Abstract
The ability to conduct advanced functional genomic studies of the thousands of sequenced bacteria has been hampered by the lack of available tools for making high-throughput chromosomal manipulations in a systematic manner that can be applied across diverse species. In this work, we highlight the use of synthetic biological tools to assemble custom suicide vectors with reusable and interchangeable DNA “parts” to facilitate chromosomal modification at designated loci. These constructs enable an array of downstream applications, including gene replacement and the creation of gene fusions with affinity purification or localization tags. We employed this approach to engineer chromosomal modifications in a bacterium that has previously proven difficult to manipulate genetically, Desulfovibrio vulgaris Hildenborough, to generate a library of over 700 strains. Furthermore, we demonstrate how these modifications can be used for examining metabolic pathways, protein-protein interactions, and protein localization. The ubiquity of suicide constructs in gene replacement throughout biology suggests that this approach can be applied to engineer a broad range of species for a diverse array of systems biological applications and is amenable to high-throughput implementation.
INTRODUCTION
The rate and depth of characterization of bacterial species have increased over the last few years due to advances in genome sequencing technology and the application of high-throughput functional genomics approaches that identify and quantify mRNA transcripts, expressed proteins, and cellular metabolites. To answer important, far-ranging questions in functional genomics (e.g., assessments of gene essentiality or cell-wide genetic interactions [epistasis] and protein-protein interactions [PPI]), experimental validation will require rapid and efficient genetic engineering of the strain of interest. Of the more than 1,400 bacterial genomes sequenced so far (3), relatively few transposon-mediated knockout libraries have been reported (15, 19, 21, 23, 24, 29, 30, 33), and systematic, large-scale, single-gene deletion collections exist only for Escherichia coli K-12 (5) and Acinetobacter baylyi ADP1 (19). Furthermore, large-scale tandem affinity purification (TAP)-based PPI identified with proteins produced from chromosomally tagged genes have been reported only for E. coli K-12 (14). Genome-wide genetic interaction screening, which requires an ordered gene knockout library, was also recently reported but was applied only for E. coli (13, 42). In summary, there is currently no systematic approach for making large-scale, targeted chromosomal manipulations that can be readily applied across a diverse range of bacteria for functional genomic studies.
Here we present a scheme for the high-throughput manipulation of bacterial genomes that is both inexpensive and flexible due to the use of interchangeable “parts” for making different kinds of chromosomal modifications, including gene deletions and tagged genes for the study of PPI and protein localization and for other applications (Fig. 1). Our goal was to create a systematic approach for chromosomal modification that could be applied to a wide range of bacteria with a minimal need for methodological alterations. For this reason, the direct use of phage-based recombination systems in candidate microbes of interest was deemed unsuitable, as this could require species-specific additional host mutations and extensive development work for individual species. Rather, we chose to leverage a common theme for chromosomal modification: the use of nonreplicating gene modification (“suicide”) constructs. Suicide constructs have been used successfully for chromosomal modification in a wide range of bacterial species and generally require only host-based RecA-mediated homologous recombination (34). Our approach for the high-throughput construction of suicide vectors was based on sequence- and ligation-independent cloning (SLIC) (28), heretofore used for plasmid-based (rather than chromosomally based) metabolic engineering and heterologous protein expression studies.
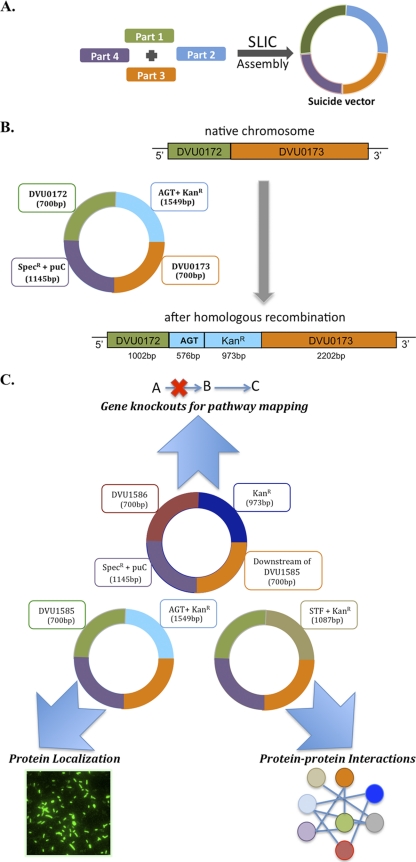
Fig. 1.
(A) The SLIC approach (for double recombinations): Suicide vectors are assembled directly from four “parts” (in purple, green, orange, and blue) using SLIC. Parts 1 and 2 (purple and orange) correspond to homology regions (HR1/HR2) of the target loci on the chromosome. Parts 3 and 4 (green and blue) correspond to an insertion part (IP) and a vector replication origin plus a selection part (RSP), respectively. Different parts may be mixed and matched depending on the choice of the application. (B) Example of a chromosomal modification in D. vulgaris Hildenborough using the SLIC approach: insertion of the SNAP tag at the 3′ end of DVU0172. The suicide construct is assembled in E. coli using the SLIC technique from the following parts: 700 bp upstream from the 3′ end of DVU0172 not including its stop codon (part 1), the AGT (visualization) tag followed by a kanamycin resistance gene (part 2), 700 bp downstream from the 5′ end of DVU0172 (part 3), and the replication origin of the vector (pUC) recognized only in E. coli, followed by a spectinomycin resistance gene (part 4). The chromosomal modification in D. vulgaris Hildenborough after the double homologous recombination of the transformed suicide vector is shown. (C) Utilizing the parts-based approach for enabling chromosomal modifications in D. vulgaris Hildenborough using DVU1585 as the target gene. A set of reusable parts (color coded) were employed for generating suicide constructs in E. coli, which were then transformed into D. vulgaris to examine the role of DVU1585 in this sulfate reducer. Results for gene essentiality, protein-protein interactions, and protein localization are discussed in the text.
We applied this approach to the sulfate-reducing bacterium (SRB) Desulfovibrio vulgaris Hildenborough. D. vulgaris has been the subject of recent functional genomics studies (7, 16, 16a, 17, 31, 37). Proposed stress response models of this bacterium are based on gene expression data alone and need to be complemented by other types of experimental data. The ability to create targeted gene deletions in a systematic manner in this organism will help fill gaps in metabolic pathways and greatly assist in the functional annotation of unknown genes. In addition, the ability to generate TAP- or visualization-tagged genes will facilitate the development of the corresponding interactome and allow the mapping of protein complex localizations within the cell. Here we compare the protocols for the facile chromosomal engineering of D. vulgaris to achieve these objectives and provide proof-of-principle data for protein complex isolation, gene deletion, and subcellular localization.
