Abstract
A decrease in ambient temperature alters membrane functionality and impairs the proper interaction between the cell and its external milieu. Understanding how cells adapt membrane properties and modulate the activity of membrane-associated proteins is therefore of major interest from both the basic and the applied points of view. Here, we have isolated multicopy suppressors of the cold sensitivity phenotype of a trp1 strain of Saccharomyces cerevisiae. Three poorly characterized genes, namely, ALY2 encoding the endocytic adaptor, CAJ1 encoding the J protein, and UBP13 encoding the ubiquitin C-terminal hydrolase, were identified as mediating increased growth at 12°C of both Trp− and Trp+ yeast strains. This effect was likely due to the downregulation of cold-instigated degradation of nutrient permeases, since it was missing from cells of the rsp5Δ mutant strain, which contains a point mutation in the gene encoding ubiquitin ligase. Indeed, we found that 12°C treatments reduced the level of several membrane transporters, including Tat1p and Tat2p, two yeast tryptophan transporters, and Gap1, the general amino acid permease. We also found that the lack of Rsp5p increased the steady state level of Tat1p and Tat2p and that ALY2-engineered cells grown at 12°C had higher Tat2p and Gap1p abundance. Nevertheless, the high copy number of ALY2 or UBP13 improved cold growth even in the absence of Tat2p. Consistent with this, ALY2- and UBP13-engineered cells of the industrial QA23 strain grew faster and produced more CO2 at 12°C than did the parental when maltose was used as the sole carbon source. Hence, the multicopy suppressors isolated in this work appear to contribute to the correct control of the cell surface protein repertoire and their engineering might have potential biotechnological applications.
INTRODUCTION
A major requirement for strain improvement involves stress tolerance and adaptability of cells to environmental stressors in industrial applications (22, 47). For example, some industrial processes involving industrial strains of Saccharomyces cerevisiae, like brewing and some wine fermentations, take place at temperatures around 10 to 12°C, which is far below the optimal temperature for this organism (∼28°C). Growth at low temperature reduces the production by S. cerevisiae of higher alcohols and increases the amount of esters (10, 20). The low fermentation temperature also has a prominent effect on primary flavors, which are retained to a greater degree. However, under these conditions, an extended lag phase before the onset of vigorous fermentation activity is observed, which reduces the cost-effectiveness and efficiency of production. In wine making, this lag phase also increases the risk of halted or sluggish fermentation (20). Hence, cold tolerance is an important biotechnological trait and there is an urgent need for strains able to ferment at low temperatures both quickly and in a reproducible way.
Like other stressors, cold influences the structural and functional properties of cellular components negatively, both physically and chemically (4). Cold modifies enzyme kinetics (28, 58) and increases the molecular order of membrane lipids, i.e., rigidification (33), affecting the membrane environment and thus the activity of membrane-associated enzymes and transporters. Key processes such as plasma membrane ATPase activity (53), the higher proton motive force (39), and the transport of various amino acids (60) depend thus on temperature-instigated changes in membrane fluidity and become limiting factors for cell growth.
In support of this view, previous reports have shown that tryptophan uptake is impaired after a downward shift in temperature (1) and that several cold-sensitive mutants are affected in tryptophan transport and biosynthesis (3, 12, 52). It has also been suggested that the sensitivity of tryptophan permeases to changes in membrane fluidity could determine or influence the growth temperature profile of tryptophan auxotroph strains of S. cerevisiae (1, 2). Bearing this is mind, we hypothesized that enhanced membrane fluidity might rescue the cold-mediated growth inhibition of Trp− yeast strains (48). Indeed, production in S. cerevisiae of sunflower desaturases (encoded by FAD2 genes) increased the content of dienoic fatty acids and fluidity of the yeast membrane; however, growth was diminished in the recombinant FAD2 strains at low temperatures (48). Thus, membrane fluidity appears to be essential in determining cold tolerance in S. cerevisiae, although its exact effect on amino acid uptake and growth is unclear.
