Abstract
pBL1 is a Lactococcus lactis theta-replicating 10.9-kbp plasmid that encodes the synthetic machinery of the bacteriocin Lcn972. In this work, the transcriptomes of exponentially growing L. lactis strains with and without pBL1 were compared. A discrete response was observed, with a total of 10 genes showing significantly changed expression. Upregulation of the lactococcal oligopeptide uptake (opp) system was observed, which was likely linked to a higher nitrogen demand required for Lcn972 biosynthesis. Strikingly, celB, coding for the membrane porter IIC of the cellobiose phosphoenolpyruvate-dependent phosphotransferase system (PTS), and the upstream gene llmg0186 were downregulated. Growth profiles for L. lactis strains MG1363, MG1363/pBL1, and MG1363 ΔcelB grown in chemically defined medium (CDM) containing cellobiose confirmed slower growth of MG1363/pBL1 and MG1363 ΔcelB, while no differences were observed with growth on glucose. The presence of pBL1 shifted the fermentation products toward a mixed acid profile and promoted substantial changes in intracellular pool sizes for glycolytic intermediates in cells growing on cellobiose as determined by high-pressure liquid chromatography (HPLC) and nuclear magnetic resonance (NMR). Overall, these data support the genetic evidence of a constriction in cellobiose uptake. Notably, several cell wall precursors accumulated, while other UDP-activated sugar pools were lower, which could reflect rerouting of precursors toward the production of structural or storage polysaccharides. Moreover, cells growing slowly on cellobiose and those lacking celB were more tolerant to Lcn972 than cellobiose-adapted cells. Thus, downregulation of celB could help to build up a response against the antimicrobial activity of Lcn972, enhancing self-immunity of the producer cells.
INTRODUCTION
Bacteriocins are ribosomally synthesized bacterial peptides with antimicrobial activity. Their production is a widespread trait in lactic acid bacteria (LAB). Bacteriocins form a rather structurally diverse group encompassing posttranslationally modified lantibiotics (class I), nonmodified heat-resistant peptides (class II), heat-labile proteins (class III), and circular bacteriocins (class IV) (references 10 and 26 and references therein). Many LAB bacteriocins are inhibitory toward a wide panel of Gram-positive bacteria, including relevant pathogen and food spoilage bacteria. Therefore, a major effort has been made in the last decades to understand their ecological role in complex ecosystems (e.g., fermented foods and the gastrointestinal tract) and the molecular basis of their inhibitory activity for rationalizing their use as natural preservatives in food or as promising anti-infectives (19, 23).
Although most LAB bacteriocins were shown to act as membrane permeabilizers when added at high concentrations, it is being recognized that bacteriocin killing is target mediated. Many bacteriocins make use of receptors or docking molecules present in the cell envelopes of susceptible strains prior to pore formation. Nisin and many other structurally related lantibiotics specifically bind to the cell wall precursor lipid II (5, 6). Moreover, several class II bacteriocins, such as lactococcin A, sakacin A, and enterocin P, form a complex with the membrane component IIC of the mannose phosphoenolpyruvate (PEP)-dependent phosphotransferase system (PTS) transporter (16, 29).
Functional PTSs consist of two general proteins, enzyme I (encoded by ptsI) and the heat-stable phosphocarrier protein HPr (encoded by ptsH), and sugar-specific permeases or enzyme II complexes. The latter catalyze the translocation and concomitant phosphorylation of several different sugars and usually consist of three or four proteins or protein domains, namely, IIA, IIB, IIC, and, when present, IID. Phosphoryl relay proceeds sequentially from PEP to EI, HPr, IIA, IIB, and the incoming sugar, which is transported across the membrane via the IIC porter (15, 39). Expression of PTS-encoding genes is tightly regulated, and that of genes encoding alternative sugars is often subject to carbon catabolite repression (14). Beyond the primary function of sugar transport, PTSs play roles in various processes central to the physiology of the cell, including a large number of mechanisms for metabolic and transcriptional regulation (15). As described above, they also play a role as receptors of some class II bacteriocins. In fact, resistance to class II bacteriocins has been often linked to the absence or repression of the genes coding for the mannose PTS, which is involved in glucose uptake (11, 16, 22, 41). Therefore, one immediate consequence of class II bacteriocin resistance is impaired growth on glucose, while utilization of other sugars is favored (28, 48).
Based on the proposed use of LAB bacteriocins as food preservatives and to provide large enough quantities for structure-function studies, several studies have focused on improving bacteriocin production and/or reducing production costs by using by-products (reference 12 and references therein). Engineering of bacteriocin immunity has been proven to increase nisin yields in Lactococcus lactis and in heterologous hosts (24, 46). Moreover, bacteriocin clusters have been suggested as a food-grade alternative to antibiotic resistance markers (47). In spite of this, little is known about the impact that synthesis of bacteriocins may have on the physiology of the producing strain. This issue is particularly relevant when bacteriocin production is seen as a valuable technological trait and needs to be transferred to industrial strains. Recently, Fallico et al. (18) reported that conjugation of the plasmid pMRC01 carrying the lacticin 3147 bacteriocin cassette imposed a metabolic burden on several L. lactis starter strains.
In this work, we have studied the impact of plasmid pBL1, encoding the bacteriocin lactococcin 972 (Lcn972), on L. lactis. Lcn972 is an atypical 66-amino-acid (aa) nonmodified bacteriocin synthesized by L. lactis IPLA972. It targets exclusively the Lactococcus genus and thus far is the only nonlantibiotic that binds to lipid II, inhibiting cell wall biosynthesis, without disrupting membrane integrity (34, 36). The genes for the synthetic machinery are carried on the 11-kbp plasmid pBL1 (GenBank accession number AF242367.1) and consist of a structural gene and two other open reading frames (ORFs) encoding a putative ABC transporter, which is presumably involved in self-immunity. Introduction of pBL1 in L. lactis rendered strains that were able to produce Lcn972 and that were immune to it, without showing other particular traits (34, 36). In this study, however, genome-wide transcriptomics revealed changes in the expression of genes directly related to oligopeptide and sugar uptake. Bearing in mind the roles of sugar PTSs in the mode of action of other bacteriocins and in bacterial metabolism, we have characterized in more detail the response of L. lactis to the presence of the bacteriocin-encoding plasmid pBL1.
MATERIALS AND METHODS
Bacteria, plasmids, and culture conditions.