MATERIALS AND METHODS
Mutant strain generation.
The design principles for the three schemes are shown in Fig. S1 in the supplemental material. Laboratory Information Management System (LIMS) tools were developed for the Gateway and SLIC schemes, as detailed in the supplemental material. The Gateway scheme involves the generation of a library of entry vectors which serve as the source of mobile DNA fragments that are directionally incorporated into the destination vectors of choice after the LR reaction. Entry vectors in this study were generated by the directional TOPO cloning of desired DNA fragments into pENTR/dTOPO (Invitrogen Inc., Carlsbad, CA) and transformed into One Shot Top10 chemically competent E. coli cells (Invitrogen) according to the manufacturer's instructions. The DNA fragments for TOPO cloning were generated by PCR amplification of the respective regions from genomic DNA of wild-type D. vulgaris Hildenborough using a proofreading DNA polymerase, Pfu Turbo (Stratagene Inc., La Jolla, CA). All plasmid extractions were performed by using QIAprep Spin Miniprep kits (Qiagen Inc., Valencia, CA). Design rules for primers used for the amplification of desired DNA fragments are described in the supplemental material. Amplified targets were visualized on E-gels (Invitrogen) before proceeding with dTOPO cloning. Up to three colonies were picked from ampicillin (100 μg/ml)-containing LB agar plates bearing the transformed cells for the sequence verification of entry vectors. Sequence-verified constructs were used in the subsequent LR recombination step (according to the manufacturer's protocol), along with custom destination vectors. The design strategy for custom destination vectors is described in the supplemental material. Sequence-verified dTOPO constructs were coupled with custom destination vectors through LR recombination to generate Gateway constructs that carried the desired DNA fragments with the corresponding tag sequence appended at their 3′ ends. Products of the LR recombination reaction were transformed into One Shot Top10 chemically competent E. coli and plated onto ampicillin (100 μg/ml)- and kanamycin (50 μg/ml)-containing LB agar plates. Two colonies were randomly picked from the resulting plates for the sequence verification of the Gateway constructs. Sequence-verified suicide constructs were transformed into competent D. vulgaris cells for chromosomal integration through homologous recombination, as described below.
The λred recombination system has been utilized to perform both chromosomal and plasmid modifications in E. coli (18, 40, 47). Here we utilized E. coli strain SW105, which expresses λred recombination functions when subjected to heat shock, to enable the recombination of a PCR product into plasmids carrying fragments of D. vulgaris genomic DNA (44). For our desired application, this PCR product encoded a sequential peptide affinity (SPA) tag and a kanamycin resistance gene and was identical to that used to create chromosomal SPA-tagged genes in E. coli (50). Five initial D. vulgaris target genes present on an 8.7-kb fragment spanning the entire apsBA-qmoABC-DVU0857 gene region were provided by pMO9034 (J. Wall, University of Missouri), which is a derivative of pCR8/GW/TOPO (49). A further 18 D. vulgaris genes were provided by three genomic DNA fragments each cloned into pUC19 and designated pDVH2-36, pDVH2-37, and pDVH2-39. Details on the PCR products employed and subsequent transformation in E. coli are described in the supplemental material. Transformed cells were plated onto selective LB medium containing kanamycin, and colonies were isolated. Isolates were then cultured, and plasmid DNA was prepared and subjected to restriction analysis to confirm the integration of the PCR product at the correct locus in the plasmid target construct.
The SLIC technique was employed as described previously (28). Specifically, four DNA fragments were pieced together to form the final suicide constructs. The same inserted part (IP) (composed of the tag and one selection marker) and selection part (SP) (composed of a plasmid origin of replication and a second selection marker) were used for a given library of insertion or deletion suicide constructs. The other two variable regions were generated by PCR amplification of the respective regions from genomic DNA of wild-type D. vulgaris Hildenborough using a proofreading DNA polymerase, Phusion High Fidelity DNA polymerase (Finnzymes Inc., Woburn, MA). Design rules for primer design are detailed in the supplemental material. The sizes of the amplified PCR products were verified by using agarose gels, and confirmed products were purified by using the QIAquick PCR purification kit (Qiagen) and subsequently quantified by using a NanoDrop 2000 instrument (NanoDrop Products, Wilmington, DE). One microgram of each part was treated with 1 μl of 0.5 U T4 DNA polymerase in 1× buffer 2 (NEB) and 1× bovine serum albumin (BSA) buffer (NEB) in a 20-μl reaction mixture at room temperature for 30 min. The reaction was stopped by the addition of a 1/10 volume of 10 mM dCTP to the mixture, followed by an annealing step for all four parts at 37°C for 30 min. The annealed mixture was next chemically transformed into DH10B competent cells and plated onto agar plates bearing spectinomycin and kanamycin (both at 50 μg/ml) antibiotic resistance markers. Two colonies were picked after an overnight incubation (37°C) for the sequence verification of suicide constructs.
Transformation of D. vulgaris strains.
