extracellular bacteria in order to determine if bacteria are located within amoebae. As the bacterial population shows viability at 10 days only when bacteria are cocultured with amoebae, the antibiotic was added at day 10. The addition of gentamicin had no significant effect on the growth of amoebae (Fig. 4A). However, no bacteria were detected at 14 days after gentamicin was added to the coculture (Fig. 4B). Then, to investigate if bacterial survival was caused by direct cell-cell contact or by soluble substances secreted by the amoebae, transwell assays were employed. Therefore, coculture experiments were repeated, but the bacteria were separated by a membrane with 0.4-μm-sized pores. Presence of bacteria in the upper compartment did not affect growth of amoebae (Fig. 4C). Bacterial survival was observed after 10 days of coculture in the presence of amoebae (Fig. 4D), suggesting that no physical contact between the microorganisms is needed for survival. As observed in other cocultures, S. Typhi continued to survive up until 14 days and was not detected after 10 days when grown without amoebae (Fig. 4D).
Fig. 4.
S. Typhi survival does not involve an intracellular state. (A) Effect of antibiotic added at day 10 to evaluate growth of A. castellanii grown by itself (square), with S. Typhi (circle), or with S. Typhi and gentamicin addition (triangle). (B) Growth of S. Typhi alone (square) or with A. castellanii and gentamicin added at day 10 (triangle). Survival by direct cell-cell contact was evaluated by transwell experiments. S. Typhi was inoculated in the upper compartment, and A. castellanii was inoculated in the lower compartment. (C) Growth of amoebae with (triangle) or without (square) bacteria in the upper compartment. (D) Growth of bacteria with (triangle) or without (square) amoebae in the lower compartment. Results are the means ± standard errors of the means of three independent experiments. Asterisks indicate a significant difference (P < 0.05).
Coculture of various S. Typhi strains with A. castellanii.
In order to characterize the persistence mechanisms of bacteria in the presence of amoebae, we evaluated cocultures for up to 21 days in addition to testing an isogenic phoP mutant of S. Typhi, strain ISP1820, known to be involved in intracellular survival in macrophages (16). We also tested the wild-type S. Typhi strain Ty2 (11). Growth of amoebae and bacteria was evaluated exactly as in cocultures above. Similar growth profiles of amoebae were observed in the presence of any of these S. Typhi strains and consisted of a significant (P < 0.05) initial drop in amoeba number that was more dramatic but nonsignificant (P = 0.102) at day 4 with strain Ty2. A similar number of amoebae were present at day 10, with or without culture with S. Typhi (Fig. 5A). As expected, none of the bacterial strains survived alone in the medium more than 10 days (Fig. 5B). In the presence of amoebae, bacterial counts attained their lowest level at day 10 and then multiplied to high density, reaching more than 108 bacteria per ml. After 21 days, many bacteria were extracellular (data not shown). The phenotype was similar for all the tested strains (Fig. 5B).
pH of culture medium.
The pH of the medium containing only bacteria became rapidly acidic and stabilized at pH 4.5, even after bacteria were noncultivable or dead (Fig. 5C). The pH of the medium containing only amoebae was initially neutral (pH 6.9) and became slightly basic with time (pH 7.6). The pH of the coculture medium became rapidly acidic, similarly to medium containing bacteria alone. After 4 days, however, the pH gradually increased to reach a pH similar to that of the medium used when amoebae were grown alone (Fig. 5C).
DISCUSSION
Typhoid fever is acquired via consumption of water or food contaminated by S. Typhi. No bacterial reservoir outside humans is known; thus transmission occurs mainly by oral-fecal contamination with feces of an infected person or from a chronic carrier. The infectious dose of S. Typhi has been estimated to 100,000 organisms, suggesting potential survival mechanisms in the environment prior to transmission to other humans. S. Typhi is not known to persist in the environment, and it was previously demonstrated that S. Typhi strains were extremely sensitive to chlorinated water (38). As amoebae were associated with increased survival and persistence in the environment for many bacterial pathogens, such as Shigella, Legionella, Vibrio, and Salmonella (2, 12, 17, 18, 24), the association between A. castellanii and S. Typhi was investigated.