The strains and plasmids used in this study are listed in Table 1. Lactococcal strains were routinely grown in M17 (Oxoid) with glucose (0.5%, wt/vol; ∼27.5 mM) at 30°C (the optimal growth temperature) or 37°C (when required for genetic manipulation). Escherichia coli DH10B was used for intermediate cloning and grown on 2× YT (44) at 37°C with shaking. For physiological characterization, lactococcal cultures were grown statically and without pH control (initial pH, 6.5), at 30°C, in chemically defined medium (CDM) containing 1% (wt/vol) glucose (∼55.5 mM) or cellobiose (∼29.2 mM), in rubber-stoppered bottles, as described previously (8). Growth was started by addition of a preculture (inoculum) in early stationary phase to an initial optical density at 600 nm (OD600) of approximately 0.05. Precultures were grown in glucose-CDM, except for adapted cells, for which the substrate was the same as for the culture; all other conditions were as described above. When necessary, erythromycin was used at a final concentration of 5 μg ml−1 and ampicillin was used at 100 μg ml−1. Growth was monitored by measuring the optical density at 600 nm. Growth rates (μ) were calculated through linear regressions of the plots of ln OD600 versus time during the exponential growth phase. Growth rates and other growth parameters were analyzed using the Instat software (GraphPad Software).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant phenotype or genotypea | Reference(s) or source |
|---|---|---|
| Strains | ||
| L. lactis | ||
| MG1363 | Plasmid-free derivative of NCDO712 | 20 |
| MG1363/pBL1 | MG1363 carrying pBL1, lcn972+ Lcn972r | This work |
| MG1363/pBL1E | MG1363 carrying pBL1E, lcn972+ Lcn972r Emr | This work |
| MG1363 ΔcelB | MG1363; chromosomal deletion of celB | This work |
| MG1614 | Strr Rifr derivative of MG1363 | 20 |
| MG1614.2 | MG1614 carrying pBL1 | 35 |
| MG1363 ΔcelB/pBL1E | MG1363 ΔcelB carrying pBL1E, Emrlcn972+ Lcn972r | This work |
| E. coli DH10B | Plasmid free | 21 |
| Plasmids | ||
| pBL1 | Lcn972-encoding plasmid, 10.9 kbp | 35, 45 |
| pBL1E | erm from pNG8048 cloned in the unique EcoRV of pBL1_orf4 | This work |
| pCR2.1 | Cloning of PCR products, Apr | Invitrogen |
| pCR::celB4-1 | celB and flanking regions cloned in pCR2.1 | This work |
| pCR::dcelB | pCR::celB4-1 with a 1,019-bp deletion in celB | This work |
| pGhost9 | Thermosensitive, Emr | 33 |
| pGhost::dcelB | Incomplete celB cloned in pGhost9 | This work |
Str, streptomycin; Rif, rifampin; Em, erythromycin; Ap, ampicillin.
Transcriptome analysis.
Genome-wide transcriptional experiments were performed using DNA microarrays containing 2,457 annotated genes in the genome of L. lactis MG1363 and were carried out essentially following the methods for cell disruption, RNA isolation, RNA quality control, cDNA synthesis, indirect labeling, hybridization, and scanning described previously (50). RNA from three biological replicates was extracted from exponentially growing (OD600 of 0.4) L. lactis MG1614 and MG1614.2 cultures in GM17 at 30°C. Data were processed as described previously (37).
Construction of L. lactis MG1363 ΔcelB and pBL1E.
Standard molecular cloning techniques were followed as described elsewhere (44). Restriction enzymes were purchased from Takara (Otsu, Shiga, Japan) and T4 ligase from Invitrogen (Barcelona, Spain). Oligonucleotides were supplied by Sigma (Madrid, Spain) and are shown in Table 2. PCRs were carried out using PuRe Taq Ready-to-Go PCR beads (GE Healthcare, Buckinghamshire, United Kingdom). The celB-1 and celB-4 primers were used to amplify a 2.4-kbp chromosomal fragment containing celB plus 0.9-kbp flanking regions. PCR conditions were as follows: 95°C for 5 min (1 cycle); 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min (35 cycles); and 72°C for 10 min (1 cycle). The resulting amplicon was cloned in the E. coli plasmid pCR2.1, generating pCR::celB4-1. This plasmid was HincII digested and religated, generating pCR::dcelB, lacking a 1.0-kbp HincII internal fragment. The incomplete celB gene plus flanking regions was released from pCR::dcelB as a 1.4-kbp XbaI-SpeI fragment and subsequently cloned in the thermosensitive L. lactis plasmid pGhost9 digested with SpeI to obtain pGhost::dcelB. L. lactis MG1363 was transformed with pGhost::dcelB, and first and second recombination events were followed essentially as previously described (33). celB deletion was confirmed by PCR with appropriate primer pairs and DNA sequencing.
Table 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ → 3′)a | Function |
|---|---|---|
| celB-qF1 | ATTTGGCCCGTGCTTACG | qRT-PCR, celB |
| celB-qR1 | TTTGGCAAACCTGCAAATAGG | |
| QptcC-F | CGTGTTCGGTATTGCTTACG | qRT-PCR, ptcC |
| QptcC-R | TGTTAAACCAGCGGGTACTC | |
| qBglS-F | TACACCGCAGTATGCTAAGG | qRT-PCR, bglS |
| qBglS-R | TTGGCCGACTTCAAGAGTTC | |
| Tuf-F | GGTAGTTGTCGAAGAATGGAGTGTGA | qRT-PCR, internal control |
| Tuf-R | TAAACCAGGTTCAATCACTCCACACA | |
| celB-1 | AACTCtAGATGGCCTTTGTA (XbaI) | Cloning and disruption of celB |
| celB-4 | gAagAtctAAGACAGCCGCTCC (BglII) |
Changes introduced to generate restriction sites (underlined and shown in parentheses) are indicated in lowercase.
To construct the recombinant plasmid pBL1E, the erythromycin resistance gene erm was excised from pNG8048 (51) by SmaI-EcoRV restriction and ligated to the unique EcoRV site present in pBL1_orf4. L. lactis MG1363/pBL1E transformants were selected on GM17 with erythromycin at 5 μg ml−1.
RT-qPCR.
RNA was extracted using the Illustra RNAspin Mini RNA isolation kit (GE Healthcare), and the RNA concentration was determined by absorbance at 260 nm. cDNA was generated with the iScript cDNA synthesis kit (Bio-Rad). PCR amplification was performed in a 7500 Fast real-time PCR system (Applied Biosystems, Warrington, United Kingdom). Primers used for reverse transcriptase quantitative PCR (RT-qPCR) are listed in Table 2. Amplification was carried out in 25 μl containing either 0.01 or 0.002 μg cDNA (according to the expected expression levels), 1× Power SYBR green (Applied Biosystems), and each primer at a concentration of 0.56 μM. After incubation at 95°C for 10 min, amplification proceeded with 40 cycles of 95°C for 15 s and 60°C for 1 min. Standard curves were generated by plotting the cycle threshold (CT) values of reactions performed on serial dilutions of cDNA against the logarithm of cDNA concentrations. cDNA concentrations were correlated to quantify relative gene expression levels. The housekeeping gene tuf, encoding the elongation factor Tu, was used to normalize (43).
ELISA detection of Lcn972.
The bacteriocin Lcn972 in culture supernatants was quantified by a noncompetitive enzyme-linked immunoassay (NCI-ELISA) with rabbit polyclonal antibodies against Lcn972 (1). Primary and secondary antibodies were used at 1:1,000 and 1:40,000 dilutions, respectively. Pure Lcn972 (15 to 1 μg/ml) was used as standard.
Lcn972 susceptibility tests.