For transformations, cells were grown in MOYLS4 medium in an anaerobic growth chamber (Coy Laboratory Products Inc., Grass Lake, MI) with an atmosphere composed of nitrogen, <5% hydrogen, and <2% oxygen at 33°C. MOYLS4 medium contained 8 mM magnesium chloride, 20 mM ammonium chloride, 0.6 mM calcium chloride, 1 mM potassium phosphate (dibasic), 1 mM sodium phosphate (monobasic), 0.06 mM ferrous chloride, 0.12 mM EDTA, 30 mM Tris (pH 7.4), 60 mM sodium lactate, 30 mM sodium sulfate, 1 ml/liter Thauer's vitamin solution (12), 6 ml/liter trace element solution, and 1.0 g/liter yeast extract. The trace element solution was modified by the addition of 2.5 mM manganese chloride, 1.26 mM cobaltous chloride, 35 μM sodium selenite, and 24 μM sodium tungstate. The pH of MOYLS4 medium was adjusted to 7.2 with 12 M HCl, and 1.2 mM sodium thioglycolate was added as a reductant prior to sterilization in the autoclave. For plates, 15 g/liter agar was added to MOYLS4 medium before sterilization. Immediately prior to pouring plates of molten MOYLS4 medium, 0.38 mM titanium citrate, prepared and stored anaerobically, was added along with antibiotics as appropriate. To prepare D. vulgaris cells for transformation, 1.0 ml of a freezer stock (early-stationary-phase cells in MOYLS4 medium containing 10% [vol/vol] glycerol) was added to 10 ml MOYLS4 medium and allowed to grow overnight. The 10-ml culture grown overnight was diluted to 100 ml in MOYLS4 medium and allowed to grow to an optical density at 600 nm (OD600) of between 0.3 and 0.7 at 33°C. The culture was harvested by centrifugation for 12 min at 3,000 × g at 4°C, the supernatant was discarded, and the cells were resuspended in 50 ml of chilled, sterile wash buffer (30 mM Tris-HCl buffer [pH 7.2], nondegassed). A second centrifugation, under the same conditions, was used to wash the cells, and the supernatant was discarded. The pellet of electrocompetent cells was resuspended in 1.0 ml wash buffer, and a 100-μl aliquot was used for electroporation. About 1 μg (10 μl) of the plasmid was added to the cells and mixed, and 100 μl of the mixture was transferred into a 1-mm gapped electroporation cuvette (Molecular BioProducts, San Diego, CA). The cuvette was transferred to the ECM 630 electroporator (BTX, Holliston, MA) and electroporated at 1,500 V, 250 Ω, and 25 μF. The electroporated cells were diluted into 1 ml MOYLS4 medium containing 0.1% (wt/vol) yeast extract. The putative transformants were transferred into the anaerobic chamber, opened momentarily for headspace exchange, and allowed to recover overnight at 33°C. Aliquots (100 μl and 900 μl) of the recovered cells were then added to petri plates, followed immediately by the addition of ∼25 ml of reduced, molten MOYLS4 medium with 15 g/liter agar containing the kanamycin analogue G418 (400 μg/ml; RPI Corp., Mt. Prospect, IL) in the anaerobic chamber. Two reductants were used. Sodium thioglycolate (1.2 mM) was added prior to autoclaving, while titanium citrate (1.2 mM, prepared under nitrogen and stored anaerobically) was added just prior to pouring the medium over the cells in the petri plates. Colonies were typically observed after 5 days of incubation. The selection and storage of mutants as well as protocols for Southern blotting and immunoprecipitation (IP) Western analysis to confirm chromosomal manipulations are described in the supplemental material.
Protein-protein interactions determined by using tandem affinity purification.
Affinity-tagged D. vulgaris strains were cultured in 1 liter of 2× LS4D medium (7) in stationary bottles present in an anaerobic chamber. Sequential peptide affinity purification was performed by using tagged D. vulgaris strains exactly as previously reported for E. coli (50). The alternative streptavidin-TEV-FLAG (STF) tag purifications were performed by using the SPA protocol (14) with the following modifications. The STF purification was identical to the SPA procedure until immediately following TEV incubation and the removal of tagged proteins from the anti-FLAG beads. For STF purification, after the TEV eluate was drained into a new column, 100 μl of a 50% slurry of Streptactin Superflow beads (IBA GmbH) was added and incubated for 3 h with rotation at 4°C (Chhabra et al., submitted). The beads were then washed with 1.4 ml wash buffer I (100 mM Tris-Cl [pH 7.9], 150 mM NaCl, 1 mM EDTA, 0.1% Triton X-100, 10 mM 2-mercaptoethanol, Roche Complete protease inhibitor), followed by 400 μl of the wash buffer without Triton X-100, and proteins were eluted by using 300 μl of elution buffer containing 2.5 mM desthiobiotin (Sigma). Purified eluates were subsequently digested with trypsin and analyzed by mass spectrometry to obtain protein identifications, as detailed in the supplemental material.
Growth studies on knockout mutants for examining methionine biosynthesis.
Growth study manipulations were done in an anaerobic growth chamber (Coy Laboratory Products Inc.), which contained approximately 95% N2 and 5% H2. D. vulgaris wild-type and mutant strains were grown at 37°C from freezer stocks to early stationary phase in 5 ml MO medium (49) supplemented with 60 mM lactate, 30 mM sulfate, and 0.1% (wt/vol) yeast extract [MOYLS4(60/30)]. DVU0171 and DVU1585 gene deletion mutants were grown in MO medium supplemented with 60 mM lactate, 30 mM sulfate, 40 mM sulfite, and 0.2% (wt/vol) yeast extract. To select for the growth of the mutant strains, the media were supplemented with G418 antibiotic at 400 μg/ml. To impose starvation conditions, two consecutive triplicate 2% subcultures were then grown in 5 ml defined medium supplemented with 60 mM lactate and 30 mM sulfate [MOLS4(60/30)] (49) plus G418 at 400 μg/ml for mutant selection. As the DVU0890 deletion mutant exhibited little or no growth in subcultures, the screening of the DVU0890 deletion mutant was performed without first subculturing in defined medium. To screen for auxotrophic mutant strains, triplicate 15-ml culture tubes containing 5 ml MOLS4(60/30) medium amended with 0.3 mM amino acids were inoculated with 5% (vol/vol) of the strains and sealed with rubber stoppers. Controls were prepared by the inoculation of MOLS4(60/30) and MOYLS4(60/30) media. For comparisons of growth, ODs were monitored at 600 nm, and final total protein concentrations were measured by using the Bradford assay (9). LC-glucose plates were streaked to check for aerobic contamination of the initial growth culture and periodically of the subcultures. Gene deletion mutations were verified by PCR prior to and again upon the completion of the growth studies.
Labeling of AGT-tagged proteins with the SNAP fluorophore.