The impact on yeast physiology of cold stress also depends on how low temperature affects the composition of membrane proteins. It has been reported that cold triggers the degradation of the tryptophan transporter Tat2p via the ubiquitination pathway (2, 42). Ubiquitination is a reversible posttranslational modification of cellular proteins, playing a central role in the regulation of protein degradation and trafficking (31, 41). In particular, Rsp5p-dependent ubiquitination of Tat2p has been observed in starved yeast cells (8). The yeast ubiquitin ligase Rsp5p (a homolog of mammalian Nedd4 family proteins) controls most trafficking-related ubiquitination events at the plasma membrane and at other membranes (9). In agreement with this, overproduction of Tat2p, or mutation of DOA4, UBP6, or UBP14, encoding ubiquitin-specific proteases, confers increased cold growth to Trp− yeast cells (1, 38). Altogether, these results suggest that Rsp5p may play a role in the degradation of Tat2p in response to low temperature, although no direct evidence of this function has been reported. Likewise, the putative adaptor proteins involved in this regulatory process are unknown, nor is it known whether cold may trigger the downregulation of other membrane proteins.
Here, we have performed a multicopy suppressor analysis of the cold sensitivity phenotype of a trp1 mutant strain of S. cerevisiae. Our hypothesis was that this screening could reveal, among others, genes governing plasma membrane properties. However, all the genes identified encode proteins directly or indirectly involved in ubiquitination machinery. We have exploited these effects to identify previously uncharacterized actors and genetic interactions that appear to be important in remodeling the membrane protein repertoire at low temperature. Finally, we demonstrate that engineering the ubiquitination machinery can potentially reverse growth inhibition associated with cold stress in industrial strains.
MATERIALS AND METHODS
Strains, culture media, and general methods.
The laboratory strains used in this study are described in Table 1. A haploid derivative (ho) of the diploid wine yeast strain QA23 (Lallemand, Montreal, Canada) was kindly provided by J. M. Guillamón. Auxotrophic mutants of the QA23 ho strain, MJHL201 (Ura−) and MJHL213 (Ura− Trp−) were constructed as described below. Yeast cells were cultured at 30 or 12°C in YPD media (1% yeast extract, 2% peptone, and 2% glucose) or SCD (0.67% yeast nitrogen base without amino acids [Difco] plus 2% glucose) supplemented with the appropriate amino acid dropout (ForMedium, England). Yeast transformants carrying the Geneticin (kanMX4)-, nourseothricin (natMX4)-, and hygromycin (hphMX4)-resistant module were selected on YPD agar plates containing 200 mg/liter of G-418 (Sigma), 50 mg/liter of nourseothricin (clonNAT; Werner Bioagents, Germany), and 300 mg/liter of hygromycin B (ForMedium), respectively (25, 61). Ura− derivatives were selected by platting an unmutagenized population of cells on SCD agar supplemented with uracil (10 mg/liter), proline (1 g/liter), and 5-fluoroorotic acid (5-FOA; 1 g/liter) (37). Auxotrophs for tryptophan were recovered as 5-fluoroanthranilic acid (FAA)-resistant mutants. FAA-containing SCD medium was prepared by the addition of 0.5 mg/ml FAA (10% [wt/vol] in ethanol) as previously described (59). The Escherichia coli DH5α host strain was grown in Luria-Bertani (LB) medium (1% peptone, 0.5% yeast extract, and 0.5% NaCl) supplemented with ampicillin (50 mg/liter). All amino acids, sugars, and antibiotics were filter sterilized and added to autoclaved medium. Solid media contained 2% agar. Yeast cells were transformed by the lithium acetate method (29). E. coli was transformed by electroporation following the manufacturer's instructions (Eppendorf). A stock solution of 25 mM phytosphingosine (PHS) was prepared in ethanol, sampled in small volumes, and stored at −20°C until use. For plate phenotype experiments cultures were diluted to an optical density at 600 nm (OD600) of 0.8 and 10-fold serial dilutions spotted (3 μl) onto SCD or YPD agar solid medium. In some experiments, maltose was used instead of glucose as the sole carbon source (SCM medium). Unless otherwise indicated, colony growth was inspected after 2 to 4 days of incubation at 30°C. Cold growth experiments were carried out at 12°C for 8 to 12 days.
Table 1.