MICs were assayed by the broth microdilution method in GM17 as described elsewhere (34). Dose-response curves were determined in CDM in microtiter plates. Overnight cultures in CDM-glucose or CDM-cellobiose were adjusted to an OD600 of 0.1, and 100 μl was used to inoculate wells containing Lcn972 from 9.6 to 0 μg/ml. Growth was followed in a microtiter reader (Bio-Rad) at 30°C until control cultures without Lcn972 reached an OD600 of 0.7 to 0.8, which was taken as 100% growth. For challenge tests, exponentially growing cultures in CDM-cellobiose at an OD600 of 0.2 were treated with Lcn972 at 0.1 μM (5 times the MIC). Sodium phosphate buffer (50 mM, pH 6.8) was used as a control. Cultures were incubated for 1 h at 30°C, and appropriate 10-fold dilutions were plated on GM17 agar plates for counting. Survival (percent) was defined as CFU/ml in treated cultures divided by CFU/ml in the control samples.
Quantification of fermentation products during growth.
Samples (2 ml) were taken at different growth stages, centrifuged (13,200 × g, 5 min, 4°C), and filtered (0.22 μm), and the supernatant solutions were stored at −20°C until analysis by high-performance liquid chromatography (HPLC). Substrate and fermentation end products (lactate, acetate, ethanol, formate, acetoin, 2,3-butanediol, and pyruvate) were quantified in an HPLC apparatus equipped with a refractive index detector (Shodex RI-101; Showa Denko K. K., Japan) using an HPX-87H anion-exchange column (Bio-Rad Laboratories Inc., Hercules, CA) at 60°C, with 5 mM H2SO4 as the elution fluid and a flow rate of 0.5 ml min−1. Cellobiose was similarly quantified in an ICSep ION-300 column preceded by an ICSep ICE-GC-801 precolumn (Transgenomic, San Jose, CA), at 65°C and 0.4 ml min−1 in 8.5 mM H2SO4, using a refractive index detector RI2414 (Waters, Milford, MA).
For the yield calculation, two time points, one immediately after inoculation and the other at the time of growth arrest (T30, nonadapted cells), were considered. ATP production was calculated from the fermentation products, assuming that all ATP was synthesized by substrate-level phosphorylation. The average specific consumption rates of cellobiose were estimated from a first-order derivative of a polynomial fit of the observed substrate consumption time series.
Determination of intracellular metabolites during growth.
Ethanol extracts for analysis by 31P nuclear magnetic resonance (NMR) and quantification of phosphorylated metabolites in MG1363/pBL1 and the control strain at the mid-exponential growth phase were prepared as described elsewhere (42). The dried extracts were dissolved in 2 ml of aqueous solution containing 5 mM EDTA and 12.5% (vol/vol) 2H2O (final pH of approximately 6.5). Assignment of resonances and quantification of phosphorylated metabolites were based on previous studies (38, 42) or done by spiking the NMR sample extracts with the suspected pure compounds. Intracellular metabolite concentrations were calculated using a value of 2.9 μl mg of protein−1 for the intracellular volume of L. lactis (40). The reported values for intracellular phosphorylated compounds are averages from two independent growth experiments, and the accuracy was around 15%.
Microarray data accession number.
The DNA microarray data are available at the Gene Expression Omnibus (GEO) repository under accession number GSE30625.
RESULTS
Transcriptional analysis of L. lactis with the bacteriocin Lcn972 plasmid pBL1.
To gain a deeper insight into the impact that the presence of the Lcn972-encoding plasmid pBL1 may exert in L. lactis, a genome-wide transcriptional analysis was carried out with L. lactis MG1614.2, carrying the bacteriocin plasmid pBL1, compared to the parental strain L. lactis MG1614 when growing under standard laboratory conditions in GM17. A relatively discrete transcriptional response was observed, with a total of 10 genes showing significantly (P < 0.001) changed expression of over a factor of two (Table 3). Besides those coding for proteins of unknown function, upregulation of the lactococcal oligopeptide uptake system was observed. oppA and oppB were clearly overexpressed, but other members of the system (oppF, oppC, oppD, and the endopeptidase gene pepO) were also upregulated, although just below the established cutoff levels (see data under GEO accession number GSE30625). Overexpression of genes coding for proteins involved in DNA rearrangement/mobilization was also noted. This could be due to cross-hybridization with transposase genes present in the composite plasmid pBL1. Indeed, pBL1_orf9 shares 86% and 99% identity at the nucleotide level with llmg0674_tnp1297 and llmg0717_tnp946, respectively, according to BLASTN (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The same could have accounted for the hypothetical acetyltransferase gene llmg0676, to which pBL1_orf1 shows 99% identity.
Table 3.
Significant changes in gene expression induced by the presence of the Lcn972-encoding plasmid pBL1 in L. lactis MG1614 cultures growing exponentially in GM17 broth
| Gene | Ratioa | Bayesian Pb | Annotationc |
|---|---|---|---|
| Upregulated | |||
| llmg0701_oppA | 4.83 | 9.50E−08 | Oligopeptide-binding protein OppA |
| llmg1012 | 4.73 | 4.00E−06 | Putative ABC transporter substrate-binding protein |
| llmg0676 | 3.14 | 1.28E−05 | Hypothetical acetyltransferase |
| llmg0699_oppB | 2.85 | 2.31E−06 | Peptide transport system permease protein OppB |
| llmg0642 | 2.41 | 5.22E−05 | Hypothetical protein |
| llmg0711_tnpR | 2.40 | 5.13E−04 | DNA-invertase/resolvase |
| llmg2348 | 2.36 | 7.65E−05 | Hypothetical protein |
| llmg0674_tnp1297 | 2.03 | 3.28E−06 | Transposase for insertion sequence element IS1297 |
| Downregulated | |||
| llmg0186 | −4.93 | 5.49E−10 | Conserved hypothetical protein |
| llmg0187_celB | −4.59 | 4.53E−04 | Cellobiose-specific PTS system IIC component |
The cellobiose-specific PTS permease (IICCel domain) gene celB and the upstream llmg0186 were the only genes downregulated in the presence of pBL1 (Table 3). llmg0186 codes for a conserved hypothetical protein, also identified in L. lactis IL1403 (ybhE), which might be functionally related to celB, as the two genes seem to form a single transcriptional unit (3). An absence of specific PTS permeases has been phenotypically linked to resistance to several class II bacteriocins. Moreover, PTSs are main players in bacterial metabolism mediating sugar uptake, the main energy source in Lactococcus. Thus, we proceeded to further investigate celB repression in the presence of pBL1.
Expression of cellobiose-related genes in the presence of pBL1.
It has been recently shown that point mutations, which might be selected within very closely related L. lactis strains, may have a pronounced effect on gene expression and, subsequently, on particular phenotypes. This has been exemplified by comparative transcriptomics of L. lactis MG1363 and its derivative L. lactis NZ9000, generated by the insertion of the two-component system nisKR, which allows nisin-inducible gene expression. Specific point mutations in the latter strain accounted for altered expression of several genes involved in carbohydrate metabolism, which translated into different growth rates on specific sugars (31). The reference strain that we used for the transcriptional analysis was L. lactis MG1614, an antibiotic-resistant derivative of L. lactis MG1363 which is poorly characterized and whose genome sequence is unknown (20). To avoid possible interferences and to unequivocally confirm by RT-qPCR that repression of celB is a direct consequence of the presence of pBL1, we transformed this plasmid into L. lactis MG1363 (Table 1). As shown below (see Table 5), L. lactis MG1363/pBL1 transformants were able to synthesize Lcn972 and were resistant to it. Furthermore, we generated a celB knockout mutant, MG1363 ΔcelB, to evaluate the consequences of celB repression.