The AGT tag or the SNAP tag (New England BioLabs, Ipswich, MA) is a highly engineered modified version of AGT (O6-alkylguanine-DNA alkyltransferase), a human DNA repair protein with a molecular mass of 20 kDa. It is a visualization tag similar to green fluorescent protein (GFP) but, unlike the latter, has been shown to work effectively under anaerobic conditions. It forms a highly stable, covalent thioether bond with fluorophores or other substituted groups when appended to benzylguanine. This reaction is highly specific; i.e., expressed SNAP tag fusion proteins can be labeled even in the presence of complex protein mixtures such as those found in cells or in cleared bacterial lysates. Anaerobically grown cell cultures were harvested at the mid-logarithmic phase at an OD600 of 0.3 to 0.4 and centrifuged under anaerobic conditions at room temperature for 10 min at 5,000 × g. The pellet was resuspended under anaerobic conditions in sterile LS4D medium in the range of 400 μl to 600 μl at an OD600 of 0.5 (±0.05) in order adjust each sample to the same relative cell density, as determined by initial optical density readings. The SNAP fluorophore reagent (New England BioLabs, Ipswich, MA) in dimethyl sulfoxide (DMSO) was added to the cell suspensions to reach a final reagent concentration of 5 μM. Cell solutions with the fluorophore reagent were protected from light and incubated at 30°C for at least 60 min to ensure that all cells were exposed to the labeling reagent. Following the incubation period, the cells were centrifuged at room temperature twice at 5,000 × g for 10 min and resuspended in sterile LS4D medium to remove any excess fluorophore reagent from the cells.
SDS-PAGE and in-gel fluorescence detection.
Labeled and washed cells were subjected to Pefabloc protease (Sigma-Aldrich Co., St. Louis, MO) and DNase (Sigma-Aldrich Co., St. Louis, MO) treatment, at concentrations of 400 μM and 0.2 mg/ml, respectively, and stored on ice. Ice-cold cell suspensions were lysed by using a Branson sonicator (Branson Ultrasonics, Danbury, CT) for 1 to 2 min at a 40% duty cycle with an output of 3. After lysis, the samples were flash-frozen in liquid nitrogen and stored at −20°C. For SDS-PAGE analysis, samples were mixed with 5× SDS loading buffer stock and boiled for 1 min prior to loading into the pockets of a precast 4 to 20% Tris-HCl SDS-PAGE well (Thermo Fisher Scientific Inc., Pittsburgh, PA) and run at 140 V for approximately 50 min. A Pageruler prestained protein marker (Fermentas Inc., Glen Burnie, MD) was run on the gels, as this marker has a 72-kDa protein that emits a fluorescent signal at a 488-nm excitation wavelength and therefore allowed an easy correlation of in-gel fluorescence detection and Coomassie-stained gels. The gels were imaged for fluorescence by using a Bio-Rad Molecular PhosphorImager with an external laser source using Alexa 488 filter settings (Bio-Rad Laboratories, Hercules, CA), followed by Coomassie staining (0.5% Coomassie brilliant blue R-250, 30% ethanol, and 10% glacial acetic acid in double-distilled water [ddH2O]) and destaining in a solution containing 30% ethanol and 10% glacial acetic acid in ddH2O before imaging on a light box with a Canon A540 digital camera. For selected samples, both intact cells as well as cell lysates were labeled, and their in-gel fluorescence values were compared to ensure that the labeling reagent had ready in vivo access to all tags. Details on cellular imaging using epifluorescence and deconvolution microscopy are provided in the supplemental material.
RESULTS AND DISCUSSION
Development and application of schemes for high-throughput generation of suicide constructs.
Traditional mutagenesis approaches with suicide constructs have generally been regarded as cumbersome due to difficulties with vector construction (i.e., cloning of large sections of homologous DNA from either side of the locus to be modified) (34). We therefore tested three recombination-based approaches, the Gateway system (Invitrogen, Carlsbad, CA), “recombineering” (18, 46), and sequence- and ligation-independent cloning (SLIC) (28), for the generation of suicide constructs in a high-throughput manner (see Fig. S1 in the supplemental material). In all examples, suicide vectors designed for modifying the D. vulgaris host chromosome were first generated in E. coli and then transformed into D. vulgaris, resulting in a modified host chromosome through single or double homologous recombination events integrating all or part of the nonreplicating delivery vector.
Suicide vector construction via the Gateway scheme was realized through two steps (see Fig. S1A in the supplemental material). In the first step, the sole homology region of the target locus was directionally cloned into a TOPO entry vector. The second step involved an LR recombination reaction with the directional placement of the homology region from the entry clone into a custom-designed destination vector. The destination vector included the sequences for the modification of the host cell and a suitable origin of replication. Destination vectors with different insertion sequences, such as TAP tags for elucidation of PPI, or visualization tags, such as SNAP (O6-alkylguanine-DNA alkyltransferase; New England BioLabs, Ipswich, MA), that allow protein localization may be used with a given library of entry clones. This powerful attribute of the Gateway scheme allows the facile exchange of the tags once a library of TOPO entry clones has been constructed.
Importantly, the introduction of a single region of homologous DNA in the construction of the entry clones allows only a single recombination event with the host chromosome that incorporates the entire plasmid. When creating tagging constructs for genes located at the beginning of their operons, we incorporated the native promoter sequence in addition to the target gene to be modified in the suicide vector to allow the expression of downstream genes. Necessary promoter sequences for each gene were assumed to be present within 300 bp upstream of the target gene. From a practical standpoint, this scheme is therefore limited to genes located at the terminal ends of their respective operons, where downstream polarity effects can be minimized. These caveats render the Gateway scheme useful for the rapid modification of a select class of target genes with a range of fusion tags. In order to be able to modify genes in a locus-independent manner, however, we leveraged two other schemes, “recombineering” and SLIC, to generate suicide vectors with two homology regions that permitted marker exchange.
In the recombineering approach, we utilized the bacteriophage lambda general recombination system (λred) (18) to modify genes carried on recombinant plasmids selected from an ordered genomic library of D. vulgaris. The expression of λred in E. coli has been shown to mediate the efficient integration of linear DNA molecules into the host chromosome or plasmids through short regions (∼40 bp) of sequence homology (46). In this scheme (see Fig. S1B in the supplemental material), a linear DNA molecule that contained homology regions 1 and 2 (HR1/HR2) flanking the marker to be exchanged or the part to be inserted (insertion part [IP]) was generated by PCR. In the example shown, the IP is an affinity purification tag and a kanamycin resistance cassette expressed from its own promoter. Plasmid constructs and an HR1-IP-HR2-containing PCR product were transformed together into an E. coli strain in which the λred system was induced. The λred recombinase facilitates recombination between the short 40-bp homology regions of the target loci flanking the IP and identical regions in the plasmid containing the fragment of chromosomal DNA with the genes to be modified. The length of the chromosomal regions of homology available for double-crossover events between modified plasmid constructs and the host chromosome varies with the location of the target gene within the genomic DNA insert in the suicide vector but is generally sufficient for detectable recombination with the genome being modified, with the exception of genes located at the termini of the insert DNA in the suicide vector. During the development of this modification procedure, it was noted that many isolated strains contained both a modified plasmid, into which the IP fragment had integrated, and an unmodified plasmid. Furthermore, when higher plasmid concentrations were used, multimeric plasmids were often isolated, a phenomenon previously reported when plasmids were used with the λred system (39). These factors led to increased processing requirements to generate a pure modified plasmid construct. Therefore, while suicide constructs made by this approach can be used for inserting or deleting sequences through marker exchange by double crossovers, the ready availability of a comprehensive ordered genomic library is an essential prerequisite. Due to the lack of such a comprehensive set of library constructs for D. vulgaris and the inefficiencies of isolating recombineered plasmid constructs (compared to SLIC), this approach was not considered further in this study.