S. cerevisiae strains used in this study
| Strain | Relevant genotype/phenotype | Reference or source |
|---|---|---|
| CEN.PK2-1C | MATaura3-52 his3-Δ1 leu2-3,112 trp1-289 MAL 2-8c SUC2 | 21 |
| CEN.PK2-1C aly2Δ | CEN.PK2-1C, aly2 Δ::kanMX4 | This study |
| BY4741 | MATahis3D1 leu2Δ0 met15Δ0 ura3Δ0 | EUROSCARF |
| BY4741 aly2Δ | BY4741, aly2Δ::kanMX4 | EUROSCARF |
| BY4741 caj1Δ | BY4741, caj1Δ::kanMX4 | EUROSCARF |
| BY4741 ubp13Δ | BY4741, ubp13Δ::kanMX4 | EUROSCARF |
| 27061b | MATaura3 trp1 | 23 |
| 27064b | 27061b, npi1 | 23 |
| 27064b aly2Δ | 27064b, aly2Δ::kanMX4 | This study |
| YPH499 | MATaura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 | 55 |
| DD12 | YPH499, cnb1::hisG | 16 |
| ASY472 | YPH499, crz1::loxP-kanMX-loxP | 57 |
| MCY300 | YPH499, cna1Δ1::hisG cna2Δ1::HIS3 | 17 |
| QA23 ho | ho | J. M. Guillamón |
| MJHL201 | QA23 ho, 5-FOAr Ura− | This study |
| MJHL213 | QA23 ho, 5-FOAr Ura− FAAr Trp− | This study |
For galactose induction of green fluorescent protein (GFP)-tagged genes, cells were pregrown overnight in SCD and then refreshed (OD600 of ∼0.25) in SC containing galactose as the sole carbon source (SCG). After 90 min, 2% glucose was added and the culture was transferred to a 12°C chilled water bath for the times indicated.
Screening of multicopy suppressors, DNA manipulations, and sequencing.
Cold multicopy suppressors were isolated from an S. cerevisiae genomic library constructed in the vector YEp24 (URA3/2 μm; kindly provided by B. A. Morgan). Transformants were selected on YPD medium containing 25 μM PHS at 12°C. Plasmid DNA was rescued, amplified in E. coli, and tested according to standard manipulations (50). The insert of each plasmid conferring cold tolerance was sequenced using YEp24 forward and reverse primers (see Table S1 in the supplemental material). DNA sequencing was performed by the dideoxy-chain termination procedure (51). Analysis of sequence data was carried out using DNAMAN sequence analysis software (Lynnon Biosoft). Similarity searches were performed at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) using BLAST software (5). A search of conserved domains was carried out by scanning the sequences against Conserved Domain Databases (CDD) at NCBI (35). Alternatively, protein domains were searched against the PROSITE database of protein families (54) at the Swiss Institute of Bioinformatics, SIB (http://www.isb-sib.ch). Sequence alignment was done using the ALIGN software at the GENESTREAM network server (http://xylian.igh.cnrs.fr/bin/align-guess.cgi).
Plasmid and disruption strain construction.
The YCplac33 (URA3; 24)-based plasmids p3HA-TAT1c and p3HA-TAT2c (24), encoding N-terminally hemagglutinin (HA)-tagged Tat1 and Tat2 proteins, respectively (2), were a gift from Fumiyoshi Abe. The plasmid YCpJ25, containing a GAP1 fusion with GFP under the control of pGAL1 (18), was provided by Sébastien Léon. PCR-amplified fragments containing the whole sequence of ALY2, EXO70, TRL1, YJL086C, CAJ1, ISD11, TPA1, AST1, PRS4, KTI11, and UBP13 genes, including its own promoter and terminator, were obtained with specific synthetic oligonucleotides (see Table S1 in the supplemental material) and genomic DNA as the template. Amplification was carried out under standard conditions. The corresponding fragments were digested with the appropriate set of enzymes, KpnI/PstI (ALY2), SalI/XbaI (EXO70, ISD11, PRS4), EcoRI/XbaI (TRL1, YJL86C, KTI11), EcoRI/PstI (CAJ1), SalI/PstI (TPA1), EcoRI/SacI (AST1), or XbaI/PstI (UBP13) and cloned into the plasmid YEplac195 (URA3; 24) digested with the same set of enzymes. The marker swap plasmid pUL9 (14) cut with SmaI was used in some cases in order to change the URA3 marker to LEU2. Disruption-deletion cassettes for ALY2, TAT1, and TAT2 were prepared by PCR using specific synthetic oligonucleotides (see Table S1) and plasmids pFa6A (kanMX4), pAG25 (natMx4), and pAG28 (hphMX4) as templates, respectively (25, 61). Correct gene disruption was detected by diagnostic PCR (27), using a set of oligonucleotides (see Table S1), designed to bind outside of the replaced gene sequence and within the marker module (data not shown).