We have demonstrated that S. Typhi survives significantly longer in the presence of amoebae while it persists less than 10 days when grown without amoebae. Therefore, the interaction between A. castellanii and S. Typhi promotes persistence of S. Typhi. The number of bacteria during coculture reached a minimum number around 10 days, and then increased and reached an equilibrium state. The acidification of the environment correlated with the initial bacterial growth and was probably due to sugar fermentation. Amoebae, however, did not acidify the medium, and cultures even became slightly basic after 14 days of incubation. The pH of the coculture medium was less acidic than the medium of bacteria grown alone after 14 days. This fact may show that amoebae act as a buffer by limiting acidification. The persistence of S. Typhi was independent of intracellular strategies required for survival within macrophages. This is shown by the phoP mutant, which is unable to survive in macrophages but survives just as well as the other strains tested in coculture. Moreover, addition of gentamicin after 10 days resulted in a greatly decreased bacterial population (Fig. 4), suggesting that persisting bacteria are extracellular. Moreover, when amoebae and bacteria were cocultured in chambers separated by a membrane allowing nutrient transfer but no physical association, survival of bacteria was still observed (Fig. 4). Persistence of bacteria in the environment may be favored by amoebae and other protists that provide nutrients. Nutrients may derive from dead amoebae or from fecal pellets. In fact, it has been shown that amoebae egest undesired or undigested food rapidly (27). Alternatively, S. Typhi may survive digestion by amoebae. It was previously shown that bacteria that resist grazing by protozoa show increased environmental fitness (20). Further, bacteria, such as S. Typhimurium, that remain undigested following uptake by protozoa were still viable when excreted in fecal pellets (7). An association between amoebae and bacteria that increases persistence without involving an intracellular cycle was observed between A. castellanii and Vibrio parahaemolyticus (28).
Cell population analyses by cytometry confirmed a decline in the bacterial population, clearly seen over time by the loss of fluorescence when bacteria were grown in pure culture (Fig. 3). It was only in the presence of amoebae that fluorescence was observed after 14 days (Fig. 3). This result confirms what earlier cocultures have shown by viable plate counts, i.e., survival of S. Typhi at 14 days. This procedure, however, did not allow the colocalization of bacteria with amoebae. Analysis of the amoeba-S. Typhi interactions by microscopy using fluorescent bacteria revealed that within minutes after infection, brightly fluorescent vacuoles were visible inside amoebae in coculture with bacteria (Fig. 3B). This suggested that amoebae either ingested bacteria or that S. Typhi was able to invade the amoebae. After 24 h of coculture, many bacteria were found associated with amoebae, with bacteria clearly visible around the amoebae, while some bacteria seemed to be located inside the amoebae (Fig. 3B). In contrast, most of the bacteria found after 14 days were free in the medium and only a few were associated with amoebae (Fig. 3B). This pattern seems to indicate that S. Typhi is ingested by A. castellanii and that intracellular localization is due to grazing rather than intracellular survival mechanisms. Under coculture conditions, the presence of bacteria initially affects the growth of amoebae but does not seem to affect their growth in the long term as a similar number of amoebae were found after 14 days, with or without bacteria. Therefore, it is unlikely that S. Typhi is cytotoxic toward amoebae. However, a change in amoeba population was induced by the presence of bacteria as amoebae rapidly formed cysts in the presence of S. Typhi, whereas the majority of the amoeba population was comprised of trophozoites when amoebae were grown in pure culture (data not shown). It was previously demonstrated that amoebae transform rapidly into cysts in contact with intracellular bacteria such as Francisella (13).
This study has shown that association with amoebae promotes persistence of S. Typhi. The nature of amoeba-S. Typhi interactions does not depend on cell-cell contact. Intracellular growth, as observed for other intracellular bacteria, such as Legionella (37) or Francisella (1), was not observed for S. Typhi. Fluorescence microscopy showed evidence of adherent or intracellular bacteria, but a gentamicin protection assay failed to show any survival, suggesting no protection by an intracellular state. There was no difference observed between the wild-type and a mutant involved in intramacrophage survival. Moreover, when amoebae in coculture were lysed to release intracellular bacteria for CFU counts, the number of recovered bacteria was similar with or without lysis treatment (data not shown). Therefore, intracellular bacteria may represent only a transient state during initial coculture. The association of Salmonella serovar Typhi with Acanthamoeba was also different than that of S. serovar Typhimurium, which resulted in death of amoebae, corresponding more to a parasitic relationship (15, 17, 40). Even if these two Salmonella serovars share many virulence determinants, phenotypic differences have been demonstrated (16, 36).