The third approach involves the de novo assembly of suicide vectors by the SLIC procedure (28). Vectors assembled by this technique are composed of four parts: two corresponding to the homology regions (HR1/HR2) from the host chromosome, an IP dictated by the application of choice, and a vegetative origin of replication and selection part (RSP). To obtain a suicide vector, the replication origin was functional only in E. coli but not in the strain targeted for manipulation (43) (see Fig. S1C in the supplemental material). The advantage of this approach lies in the reusability of parts for various applications. The IP and RSP regions remain constant for each specific application, whereas the HR1 and HR2 regions vary. Alternative IP regions, such as molecular barcodes, purification tags, antibiotic markers, and origins of replication, are incorporated into vector constructions depending on the downstream application. The RSP region is the most generic part used for suicide vector construction. However, a modification of the RSP to include an oriT (origin of transfer) sequence is possible if the suicide vectors are to be transferred by conjugation from E. coli to the target microbe.
Chromosome modifications by suicide vectors may be characterized as “marked” or “unmarked” depending on the presence or absence of suitable selection markers in the host chromosome after the modifications have been introduced. In either case, chromosomal modifications at the 3′ end of a target gene may alter the expression or translation of co-operonic downstream genes. In the case of the marked approach, the incorporation of a selection marker and its cognate promoter may introduce a second transcription initiation site. For genes located in close proximity to each other, one must also consider the possibility of a displaced ribosomal binding site (RBS). The SLIC approach, through appropriate design, may be used to correct these problems for both marked and unmarked approaches. Systems for unmarked modifications, such as the Cre-lox recombination system (6, 26) or the levansucrase-dependent sucrose sensitivity system (11), could easily be implemented by the incorporation of the respective parts (loxP sites and sacB) into IP or SP regions of SLIC-generated suicide vectors. In hosts where sucrose sensitivity is not a strong selection factor or a residual “scar” is not desired, alternative counterselection systems are available. For D. vulgaris, sensitivity to the toxic pyrimidine analogue 5-fluorouracil allows selection against the expression of upp, encoding the salvage enzyme uracil phosphoribosyl transferase. This marker has been successfully used in a number of microbes (25, 27).
We compared 550 distinct target genes for tagging with suicide vectors assembled by the Gateway and SLIC strategies and generated 297 (54%) and 468 (85%) sequence-verified plasmids, respectively. In general, we observed that the SLIC strategy yielded a higher percentage of confirmed D. vulgaris mutant strains (304 strains/468 plasmids constructed; ∼65%) than the Gateway scheme (70 strains/297 plasmids constructed; ∼24%). For the recombineering strategy, we generated 18 sample suicide vectors, which resulted in 9 confirmed mutant strains. Given the apparent superiority of the SLIC approach, it was the method of choice for the further manipulation of the D. vulgaris chromosome to study the effects of gene deletions, to identify physical interactions of proteins, and to localize selected tagged proteins. To date, we have generated a library of over 700 engineered strains of D. vulgaris using the methodologies described in this work.
Screening for protein-protein interactions by tandem affinity purification.
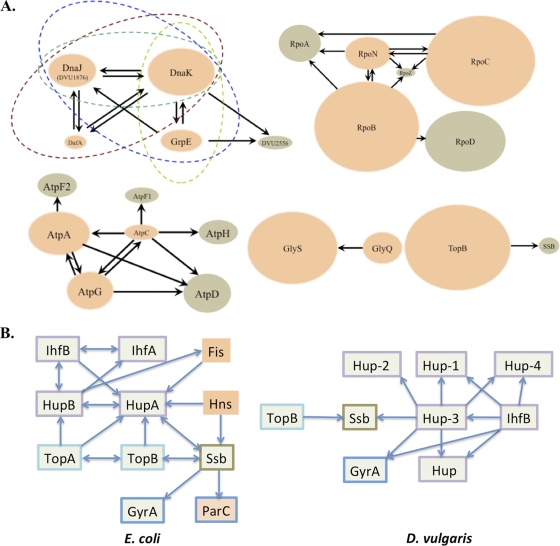
With plasmids constructed by the SLIC procedure, we introduced sequences into the chromosome encoding TAP tags in frame at the 3′ ends of several genes from D. vulgaris. Engineered strains expressing native levels of C-terminally TAP-tagged fusion proteins were used to examine the protein complexes isolated with the tagged baits inferred to represent functional PPI. We validated conserved interactions in several essential complexes, such as the F1-ATPase, the RNA polymerase, the chaperone DnaK, and others (Fig. 2A and see Table S1 in the supplemental material).
Fig. 2.
Conserved protein-protein interactions observed in this study. Chromosomally tagged (STF) baits are shown in orange, and prey proteins are shown in brown. The relative sizes of interacting pairs are roughly proportional to their molecular masses, and arrows point from tagged baits to the respective prey. (A) Conserved interactions from the following complexes are shown: (i) the chaperonin complex composed of the heat shock proteins DnaK (DVU0811), DnaJ (DVU1876 and DVU3243), DafA (DVU1875), and GrpE (DVU0812) and a hypothetical protein (DVU2556); (ii) the ATP synthase complex composed of the α subunit (AtpA [DVU0777]), β subunit (AtpD [DVU0775]), γ subunit (AtpG [DVU0776]), δ subunit (AtpH [DVU0778]), and ε subunit (AtpC [DVU0774]); (iii) the RNA polymerase complex composed of the α subunit (DVU1329), β subunit (DVU2928), β′ subunit (DVU2929), and σ54 factor (DVU1628); (iv) the glycyl-tRNA synthetase complex composed of the α subunit (GlyQ [DVU1898]) and β subunit (GlyS [DVU1897]); and (v) the binary interaction between DNA topoisomerase III (TopB [DVU2316]) and single-strand binding protein (SSB) (DVU0222). (B) Comparison of interacting partners of DNA-binding proteins (COG766 and COG550) from E. coli and D. vulgaris. Arrows point from chromosomally tagged baits in each organism to the respective prey. Green boxes indicate the presence of orthologs in both organisms. Orange boxes indicate proteins unique to E. coli. Border colors represent proteins from the same COG category.