Table 5.
Production of Lcn972 by L. lactis strains in CDM-glucose and CDM-cellobiose
| L. lactis strain | Lcn972 production (μg/OD unit)a in: |
|
|---|---|---|
| CDM-glucose | CDM-cellobiose | |
| MG1363/pBL1 | 5.4 ± 0.0 A | 6.6 ± 0.0 A |
| MG1363/pBL1E | 4.7 ± 0.1 AB | 6.9 ± 0.1 A |
| MG1363 ΔcelB/pBL1E | 4.4 ± 0.3 B | 11.1 ± 1.1 B |
Samples were taken at the transition to stationary phase. Lcn972 was quantified by ELISA using rabbit polyclonal Lcn972 antibodies and corrected by OD600. Means and standard deviations for two independent cultures are shown. Letters indicate significant differences (P < 0.05) between strains growing in the same medium.
Initially, the celB operon structure was investigated by RT-PCR. The genomic context of celB in MG1363 is similar to that described in L. lactis IL1403 (3). celB is flanked upstream by the conserved gene llmg0186 and downstream by two small ORFs followed by bglS, potentially encoding a phospho-β-glucosidase involved in cellobiose hydrolysis. The presence of mRNA encompassing llmg0186 and celB in L. lactis MG1363 cells growing on cellobiose was confirmed (data not shown). This result is in agreement with the codownregulation of both genes noted in the transcriptomic analysis (Table 3). In contrast, we failed to detect a putative celB-bglS mRNA by RT-PCR.
The expression of cellobiose-related genes, i.e., celB, ptcC, and bglS, in the presence of pBL1 was determined by RT-qPCR (Table 4) using total RNA isolated from glucose- and cellobiose-growing cultures in CDM. ptcC was also included as a cellobiose/glucose IIC porter (8, 30). In the presence of cellobiose, the three analyzed genes were induced in MG1363, but celB showed the highest fold induction which supports its role as a main cellobiose uptake system in L. lactis (Table 4). Interestingly, loss of CelB (MG1363 ΔcelB) resulted in a higher expression ratio of ptcC, possibly as a way to counteract the lack of celB and facilitate cellobiose uptake. In contrast, induction of celB and bglS was inhibited 5-fold and that of ptcC 3-fold by the presence of pBL1, confirming the hypothesis raised from the transcriptomic results that pBL1 downregulates celB.
Table 4.
Expression ratios of cellobiose-related genes in exponentially growing L. lactis MG1363, MG1363/pBL1, and MG1363 ΔcelB grown in CDM-cellobiose relative to growth in CDM-glucose, determined by RT-qPCR
| L. lactis strain | Expression ratio for target gene |
||
|---|---|---|---|
| ptcC | celB | bglS | |
| MG1363 | 3.0 | 216.3 | 53.5 |
| MG1363/pBL1 | 1.2 | 43.5 | 10.2 |
| MG1363 ΔcelB | 7.6 | —a | 16.7 |
—, celB is not present in this strain.
pBL1 impairs growth on cellobiose.
Based on the differential expression of cellobiose-related genes in the presence of pBL1, we proceeded to determine possible effects on the catabolism of relevant sugars. As celB encodes the cellobiose PTS porter, cellobiose was an obvious choice to use as a carbon source in physiological studies. Glucose was used as control sugar.
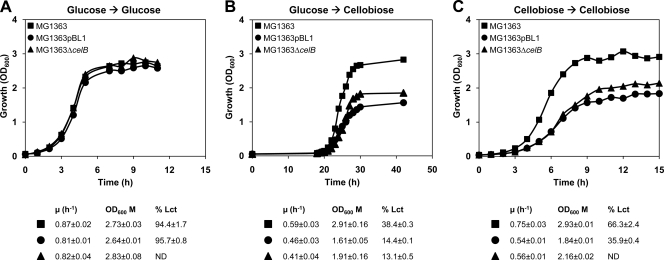
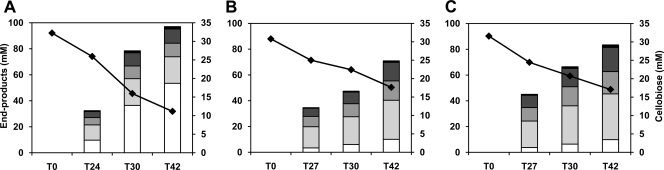
Small differences in growth, although not statistically significant (P > 0.05), were observed for glucose-grown cells subcultured in fresh glucose-CDM, as the growth rates of MG1363/pBL1 and MG1363 ΔcelB were slightly reduced compared to that of MG1363 (Fig. 1A). Similarly, pBL1 did not affect the homolactic behavior of MG1363. In contrast, subculturing glucose-grown cells in CDM supplemented with cellobiose (i.e., nonadapted cells) resulted in major and very significant (P < 0.01) differences in the growth profiles. Cultures of MG1363 and derivatives were characterized by long lag phases, and exponential growth started only 18 to 19 h after inoculation (Fig. 1B). This was not surprising, as this behavior was already reported for L. lactis MG1363 (31). The growth rate and the maximal biomass produced were substantially lower for strains MG1363/pBL1 and MG1363 ΔcelB than for the control strain. These remarkable differences led us to examine in greater detail the kinetics of substrate consumption and fermentation product formation in nonadapted cells growing on cellobiose (Fig. 2). The three strains showed a mixed acid fermentation profile, but the product distribution was markedly different in the parent strain compared with the strain carrying pBL1 or MG1363 ΔcelB. In the manipulated strains, formate, acetate, and ethanol (2:1:1) were the major products, while lactate accounted for less than 20% of the substrate consumed. In contrast, lactate was the main product from cellobiose metabolism in L. lactis MG1363, and curiously, its yield increased dramatically during growth, from 0.4 at mid-exponential phase (T24) to 1.3 in late stationary phase (T42). At the time of growth arrest (T30), MG1363 had consumed about 50% of the initial substrate, whereas MG1363/pBL1 and MG1363 ΔcelB used only 30% of the cellobiose in the medium. The biomass yield was, however, similar in MG1363 and the ΔcelB mutant (about 17 g mol−1 of substrate) and slightly lower in the presence of pBL1 (about 16.2 g mol−1 of substrate), correlating well with the OD600 values determined (Fig. 1B). In view of these data, it is reasonable to speculate that the observed differences in growth rate and maximal biomass in MG1363 compared to MG1363/pBL1 and MG1363 ΔcelB are directly associated with cellobiose transport. In line with this, the average cellobiose consumption rate during exponential growth was higher in MG1363 (3.6 mmol g−1 h−1) than in the presence of pBL1 (2.1 mmol g−1 h−1) or in the absence of celB (2.3 mmol g−1 h−1).
Fig. 1.
Growth profiles of L. lactis MG1363 and its isogenic strains MG1363/pBL1 and MG1363 ΔcelB on glucose or cellobiose. Cultures were grown in CDM supplemented with 1% (wt/vol) sugar substrate at 30°C in rubber-stoppered bottles without pH control (initial pH of 6.5). (A) Preculture and culture grown on glucose. (B) Preculture grown on glucose and culture grown on cellobiose (nonadapted cells). (C) Preculture and culture grown on cellobiose (adapted cells). The growth rate (μ), maximal OD600, and percentage of lactate (% Lct) at an OD600 of 0.7 to 0.9 formed from the substrate consumed are also shown for each condition tested. Growth curves are from a representative experiment. Growth was repeated at least twice, and the values are averages for each condition.