Preparation of protein extracts and Western blot analysis.
For crude extracts, 10 OD600 units of cells were harvested by centrifugation at 1,090 × g and resuspended in 300 μl of lysis TNE buffer (50 mM Tris-HCl, 150 mM NaCl, 5 mM EDTA; pH 7.5) supplemented with a protease inhibitor cocktail (catalog number [cat#] 1861278; Roche). Then, glass beads (acid washed, 0.4 mm) were added, and the mixture was vortexed three times for 1 min, with 1 min on ice between each mixing. Cell debris and unbroken cells were removed by centrifugation at 500 × g for 5 min, and the supernatant was incubated with SDS loading buffer (cat# R0891; Fermentas) at 95°C for 10 min. Finally, 10 μl of each sample was separated by SDS-PAGE and blotted onto nitrocellulose membranes, and filters were blocked with 5% milk in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.1% Tween 20).
Gap1-GFP was detected by using a monoclonal anti-GFP antibody (clones 7.1 and 13.1; Roche Diagnostics). HA-Tat1p and HA-Tat2p were visualized with an anti-HA rabbit polyclonal antibody (cat# sc-805; Santa Cruz Biotechnology). A specific anti-Hxk2p serum raised in rabbits by immunization with a purified fraction of hexokinase PII (46) was used as the loading control of total protein. The antisera were applied at 1:3,000 dilutions. As the secondary antibody, we used horseradish peroxidase-conjugated goat anti-rabbit (1:2,000, cat# 7074; Cell Signaling, Danvers, MA) or rabbit anti-mouse (1:5,000, cat# P0260; Dako). Blots were developed using the ECL Western blotting detection kit by Pierce (Rockford, IL) or the ECL Advance system by GE Healthcare (Waukesha, WI). Images were captured with the Las-1000 Plus imaging system (Fuji, Kyoto, Japan).
Fermentative performance.
The fermentation capacity of the yeast strains under study was evaluated by measuring CO2 production. Yeast biomass from QA23 transformants was prepared by cultivating cells o/n at 30°C in liquid SCD-Ura. Then, cells were collected by centrifugation, washed with water, and finally resuspended (OD600 = 1.0) in 12°C prechilled YP containing maltose as the sole carbon source (YPM). Eighty milliliters of the yeast mixture was poured into a screw-cap bottle and placed in a 12°C water bath. CO2 production was recorded for 42 h in a Fermograph II apparatus (Atto Co., Ltd., Tokyo, Japan). Values are expressed as milliliters of CO2 and represent the total volume of gas produced after 24 or 42 h of fermentation or are shown as the time course graph of total gas. At least three independent experiments were conducted for each yeast strain.
RESULTS AND DISCUSSION
Screening of multicopy suppressors.
The cold-sensitive Trp− S. cerevisiae strain CEN.PK2-1C was transformed with a high-copy-number yeast genomic DNA library. In order to prevent excessive basal growth, the screening was carried out at 12°C on complex YPD medium containing 25 μM phytosphingosine (PHS). The addition of PHS to yeast cells promotes cellular stress responses (19), among them the stimulation of ubiquitin-dependent turnover of amino acid transporters (11, 13, 56). Thus, exposure to this sphingoid long-chain base restricts further yeast cell growth at low temperature, an effect that can be counteracted by increased tryptophan availability in the culture medium or overexpression of the high-affinity tryptophan transporter Tat2p (56).