Next, we examined potential interacting partners of proteins associated with the D. vulgaris nucleoid (Fig. 2B). Well-known components of the E. coli nucleoid include DNA-binding proteins such as Fis, HNS, Dps, IHF (IhfAB), and HU (HupAB). By the very nature of their inherent DNA-binding capabilities, these highly abundant proteins are involved in the modulation of cellular processes such as transcriptional regulation, maintenance of DNA architecture, replication, recombination, and stress protection (1, 2, 10, 20).
Given the common set of functions attributed to these proteins, it is not surprising that they exhibit a high level of interactions with each other. Indeed, proteins precipitated with TAP-tagged baits of HU and IHF from D. vulgaris suggest a closely knit interaction subnetwork comprising many of these DNA-binding proteins. Intriguingly, the D. vulgaris genome appears to lack the diversity of nucleoid protein domains reported for E. coli, such as Fis (COG2901), HNS (COG2916), and Dps (COG783) and their corresponding interacting partners (8). In contrast, D. vulgaris encodes twice as many proteins with the “bacterial nucleoid DNA-binding protein” domain, COG776, as those found in E. coli (3). In order to compare the E. coli and D. vulgaris subnetworks associated with COG776 family proteins, we identified interacting partners of D. vulgaris-tagged baits, Hup-3 and IhfB. With the exception of DVUA0004 and DVU1134, all members of the COG776 family appeared to interact with the tagged baits and potentially with each other (Fig. 2B and see Table S2 in the supplemental material).
Unlike topoisomerases from E. coli, members of the D. vulgaris “topoisomerase” family (TopA and TopB) did not appear to copurify with the tagged HU proteins. This was also confirmed when TopB was used as the bait and none of the COG776 family proteins were observed as interacting partners. In E. coli, HU (HupAB) was reported previously to introduce negative supercoiling in covalently closed circular DNA in the presence of topoisomerase I (TopA) (36). From these results, it appears that mechanisms of DNA architecture maintenance and global regulatory controls in D. vulgaris may differ from those in E. coli.
Gene deletions: examining the methionine biosynthesis pathway of D. vulgaris.
Although the genome sequence of D. vulgaris was published in 2004, several amino acid biosynthesis pathways in this SRB remain to be elucidated. In this study, we examined putative alternative steps in methionine biosynthesis. At least 18 variant methionine pathways have been proposed to originate from the common precursor, homoserine (22). In examining the D. vulgaris genome for all known variant genes related to the three major steps of methionine synthesis, (i) homoserine activation, (ii) sulfur incorporation, and (iii) methylation, homologs corresponding only to step 3 were apparent: vitamin B12-dependent methionine synthase (DVU1585 [metH]) and methionine synthase II (cobalamin independent) (DVU3371 [metE]). We tested these and other genes putatively involved in the production of the methionine precursor homoserine from l-aspartate. These included a putative aspartate kinase (DVU1913 [lysC]), homoserine dehydrogenase (DVU0890 [hom]) (probable counterparts to bifunctional aspartate kinase II/homoserine dehydrogenase from E. coli), a putative beta-cystathionase (DVU0171, similar to patB [4]), and a protein with predicted methyltransferase activity (DVU3369, similar to metW [19]).
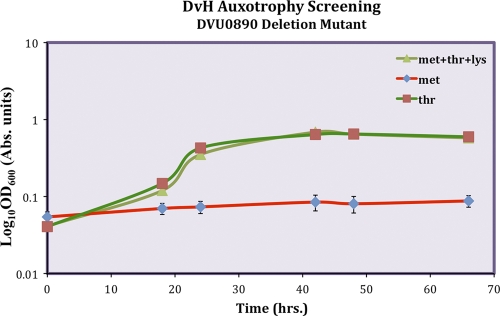
We verified all gene deletion mutations by PCR as well as Southern blot analysis. These gene deletion studies revealed that a majority of the putative methionine biosynthesis pathway knockouts (DVU1585, DVU3371, DVU0890, DVU0171, and DVU3369) did not result in methionine auxotrophy. A surprising result of this study was that the mutant deleted for DVU0890, Δhom, was found to be auxotrophic for threonine but not methionine (Fig. 3). This unexpected phenotype and the difficulty encountered in the isolation of a deletion of DVU1913 were interpreted to indicate that an unusual pathway for methionine biosynthesis might be operational in this SRB. Further studies in this direction are under way.
Fig. 3.
Optical density (600 nm) growth curve data for the D. vulgaris Hildenborough (DvH) ΔDVU0890 strain. The growth of the mutant strain was restored in LS4 minimal medium by supplementation with threonine but not methionine.
Protein localization with visualization tags.
We engineered D. vulgaris strains to express proteins bearing a SNAP tag, which is designed for subcellular visualization in anaerobic bacteria. Conventional green fluorescent protein derivatives require molecular oxygen for proper chromophore formation and hence cannot be utilized under anaerobic culturing conditions. We therefore explored the use of a modified SNAP tag that has a dead-end reaction with a modified O6-benzylguanine (BG) derivative (32, 35). To validate the use of the AGT-tag-based method for subcellular localization in anaerobic bacteria, we first compared the SNAP labelings of three AGT-tagged proteins from D. vulgaris, DsrC (DVU2776), MreB (DVU0789) (data not shown), and FtsZ (DVU2499), from the respective engineered strains to the unmodified wild-type strain. We confirmed the specific labeling of tagged proteins using two complementary methods: in-gel fluorescence detection by SDS-PAGE and fluorescence microscopy. SDS-PAGE analysis typically yielded single bands at the expected molecular masses, indicating the specific labeling of the tag, with little or no nonspecific binding. Interestingly, in our fluorescence micrographs, we found a robust cell-to-cell variability in the labeling signal. To eliminate the possibility that the labeling reagent did not reach all tagged proteins, we compared in vivo-labeled intact cells to in vitro-labeled whole-cell extracts and observed no difference in the fluorescence signals between the two, as judged by SDS-PAGE analysis. This suggested efficient reagent access and the specific labeling of intracellular AGT-tagged proteins. In the case of MreB and FtsZ, unlike DsrC, the chromosomal tagging appeared to alter the cellular morphology normally associated with the wild-type strain. Morphological changes included either the loss of the vibrio-typic cell shape (MreB-AGT) (data not shown) or extensive elongation (FtsZ-AGT) (see Fig. S2 in the supplemental material), suggesting diminished or altered protein function due to the presence of the visualization tag. Our results are comparable to those for the GFP-based protein localization of FtsZ, as demonstrated previously for E. coli (34). To our knowledge, this is the first account of specific tag-based fluorescence labeling for the purpose of protein localization in an anaerobic bacterium.