Fig. 2.
Substrate consumption and end product profiles during the fermentation of cellobiose (1%, wt/vol) by nonadapted L. lactis cells. Strains MG1363 (A), MG1363/pBL1 (B), and MG1363 ΔcelB (C) were grown overnight in CDM supplemented with glucose (1%, wt/vol) and subcultured in fresh medium containing cellobiose (1%, wt/vol) as for Fig. 1B. Supernatants obtained at different growth stages were analyzed by HPLC as described in Materials and Methods. The values are averages from at least two independent experiments, and the average accuracy was ±5%. Symbols: diamonds, cellobiose; white bars, lactate; light gray bars, formate; medium gray bars, acetate; dark gray bars, ethanol; black bars, acetoin plus 2,3-butanediol.
This view was further supported by the growth profiles obtained for adapted cells, i.e., early-stationary-phase cellobiose-grown precultures subcultured in CDM-cellobiose (Fig. 1C). Adaptation abolished the long lag phases (maximal growth after 1 h at 30°C) and resulted in improved growth rates (20 to 35% increase) for all strains. Nevertheless, both the maximal biomass produced and the growth rate were still considerably higher in strain MG1363 than in MG1363/pBL1 or MG1363 ΔcelB. Adaptation also promoted the formation of lactate in either the presence or absence of pBL1 in MG1363 (Fig. 1C).
pBL1 changes the intracellular dynamics of phosphorylated metabolites.
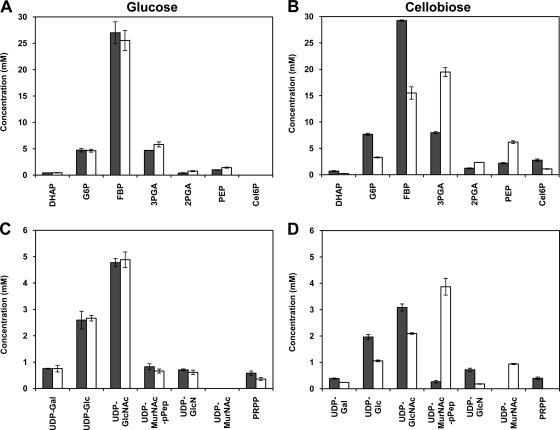
Prompted by the notable effect of pBL1 on the growth properties and end product profiles of MG1363, we asked whether intracellular metabolite levels would also be affected by the plasmid during growth on cellobiose. Glucose was used as a control condition. As expected, pBL1 had no effect on the pool sizes of glycolytic intermediates (Fig. 3A) or sugar-nucleotides (Fig. 3C) during growth on glucose as determined by 31P NMR in cell extracts. Contrastingly, pBL1 promoted substantial changes in intracellular pool sizes in cellobiose-growing cells. Fructose-1,6-bisphosphate (FBP), the major mid-exponential-phase glycolytic intermediate in glucose-growing cells and in cellobiose-growing MG1363, was reduced by about 2-fold (Fig. 3A). A similar effect was observed for other upper glycolytic metabolites (dihydroxyacetone phosphate [DHAP] and glucose-6-phosphate [G6P]). The concentration of cellobiose-6-phosphate, the product of the transport step, was also 2-fold lower in the presence of pBL1. On the other hand, the lower glycolytic metabolites 3-phosphoglycerate (3-PGA), 2-PGA, and PEP showed increased concentrations.
Fig. 3.
Effect of pBL1 on intracellular phosphorylated metabolites during growth on glucose or cellobiose. Phosphorylated metabolites were measured by 31P NMR in ethanol extracts of adapted MG1363 and MG1363/pBL1 cells (the same sugar substrate was used in precultures and cultures) grown to mid-exponential phase in CDM supplemented with 1% (wt/vol) glucose (A and C) or cellobiose (B and D). Glycolytic metabolites (A and B) and UDP-activated metabolites (C and D) are depicted. The average accuracy was ±15%. Symbols: dark gray bars, MG1363; white bars, MG1363/pBL1. DHAP, dihydroxyacetone phosphate; G6P, glucose-6-phosphate; FBP, fructose-1,6-biphosphate; 3PGA, 3-phosphoglycerate; 2PGA, 2-phosphoglycerate; PEP, phosphoenolpyruvate; Cel6P, cellobiose-6-phosphate; Gal, galactose; Glc, glucose; GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid; pPep, pentapeptide; PRPP, 5-phosphorylribose 1-pyrophosphate.
Of note was the drastic effect of pBL1 on the concentration of the cell wall cytoplasmic precursors, UDP-N-acetylmuramoyl-pentapeptide (UDP-MurNAc-pPep) and UDP-N-acetyl muramic acid (UDP-MurNAc) (Fig. 3D). The latter accumulated only in cellobiose-growing MG1363/pBL1, while UDP-MurNAc-pPep increased by about 14-fold in this strain. In contrast, the level of the other peptidoglycan cytoplasmic precursor, UDP-N-acetylglucosamine (UDP-GlcNAc), was slightly lower, as were those of all other UDP-activated sugars detected and 5-phosphorylribose 1-pyrophosphate.
Contribution of impaired growth on cellobiose to production of and resistance to Lcn972.
According to the physiological data, the presence of pBL1 correlated well with defective growth of L. lactis on cellobiose in a fashion similar to that for a nonfunctional celB. Since the main phenotype that could be attributed to pBL1 is the production of the bacteriocin Lcn972 (35, 45), we attempted to establish if there was a link between Lcn972 synthesis or immunity and impaired growth on cellobiose.
We determined Lcn972 production in supernatants from L. lactis MG1363/pBL1 grown on glucose (i.e., low celB expression) or cellobiose (i.e., high celB expression) and in the ΔcelB background, where celB is not present. In this case, we had to make use of pBL1E, in which the erythromycin resistance marker was cloned in the unique EcoRV site of pBL1, disrupting pBL1_orf4, because we were unable to recover any L. lactis ΔcelB/pBL1 transformants by Lcn972 selection. Supernatant samples were taken at the transition toward the stationary phase (OD600s of 2.8 and 1.6 in glucose and cellobiose cultures, respectively). As shown in Table 5, the mutation in pBL1E did not affect production of Lcn972, as yields similar to those with the wild-type plasmid pBL1 were detected (Table 5). No large differences in Lcn972 production were observed when the strains were growing in glucose. On the contrary, in cellobiose-growing cultures, yields were somewhat higher in the ΔcelB background, sustaining the idea that cells lacking this gene may support higher Lcn972 production levels.