The prolonged survival of S. Typhi during interaction with amoebae may have implications for the mode of transmission. Amoebae may increase S. Typhi environmental persistence and transmission as well as prime S. Typhi for enhanced survival during its journey through the human gut. The ultimate goal would be to have a broader, general understanding of the microbial ecology of S. Typhi and protozoans. Discerning survival of S. Typhi outside the human body and its interactions with protozoans and the environment could lead to a better understanding of its transmissibility in areas of endemicity.
ACKNOWLEDGMENTS
This research was supported by the Canadian Natural Sciences and Engineering Research Council grant number 251114-06.
We thank C. M. Dozois for critical reading of the manuscript, S. Gravel for her help with amoeba culture, S. Sénéchal for fluorescence-activated cell sorting analysis, and members of the laboratory for technical help.
Footnotes
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Abd H., Johansson T., Golovliov I., Sandstrom G., Forsman M. 2003. Survival and growth of Francisella tularensis in Acanthamoeba castellanii. Appl. Environ. Microbiol. 69:600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abd H., Saeed A., Weintraub A., Nair G. B., Sandstrom G. 2007. Vibrio cholerae O1 strains are facultative intracellular bacteria, able to survive and multiply symbiotically inside the aquatic free-living amoeba Acanthamoeba castellanii. FEMS Microbiol. Ecol. 60:33–39 [DOI] [PubMed] [Google Scholar]
- 3. Abd H., et al. 2008. Pseudomonas aeruginosa utilises its type III secretion system to kill the free-living amoeba Acanthamoeba castellanii. J. Eukaryot. Microbiol. 55:235–243 [DOI] [PubMed] [Google Scholar]
- 4. Abu Kwaik Y., Gao L. Y., Stone B. J., Venkataraman C., Harb O. S. 1998. Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl. Environ. Microbiol. 64:3127–3133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Amann R., et al. 1997. Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63:115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Backer H. 2002. Water disinfection for international and wilderness travelers. Clin. Infect. Dis. 34:355–364 [DOI] [PubMed] [Google Scholar]
- 7. Brandl M. T., Rosenthal B. M., Haxo A. F., Berk S. G. 2005. Enhanced survival of Salmonella enterica in vesicles released by a soilborne Tetrahymena species. Appl. Environ. Microbiol. 71:1562–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cirillo J. D., Falkow S., Tompkins L. S. 1994. Growth of Legionella pneumophila in Acanthamoeba castellanii enhances invasion. Infect. Immun. 62:3254–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cosson P., Soldati T. 2008. Eat, kill or die: when amoeba meets bacteria. Curr. Opin. Microbiol. 11:271–276 [DOI] [PubMed] [Google Scholar]
- 10. Crump J. A., Luby S. P., Mintz E. D. 2004. The global burden of typhoid fever. Bull. World Health Organ. 82:346–353 [PMC free article] [PubMed] [Google Scholar]
- 11. Deng W., et al. 2003. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 185:2330–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dey R., Bodennec J., Mameri M. O., Pernin P. 2009. Free-living freshwater amoebae differ in their susceptibility to the pathogenic bacterium Legionella pneumophila. FEMS Microbiol. Lett. 290:10–17 [DOI] [PubMed] [Google Scholar]
- 13. El-Etr S. H., et al. 2009. Francisella tularensis type A strains cause the rapid encystment of Acanthamoeba castellanii and survive in amoebal cysts for three weeks postinfection. Appl. Environ. Microbiol. 75:7488–7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Elliott R. P., Heiniger P. K. 1965. Improved temperature-gradient incubator and the maximal growth temperature and heat resistance of Salmonella. Appl. Microbiol. 13:73–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feng Y., et al. 2009. Apoptosis-like cell death induced by Salmonella in Acanthamoeba rhysodes. Genomics 94:132–137 [DOI] [PubMed] [Google Scholar]
- 16. Forest C. G., Ferraro E., Sabbagh S. C., Daigle F. 2010. Intracellular survival of Salmonella enterica serovar Typhi in human macrophages is independent of Salmonella pathogenicity island (SPI)-2. Microbiology 156:3689–3698 [DOI] [PubMed] [Google Scholar]
- 17. Gaze W. H., Burroughs N., Gallagher M. P., Wellington E. M. 2003. Interactions between Salmonella typhimurium and Acanthamoeba polyphaga, and observation of a new mode of intracellular growth within contractile vacuoles. Microb. Ecol. 46:358–369 [DOI] [PubMed] [Google Scholar]