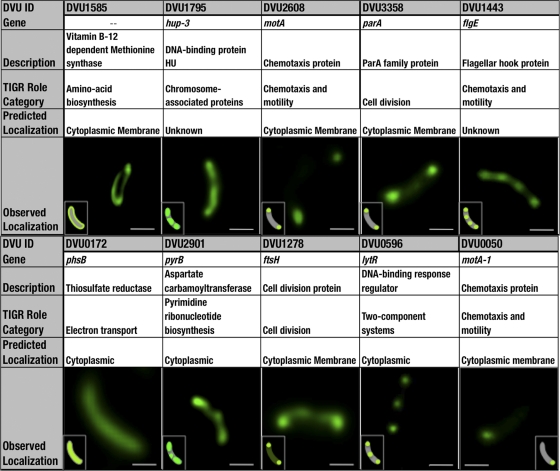
Subsequently, we expanded the method to 15 additional proteins (Fig. 4). We were able to decipher localization patterns for each of the 15 SNAP-tagged proteins, presumably reflecting their respective biological roles in this SRB. ParA, MotA-1, and MotA-3 localized exclusively to the poles, a subcellular area that has been referred to as a “localization hot spot,” whereas LytR, FtsH, FlgE, and UvrB localized at the poles as well as at additional regions in or toward the center of the cells. Hup-3 and PyrB showed a patchy or spotty distribution along the length of the cells. The remaining proteins displayed a cytoplasmically uniform distribution. Orthologous counterparts of ParA and FtsH from Caulobacter crescentus and E. coli were experimentally visualized previously (41, 45). For the remaining proteins, only theoretical in silico localization predictions have been made to date (48). In these localization studies, we consistently noted cell-to-cell variations in fluorescent signals in any given population, which may be attributed to corresponding differences in expression levels (38).
Fig. 4.
Predicted and observed localizations of AGT-tagged proteins in D. vulgaris. Each column (left to right) depicts a representative image of an observed localization pattern in 10 proteins from D. vulgaris Hildenborough bearing chromosomally inserted visualization tags (AGT) at their respective C termini. Fluorescently labeled cells were imaged by deconvolution microscopy, and images in the table represent an optical section through the middle of the three-dimensional (3D) deconvolved image stack (20 to 30 sections along the z axis). Predicted localizations were obtained from PSORTb (www.psort.org/psortb/). PhsB (DVU0172), a predicted cytoplasmic protein, is uniformly distributed intracellularly. Proteins localizing exclusively at both cell poles include MotA (DVU2608) and ParA (DVU 3358). The FlgE (DVU1443) and UvrB (DVU1605) proteins localize at four distinct locations along the length of the cell. The Hup-3 (DVU1795) and PyrB (DVU2901) proteins show a patchy or spotty distribution. FtsH (DVU1278) localizes to the polar ends, in addition to having a dispersed cytoplasmic distribution. LytR (DVU0596) displays bipolar and midband localizations. MotA-1 (DVU0050) has its localization signal restricted to one polar end of the cell. A schematic representation of the observed localization pattern is shown in the inset. Scale bars represent 400 nm for images of PhsB and MotA and 500 nm for the rest of the images.
Summary.
In this work, we successfully established the use of a “parts” approach to generate a library of over 700 engineered strains of the model sulfate reducer Desulfovibrio vulgaris Hildenborough for advanced systems biology applications. We highlighted three functional genomics tools, including (i) gene deletions to study methionine biosynthesis, (ii) protein-protein interactions associated with chaperones and nucleoid proteins, and (iii) subcellular localizations of select proteins to demonstrate the utility of our approach in this SRB, generally regarded as genetically intractable. One may extend the approach to realize applications such as synthetic genetic arrays (13), in vivo expression profiling (38), and others. The ubiquity of suicide constructs in gene replacement throughout biology suggests that our approach may be applied to engineer a broad range of species for a diverse array of systems biological applications and is amenable to high-throughput implementation.
Supplementary Material
ACKNOWLEDGMENTS
This work received support from ENIGMA under contract no. DE-AC02-05CH11231. This work conducted at the Joint BioEnergy Institute was supported by the Office of Science, Office of Biological and Environmental Research, U.S. Department of Energy, under contract no. DE-AC02-05CH11231.