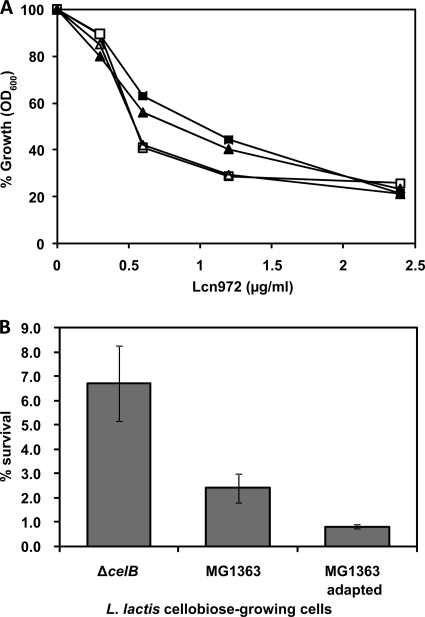
Next, we hypothesized that CelB may act as a putative receptor to facilitate Lcn972 antimicrobial activity and, consequently, that producer cells would have a tendency to suppress gene expression. According to MIC values in GM17, L. lactis MG1363 and MG1363 ΔcelB were equally susceptible to Lcn972 (MIC = 0.15 μg/ml). Moreover, dose-response curves with increasing Lcn972 concentrations were essentially identical for L. lactis MG1363 and MG1363 ΔcelB regardless of whether cultures were grown on glucose or cellobiose (Fig. 4A). In this light, in contrast to the mannose PTS, which is targeted by several class II bacteriocins and determines bacteriocin activity, CelB is unlikely to be an essential receptor for Lcn972. Interestingly, both the wild-type and ΔcelB strains were somewhat more susceptible to Lcn972 when growing on cellobiose than when growing on glucose (Fig. 4A). Therefore, we asked whether repression of celB and the subsequent metabolic changes could make lactococcal cells able to cope better with the presence of Lcn972, a scenario that Lcn972-producing cells have to face. To test this, early-exponential-phase cellobiose-growing (OD600 = 0.2) nonadapted L. lactis MG1363 and MG1363 ΔcelB cells were challenged with Lcn972 and survival was scored (Fig. 4B). The highest percentage of surviving cells was found for L. lactis ΔcelB compared to the wild-type L. lactis MG1363. Moreover, resistance to Lcn972 decreased further when L. lactis MG1363 had been previously subcultured in CDM-cellobiose for 30 generations (adapted cells) prior to the treatment. In this case, only 0.8% of the cells survived (Fig. 4B). These results sustain the hypothesis that downregulation of celB somehow triggers a defense mechanism against the antimicrobial activity of Lcn972. Furthermore, it was also confirmed by RT-qPCR that celB RNA levels in L. lactis MG1363 cellobiose-adapted cells were 6.4 times higher than those in nonadapted cells when growing on glucose (data not shown). This suggests that adaptation to cellobiose in L. lactis MG1363 may imply increased basal expression of celB in the presence of glucose.
Fig. 4.
Susceptibility of L. lactis strains to Lcn972. (A) Dose-response curves of L. lactis MG1363 (squares) and L. lactis MG1363 ΔcelB (triangles) with increasing Lcn972 concentrations growing in CDM-glucose (closed symbols) and CDM-cellobiose (open symbols) at 30°C in a microtiter plate. Growth in the absence of Lcn972 was taken as 100% as monitored by OD600. Results are mean values for triplicate wells, and standard deviations never exceeded 10% of the given value. (B) Survival of L. lactis MG1363 ΔcelB and L. lactis MG1363 nonadapted and adapted to cellobiose. Exponentially growing cells on cellobiose (OD600 = 0.2) were treated with 0.1 μM Lcn972 for 1 h at 30°C before appropriate dilutions were plated on GM17. Percent survival was defined as CFU/ml of treated cultures divided by CFU/ml of control cultures.
DISCUSSION
The plasmid pBL1 had been previously shown to encode the production of and immunity to the bacteriocin Lcn972 (35, 45). This plasmid could be transferred into the susceptible L. lactis MG1614, conferring to the new transformants the ability to produce Lcn972 while any other obvious phenotype remained elusive under standard laboratory conditions (35). However, the genome-wide transcriptional analysis carried out in this work revealed unexpected changes in gene expression that suggested that bacteriocin synthesis is not gratuitous for producing cells and may impose a metabolic burden.
Although we have not further investigated this, it seems reasonable to speculate that the overexpression of genes involved in oligopeptide transport in the presence of pBL1 responds to a higher nitrogen demand needed for Lcn972 biosynthesis. L. lactis is auxotrophic for multiple amino acids and depends on its proteolytic system and peptide uptake for growth. Moreover, the opp operon is highly repressed by the presence of peptides in the growth medium via the pleiotropic transcriptional repressor CodY, which senses the intracellular pool of branched-chain amino acids (BCAAs), which are corepressors of CodY (references 13 and 17 and references therein). The intracellular amino acid content in Lcn972-producing cultures would presumably be lower, relieving opp from CodY repression.
Another response to the presence of pBL1 deduced from the microarray data was repression of llmg0186 and celB involved in cellobiose metabolism. This PTS has been more deeply characterized in L. lactis IL1403, but our results showed that the structural organization is similar and that the two genes form an operon also in L. lactis MG1363. The specific downregulation of celB by pBL1 was further confirmed by transforming this plasmid into L. lactis MG1363 and demonstrating by RT-qPCR that celB RNA levels were lower in the presence of pBL1. In this way, the possibility of strain-to-strain variation based on nonidentified mutations prompted by the plasmid was discarded. Moreover, although to a lesser extent, induction of the other cellobiose transporter gene, ptcC, was also inhibited by the presence of pBL1, supporting the notion that cellobiose metabolism was specifically targeted by pBL1. It is worth mentioning that celB RNA levels were almost identical in cells growing on glucose regardless of the presence of pBL1 (expression ratio of 1.2 in MG1363 versus pBL1). This observation is not in agreement with the initial transcriptomic analysis carried out with cultures grown on glucose in the complex medium M17. In L. lactis IL1403, induction of celB requires the presence of cellobiose (3). Residual dextrins present in the formulation of M17 may be enough to induce celB expression in the control cells, thereby making more evident the repression posed by pBL1.
The genetic evidence that cellobiose uptake was hindered in Lcn972-producing cells was further supported by the subsequent physiological studies. All the results are in agreement with a constrained sugar uptake based on the lower growth rate in CDM-cellobiose, the more pronounced mixed acid fermentation profile, and the lower substrate consumption rate observed in L. lactis MG1363/pBL1. Moreover, these variables paralleled those defined for the celB-defective strain. This view was further supported by the differences found in the pool of internal metabolites. Overall, the pool sizes of glycolytic metabolites reflect a constriction in cellobiose utilization, which most likely arises from the transport step limitation, and are in accordance with previous studies on mutants lacking particular sugar PTSs (8).
The lower pool sizes of UDP-activated sugars and/or amino sugars might reflect rerouting of carbon flux to the production of structural or storage polysaccharides in the presence of pBL1. Moreover, since the lipid carrier C-55 is used both for exocellular polysaccharide biosynthesis and for peptidoglycan biosynthesis, a higher demand for the former would lead to the accumulation of cell wall precursors, as previously described (42). Lcn972 itself could also contribute, as it is a cell wall-active bacteriocin that binds to lipid II, precluding its incorporation into preexisting peptidoglycan (34). Recently, it has been shown that L. lactis MG1363 is able to synthesize a cell wall polysaccharide pellicle that acts as a protective barrier (9). Curiously, dense cell suspensions of MG1363/pBL1 were considerably slimier than those of MG1363 (data not shown). Whether pBL1 induces the synthesis of this protective pellicle is under investigation.