- 18. Greub G., Raoult D. 2004. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17:413–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Griffin J. L. 1972. Temperature tolerance of pathogenic and nonpathogenic free-living amoebas. Science 178:869–870 [DOI] [PubMed] [Google Scholar]
- 20. Hahn M. W., Hofle M. G. 2001. Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol. Ecol. 35:113–121 [DOI] [PubMed] [Google Scholar]
- 21. Harwood J. L. 2007. Temperature stress: reacting and adapting: lessons from poikilotherms. Ann. N. Y. Acad. Sci. 1113:52–57 [DOI] [PubMed] [Google Scholar]
- 22. Holt J. G., Krieg N. R., Sneath P. H. A., Staley J. T., Williams S. T. (ed.). 1994. Bergey's manual of determinative bacteriology, 9th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 23. Hone D. M., Harris A. M., Chatfield S., Dougan G., Levine M. M. 1991. Construction of genetically defined double aro mutants of Salmonella typhi. Vaccine 9:810–816 [DOI] [PubMed] [Google Scholar]
- 24. Jeong H. J., et al. 2007. Acanthamoeba: could it be an environmental host of Shigella? Exp. Parasitol. 115:181–186 [DOI] [PubMed] [Google Scholar]
- 25. Khan N. A. 2009. Acanthamoeba: biology and pathogenesis. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 26. King C. H., Shotts E. B., Jr., Wooley R. E., Porter K. G. 1988. Survival of coliforms and bacterial pathogens within protozoa during chlorination. Appl. Environ. Microbiol. 54:3023–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Landry M., Lehner-Fournier J., Sundstrom J., Fagerness V., Selph K. 1991. Discrimination between living and heat-killed prey by a marine zooflagellate, Paraphysomonas vestita. J. Exp. Mar Biol. Ecol. 146:139–152 [Google Scholar]
- 28. Laskowski-Arce M. A., Orth K. 2008. Acanthamoeba castellanii promotes the survival of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 74:7183–7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martinez A. J., Visvesvara G. S. 1997. Free-living, amphizoic and opportunistic amebas. Brain Pathol. 7:583–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Michener H. D., Elliott R. P. 1964. Minimum growth temperatures for food-poisoning, fecal-indicator, and psychrophilic microorganisms. Adv. Food Res. 13:349–396 [DOI] [PubMed] [Google Scholar]
- 31. Molmeret M., Horn M., Wagner M., Santic M., Abu Kwaik Y. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl. Environ. Microbiol. 71:20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Neumeister B., Schoniger S., Faigle M., Eichner M., Dietz K. 1997. Multiplication of different Legionella species in Mono Mac 6 cells and in Acanthamoeba castellanii. Appl. Environ. Microbiol. 63:1219–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Parry C. M., Hien T. T., Dougan G., White N. J., Farrar J. J. 2002. Typhoid fever. N. Engl. J. Med. 347:1770–1782 [DOI] [PubMed] [Google Scholar]
- 34. Perez-Serrano J., Martinez J., Perez B., Bernadina W. E., Rodriguez-Caabeiro F. 2000. In vitro shock response to different stressors in free living and pathogenic Acanthamoeba. Int. J. Parasitol. 30:829–835 [DOI] [PubMed] [Google Scholar]
- 35. Rasmussen M. A., et al. 2005. Exposure to rumen protozoa leads to enhancement of pathogenicity of and invasion by multiple-antibiotic-resistant Salmonella enterica bearing SGI1. Infect. Immun. 73:4668–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sabbagh S. C., Forest C. G., Lepage C., Leclerc J. M., Daigle F. 2010. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol. Lett. 305:1–13 [DOI] [PubMed] [Google Scholar]
- 37. Segal G., Shuman H. A. 1999. Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect. Immun. 67:2117–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi H., et al. 2010. Live recombinant Salmonella Typhi vaccines constructed to investigate the role of rpoS in eliciting immunity to a heterologous antigen. PLoS One 5:e11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Steinert M., Birkness K., White E., Fields B., Quinn F. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl. Environ. Microbiol. 64:2256–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tezcan-Merdol D., et al. 2004. Uptake and replication of Salmonella enterica in Acanthamoeba rhysodes. Appl. Environ. Microbiol. 70:3706–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wain J., House D., Parkhill J., Parry C., Dougan G. 2002. Unlocking the genome of the human typhoid bacillus. Lancet Infect. Dis. 2:163–170 [DOI] [PubMed] [Google Scholar]
- 42. Winiecka-Krusnell J., Linder E. 2001. Bacterial infections of free-living amoebae. Res. Microbiol. 152:613–619 [DOI] [PubMed] [Google Scholar]