We thank Steven Ruzin and Denise Schichnes of the Biological Imaging Facility at the University of California, Berkeley.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Afflerbach H., Schroder O., Wagner R. 1998. Effects of the Escherichia coli DNA-binding protein H-NS on rRNA synthesis in vivo. Mol. Microbiol. 28:641–653 [DOI] [PubMed] [Google Scholar]
- 2. Aki T., Adhya S. 1997. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 16:3666–3674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alm E. J., et al. 2005. The MicrobesOnline Web site for comparative genomics. Genome Res. 15:1015–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Auger S., Gomez M. P., Danchin A., Martin-Verstraete I. 2005. The PatB protein of Bacillus subtilis is a C-S-lyase. Biochimie 87:231–238 [DOI] [PubMed] [Google Scholar]
- 5. Baba T., et al. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Banerjee A., Biswas I. 2008. Markerless multiple-gene-deletion system for Streptococcus mutans. Appl. Environ. Microbiol. 74:2037–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bender K. S., et al. 2007. Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 73:5389–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blattner F. R., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453–1462 [DOI] [PubMed] [Google Scholar]
- 9. Bradford M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 10. Bradley M. D., Beach M. B., de Koning A. P., Pratt T. S., Osuna R. 2007. Effects of Fis on Escherichia coli gene expression during different growth stages. Microbiology 153:2922–2940 [DOI] [PubMed] [Google Scholar]
- 11. Bramucci M. G., Nagarajan V. 1996. Direct selection of cloned DNA in Bacillus subtilis based on sucrose-induced lethality. Appl. Environ. Microbiol. 62:3948–3953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brandis A., Thauer R. K. 1981. Growth of Desulfovibrio species on hydrogen and sulfate as sole energy-source. J. Gen. Microbiol. 126:249–252 [Google Scholar]
- 13. Butland G., et al. 2008. eSGA: E. coli synthetic genetic array analysis. Nat. Methods 5:789–795 [DOI] [PubMed] [Google Scholar]
- 14. Butland G., et al. 2005. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 433:531–537 [DOI] [PubMed] [Google Scholar]
- 15. Cameron D. E., Urbach J. M., Mekalanos J. J. 2008. A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 105:8736–8741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chhabra S. R., et al. 2006. Global analysis of heat shock response in Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 188:1817–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a. Chhabra S. R., et al. 2011. Towards a rigorous network of protein-protein interactions of the model sulfate reducer Desulfovibrio vulgaris Hildenborough. PLoS One 6:e21470 doi:10.1371/journal.pone.0021470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clark M. E., et al. 2006. Temporal transcriptomic analysis as Desulfovibrio vulgaris Hildenborough transitions into stationary phase during electron donor depletion. Appl. Environ. Microbiol. 72:5578–5588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. de Berardinis V., et al. 2008. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol. Syst. Biol. 4:174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Friedman D. I. 1988. Integration host factor: a protein for all reasons. Cell 55:545–554 [DOI] [PubMed] [Google Scholar]
- 21. Gallagher L. A., et al. 2007. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. U. S. A. 104:1009–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gophna U., Bapteste E., Doolittle W. F., Biran D., Ron E. Z. 2005. Evolutionary plasticity of methionine biosynthesis. Gene 355:48–57 [DOI] [PubMed] [Google Scholar]
- 23. Groh J. L., Luo Q., Ballard J. D., Krumholz L. R. 2005. A method adapting microarray technology for signature-tagged mutagenesis of Desulfovibrio desulfuricans G20 and Shewanella oneidensis MR-1 in anaerobic sediment survival experiments. Appl. Environ. Microbiol. 71:7064–7074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jacobs M. A., et al. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339–14344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Keller K. L., Bender K. S., Wall J. D. 2009. Development of a markerless genetic exchange system in Desulfovibrio vulgaris Hildenborough and its use in generating a strain with increased transformation efficiency. Appl. Environ. Microbiol. 75:7682–7691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kim J. M., Lee K. H., Lee S. Y. 2008. Development of a markerless gene knock-out system for Mannheimia succiniciproducens using a temperature-sensitive plasmid. FEMS Microbiol. Lett. 278:78–85 [DOI] [PubMed] [Google Scholar]
- 27. Kristich C. J., Manias D. A., Dunny G. M. 2005. Development of a method for markerless genetic exchange in Enterococcus faecalis and its use in construction of a srtA mutant. Appl. Environ. Microbiol. 71:5837–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li M. Z., Elledge S. J. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat. Methods 4:251–256 [DOI] [PubMed] [Google Scholar]
- 29. Liberati N. T., et al. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. U. S. A. 103:2833–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Molina-Henares M. A., et al. 2010. Identification of conditionally essential genes for growth of Pseudomonas putida KT2440 on minimal medium through the screening of a genome-wide mutant library. Environ. Microbiol. 12:1468–1485 [DOI] [PubMed] [Google Scholar]
- 31. Mukhopadhyay A., et al. 2006. Salt stress in Desulfovibrio vulgaris Hildenborough: an integrated genomics approach. J. Bacteriol. 188:4068–4078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nicolle O., et al. 2010. Development of SNAP-tag-mediated live cell labeling as an alternative to GFP in Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 59:357–363 [DOI] [PubMed] [Google Scholar]
- 33. Noble S. M., French S., Kohn L. A., Chen V., Johnson A. D. 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat. Genet. 42:590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ortiz-Martin I., Macho A. P., Lambersten L., Ramos C., Beuzon C. R. 2006. Suicide vectors for antibiotic marker exchange and rapid generation of multiple knockout mutants by allelic exchange in Gram-negative bacteria. J. Microbiol. Methods 67:395–407 [DOI] [PubMed] [Google Scholar]
- 35. Regoes A., Hehl A. B. 2005. SNAP-tag mediated live cell labeling as an alternative to GFP in anaerobic organisms. Biotechniques 39:809–810, 812 [DOI] [PubMed] [Google Scholar]
- 36. Rouviere-Yaniv J., Yaniv M., Germond J. E. 1979. E. coli DNA binding protein HU forms nucleosomelike structure with circular double-stranded DNA. Cell 17:265–274 [DOI] [PubMed] [Google Scholar]
- 37. Stolyar S., et al. 2007. Response of Desulfovibrio vulgaris to alkaline stress. J. Bacteriol. 189:8944–8952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taniguchi Y., et al. 2010. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329:533–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thomason L. C., Costantino N., Shaw D. V., Court D. L. 2007. Multicopy plasmid modification with phage lambda Red recombineering. Plasmid 58:148–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomason L. C., Oppenheim A. B., Court D. L. 2009. Modifying bacteriophage lambda with recombineering. Methods Mol. Biol. 501:239–251 [DOI] [PubMed] [Google Scholar]
- 41. Tomoyasu T., et al. 1993. Topology and subcellular localization of FtsH protein in Escherichia coli. J. Bacteriol. 175:1352–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Typas A., et al. 2008. High-throughput, quantitative analyses of genetic interactions in E. coli. Nat. Methods 5:781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vieira J., Messing J. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259–268 [DOI] [PubMed] [Google Scholar]
- 44. Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Werner J. N., et al. 2009. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc. Natl. Acad. Sci. U. S. A. 106:7858–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yu D., et al. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu D., Sawitzke J. A., Ellis H., Court D. L. 2003. Recombineering with overlapping single-stranded DNA oligonucleotides: testing a recombination intermediate. Proc. Natl. Acad. Sci. U. S. A. 100:7207–7212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu N. Y., et al. 2010. PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26:1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zane G. M., Yen H. C., Wall J. D. 2010. Effect of the deletion of qmoABC and the promoter-distal gene encoding a hypothetical protein on sulfate reduction in Desulfovibrio vulgaris Hildenborough. Appl. Environ. Microbiol. 76:5500–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zeghouf M., et al. 2004. Sequential peptide affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3:463–468 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.