For some bacteriocins, it has been shown that engineering of bacteriocin immunity leads to higher bacteriocin production yields (25, 27), showing that self-toxicity poses a burden to increase bacteriocin productivity. Moreover, downregulation of key enzymes involved in sugar metabolism has been shown to be involved in tolerance to antibiotics (4). On this background, the observed metabolic response of L. lactis to the presence of pBL1 suggests that pBL1-carrying cells seem to mount a response to counteract the antimicrobial activity of Lcn972, even in the presence of the putative immune system, and signaling might occur via downregulation of celB. In favor of this hypothesis are the facts that (i) slightly higher Lcn972 yields were obtained in a ΔcelB background, (ii) susceptibility of L. lactis to Lcn972 increases when growth is on cellobiose, and (iii) L. lactis ΔcelB is more tolerant than the wild type. Moreover, celB is also downregulated in L. lactis strains that are resistant to the bacteriocin Lcn972 (our unpublished results), supporting its role in self-protection against Lcn972. On the other hand, CelB itself might also play a role as a docking molecule to facilitate access of Lcn972 to its target lipid II. However, in contrast to the case for the mannose PTSs which are targeted by class II bacteriocins (16, 22, 29, 41), CelB seems to be dispensable because L. lactis ΔcelB is still susceptible to Lcn972.
Beyond the significance of tuning celB expression as a trigger to increase tolerance of L. lactis MG1363 to Lcn972, our results have also added some hints on cellobiose metabolism in this strain which support future research on this particular PTS. First, celB seems to be the main cellobiose transporter in this strain, as it is highly induced in cellobiose-growing cultures and at levels more than 70 times higher than those for ptcC. Moreover, coregulation of bglS, demonstrated by both induction by cellobiose and a similar inhibition rate posed by pBL1, supports its role as the putative phospho-β-glucosidase responsible for cleavage of cellobiose-6-phosphate, as described for IL1403 (2). Our data are also in agreement with a recent report showing that rapid growth of L. lactis MG1363 on cellobiose is preceded by the induction of cellobiose-specific genes (31). In this regard, our results showed that adaptation seems to rely on the higher basal expression levels of the llmg0186-celB operon, and once the cells were grown on cellobiose, the initial lag phase was no longer observed. The underlying molecular mechanism remains to be clarified.
In many references in the literature, altered carbon metabolism has been linked to class IIa bacteriocin resistance (7, 28, 48, 49). However, some other practical consequences for the field of bacteriocin production stem from the results of this work. In the case of the bacteriocin Lcn972, a choice of inexpensive or renewable sources rich in dextrins or cellobiose as substrates should be avoided, as a lower cell biomass will be reached. Furthermore, celB has been shown to be involved in lactose uptake by L. lactis lacking a lactose-specific PTS (3), meaning that Lcn972 production by the recombinant L. lactis MG1363/pBL1 would be seriously compromised in milk or dairy products where lactose is the main available carbohydrate.
ACKNOWLEDGMENTS
P. Gaspar was supported by a Fundação para a Ciência e a Tecnologia (FCT) postdoctoral grant (SFRH/BPD/31251/2006), and C. Roces holds a JAEPre-CSIC (Spain) predoctoral fellowship. B. Martínez and A. R. Neves gratefully acknowledge the Ministerio de Ciencia e Innovación (MCINN) and Conselho de Reitores das Universidades Portuguesas (CRUP) for the Luso-Spanish Integrated Action (HP2008-0042; Pt, E-67/09). This work was supported by grants BIO2007-65061 and BIO2010-17414 (MCINN-Spain) and in part by FCT grants PTDC/SAU-MII/100964/2008 and PTDC/BIA-MIC/099963/2008. The NMR spectrometers are part of the National NMR Network (REDE/1517/RMN/2005), supported by “Programa Operacional Ciência e Inovação (POCTI) 2010” and FCT.
We thank Tanja Schneider (Bonn University, Germany) for kindly providing UDP-N-acetylmuramic acid and Anne de Jong (RuG, The Netherlands) and Aldert Zomer (Radboud University, Nijmegen, The Netherlands) for handling array data.
Footnotes
Published ahead of print on 2 September 2011.
REFERENCES
- 1. Alegría A., Delgado S., Roces C., López B., Mayo B. 2010. Bacteriocins produced by wild Lactococcus lactis strains isolated from traditional, starter-free cheeses made of raw milk. Int. J. Food Microbiol. 143:61–66 [DOI] [PubMed] [Google Scholar]
- 2. Aleksandrzak-Piekarczyk T., Kok J., Renault P., Bardowski J. 2005. Alternative lactose catabolic pathway in Lactococcus lactis IL1403. Appl. Environ. Microbiol. 71:6060–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Aleksandrzak-Piekarczyk T., Polak J., Jezierska B., Renault P., Bardowski J. 2011. Genetic characterization of the CcpA-dependent, cellobiose-specific PTS system comprising CelB, PtcB and PtcA that transports lactose in Lactococcus lactis IL1403. Int. J. Food Microbiol. 145:186–194 [DOI] [PubMed] [Google Scholar]
- 4. Bizzini A., et al. 2010. A single mutation in enzyme I of the sugar phosphotransferase system confers penicillin tolerance to Streptococcus gordonii. Antimicrob. Agents Chemother. 54:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breukink E., et al. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364 [DOI] [PubMed] [Google Scholar]
- 6. Brötz H., et al. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317–327 [DOI] [PubMed] [Google Scholar]
- 7. Calvez S., Rincé A., Auffray Y., Prévost H., Drider D. 2007. Identification of new genes associated with intermediate resistance of Enterococcus faecalis to divercin V41, a pediocin-like bacteriocin. Microbiology 153:1609–1618 [DOI] [PubMed] [Google Scholar]
- 8. Castro R., et al. 2009. Characterization of the individual glucose uptake systems of Lactococcus lactis: mannose-PTS, cellobiose-PTS and the novel GlcU permease. Mol. Microbiol. 71:795–806 [DOI] [PubMed] [Google Scholar]
- 9. Chapot-Chartier M. P., et al. 2010. Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J. Biol. Chem. 285:10464–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cotter P. D., Hill C., Ross R. P. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777–788 [DOI] [PubMed] [Google Scholar]
- 11. Dalet K., Cenatiempo Y., Cossart P., Hechard Y. 2001. A sigma(54)-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology 147:3263–3269 [DOI] [PubMed] [Google Scholar]
- 12. de Arauz L. J., Jozala A. F., Mazzola P. G., Penna T. C. V. 2009. Nisin biotechnological production and application: a review. Trends Food Sci. Technol. 20:146–154 [Google Scholar]
- 13. den Hengst C. D., et al. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332–34342 [DOI] [PubMed] [Google Scholar]
- 14. Deutscher J. 2008. The mechanisms of carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 11:87–93 [DOI] [PubMed] [Google Scholar]
- 15. Deutscher J., Francke C., Postma P. W. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Diep D. B., Skaugen M., Salehian Z., Holo H., Nes I. F. 2007. Common mechanisms of target cell recognition and immunity for class II bacteriocins. Proc. Natl. Acad. Sci. U. S. A. 104:2384–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doeven M. K., Kok J., Poolman B. 2005. Specificity and selectivity determinants of peptide transport in Lactococcus lactis and other microorganisms. Mol. Microbiol. 57:640–649 [DOI] [PubMed] [Google Scholar]
- 18. Fallico V., McAuliffe O., Fitzgerald G. F., Hill C., Ross R. P. 2009. The presence of pMRC01 promotes greater cell permeability and autolysis in lactococcal starter cultures. Int. J. Food Microbiol. 133:217–224 [DOI] [PubMed] [Google Scholar]
- 19. Gálvez A., Abriouel H., López R. L., Ben Omar N. 2007. Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 120:51–70 [DOI] [PubMed] [Google Scholar]
- 20. Gasson M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grant S. G., Jessee J., Bloom F. R., Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. US. 87:4645–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gravesen A., et al. 2002. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology 148:2361–2369 [DOI] [PubMed] [Google Scholar]
- 23. Hancock R. E., Sahl H. G. 2006. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 24:1551–1557 [DOI] [PubMed] [Google Scholar]
- 24. Hansen M. E., Wangari R., Hansen E. B., Mijakovic I., Jensen P. R. 2009. Engineering of Bacillus subtilis 168 for increased nisin resistance. Appl. Environ. Microbiol. 75:6688–6695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heinzmann S., Entian K. D., Stein T. 2006. Engineering Bacillus subtilis ATCC 6633 for improved production of the lantibiotic subtilin. Appl. Microbiol. Biotechnol. 69:532–536 [DOI] [PubMed] [Google Scholar]
- 26. Heng N. C. K., Tagg J. R. 2006. What is in a name? Class distinction for bacteriocins. Nat. Rev. Microbiol. 4:doi:10.1038/nrmicro1273-c1 [Google Scholar]
- 27. Kim W. S., Hall R. J., Dunn N. W. 1998. Improving nisin production by increasing nisin immunity/resistance genes in the producer organism Lactococcus lactis. Appl. Microbiol. Biotechnol. 50:429–433 [Google Scholar]
- 28. Kjos M., Nes I. F., Diep D. B. 2011. Resistance mechanisms against bacteriocins targeting the mannose phosphotransferase system. Appl. Environ. Microbiol. 77:3335–3342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kjos M., Salehian Z., Nes I. F., Diep D. B. 2010. An extracellular loop of the mannose phosphotransferase system component IIC is responsible for specific targeting by class IIa bacteriocins. J. Bacteriol. 192:5906–5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kowalczyk M., Cocaign-Bousquet M., Loubiere P., Bardowski J. 2008. Identification and functional characterisation of cellobiose and lactose transport systems in Lactococcus lactis IL1403. Arch. Microbiol. 189:187–196 [DOI] [PubMed] [Google Scholar]
- 31. Linares D. M., Kok J., Poolman B. 2010. Genome sequences of Lactococcus lactis MG1363 (revised) and NZ9000 and comparative physiological studies. J. Bacteriol. 192:5806–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Long A. D., et al. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937–19944 [DOI] [PubMed] [Google Scholar]
- 33. Maguin E., Prévost H., Ehrlich S., Gruss A. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martínez B., et al. 2008. Specific interaction of the unmodified bacteriocin lactococcin 972 with the cell wall precursor lipid II. Appl. Environ. Microbiol. 74:4666–4670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martínez B., Fernández M., Suárez J. E., Rodríguez A. 1999. Synthesis of lactococcin 972, a bacteriocin produced by Lactococcus lactis IPLA 972, depends on the expression of a plasmidencoded bicistronic operon. Microbiology 145:3155–3161 [DOI] [PubMed] [Google Scholar]
- 36. Martínez B., Rodríguez A., Suárez J. E. 2000. Lactococcin 972, a bacteriocin that inhibits septum formation in lactococci. Microbiology 146:949–955 [DOI] [PubMed] [Google Scholar]
- 37. Martínez B., Zomer A. L., Rodríguez A., Kok J., Kuipers O. P. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64:473–486 [DOI] [PubMed] [Google Scholar]
- 38. Neves A. R., et al. 2006. The alpha-phosphoglucomutase of Lactococcus lactis is unrelated to the alpha-d-phosphohexomutase superfamily and is encoded by the essential gene pgmH. J. Biol. Chem. 281:36864–36873 [DOI] [PubMed] [Google Scholar]
- 39. Neves A. R., Pool W. A., Kok J., Kuipers O. P., Santos H. 2005. Overview on sugar metabolism and its control in Lactococcus lactis—the input from in vivo NMR. FEMS Microbiol. Rev. 29:531–554 [DOI] [PubMed] [Google Scholar]
- 40. Poolman B., Smid E. J., Veldkamp H., Konings W. N. 1987. Bioenergetic consequences of lactose starvation for continuously cultured Streptococcus cremoris. J. Bacteriol. 169:1460–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ramnath M., Beukes M., Tamura K., Hastings J. W. 2000. Absence of a putative mannose-specific phosphotransferase system enzyme IIAB component in a leucocin A-resistant strain of Listeria monocytogenes, as shown by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Appl. Environ. Microbiol. 66:3098–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ramos A., Boels I. C., de Vos W. M., Santos H. 2001. Relationship between glycolysis and exopolysaccharide biosynthesis in Lactococcus lactis. Appl. Environ. Microbiol. 67:33–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Roces C., et al. 2009. Contribution of the CesR-regulated genes llmg0169 and llmg2164-2163 to Lactococcus lactis fitness. Int. J. Food Microbiol. 133:279–285 [DOI] [PubMed] [Google Scholar]
- 44. Sambrook J., Maniatis T., Fritsch E. F. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 45. Sánchez C., et al. 2000. Nucleotide sequence and analysis of pBL1, a bacteriocin-producing plasmid from Lactococcus lactis IPLA 972. Plasmid 44:239–249 [DOI] [PubMed] [Google Scholar]
- 46. Simsek O., Con A. H., Akkoc N., Saris P. E., Akcelik M. 2009. Influence of growth conditions on the nisin production of bioengineered Lactococcus lactis strains. J. Ind. Microbiol. Biotechnol. 36:481–490 [DOI] [PubMed] [Google Scholar]
- 47. Takala T. M., Saris P. E. 2002. A food-grade cloning vector for lactic acid bacteria based on the nisin immunity gene nisI. Appl. Microbiol. Biotechnol. 59:467–471 [DOI] [PubMed] [Google Scholar]
- 48. Tessema G. T., Moretro T., Snipen L., Axelsson L., Naterstad K. 2011. Global transcriptional analysis of spontaneous sakacin P-resistant mutant strains of Listeria monocytogenes during growth on different sugars. PLoS One 6:e16192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vadyvaloo V., Snoep J. L., Hastings J. W., Rautenbach M. 2004. Physiological implications of class IIa bacteriocin resistance in Listeria monocytogenes strains. Microbiology 150:335–340 [DOI] [PubMed] [Google Scholar]
- 50. van Hijum S. A., et al. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zúñiga M., Franke-Fayard B., Venema G., Kok J., Nauta A. 2002. Characterization of the putative replisome organizer of the lactococcal bacteriophage r1t. J. Virol. 76:10234–10244 [DOI] [PMC free article] [PubMed] [Google Scholar]