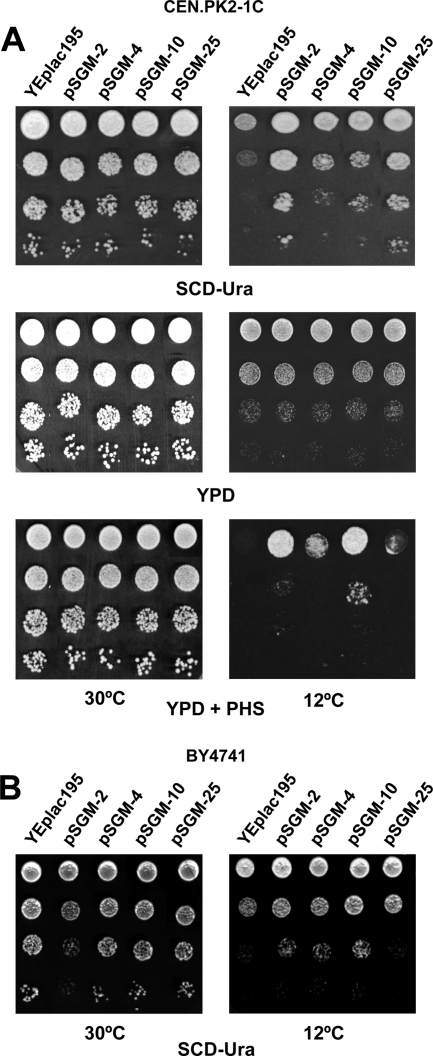
Twenty-eight transformants were initially selected, from which only four were reconfirmed as showing enhanced growth at 12°C. These were further analyzed by plasmid isolation and compared on the basis of their restriction map, which revealed four different patterns (data not shown). A representative of each group (referred to as pSGM-2/-4/-10/-25) was used to retransform the S. cerevisiae CEN.PK2-1C strain. As can be seen in Fig. 1A, all of the plasmids tested were able to confer improved cold growth on a synthetic SCD medium lacking PHS, whereas no effect was detected in the presence of PHS at 30°C. Thus, the screening appeared to select for low-temperature-sensitive suppressors. However, all transformants grew as well as the control strain on rich YPD medium at 12°C (Fig. 1A). Therefore, it seems that the 12°C growth advantage provided by the multicopy suppressors is effective only under conditions of reduced nutrient availability.
Fig. 1.
Cold sensitivity suppressors provide enhanced growth at 12°C under conditions of reduced nutrient availability. (A) Tryptophan auxotrophic (trp1) cells of the S. cerevisiae CEN.PK2-1C wild-type strain were transformed with plasmids pSGM-2, pSGM-4, pSGM-10, and pSGM-25, all of which were isolated from the screening of a high-copy-number yeast genomic DNA library for cold sensitivity suppressors, and transformants were assayed for growth at 30°C or 12°C. Cells were pregrown in SCD liquid medium at 30°C until the early exponential phase and adjusted to an OD600 of 0.8. Serial dilutions (1 to 10−3) of the adjusted cultures were spotted (3 μl) onto standard SCD-Ura, YPD, or YPD containing 25 μM phytosphingosine (PHS) solid medium. Cells transformed with plasmid YEplac195 (URA3, empty plasmid) were used as control. (B) Transformants of the Trp+ BY4741 wild-type strain were cultivated at 30°C or 12°C on SCD-Ura plates as indicated above. In all cases, a representative experiment is shown.
Subsequently, the effect of the aforementioned plasmids was analyzed in a different genetic background, specifically in the tryptophan prototroph yeast strain BY4741. TRP1 or tryptophan transporters like TAT2 (52) are most likely the genes responsible for the observed cold phenotype in the CEN.PK2-1C transformants (Fig. 1A). Accordingly, growth inhibition at low temperature should not be alleviated in Trp+ cells. Indeed, this was the case for cells containing plasmid pSGM-25 (Fig. 1B), which therefore was not studied further. However, the high copy number of pSGM-2, pSGM-4, and pSGM-10 still conferred some growth improvement in cold-exposed cells of the wild-type (wt) BY4741 strain (Fig. 1B).
Characterization of the isolated plasmids.
Sequencing of the insert contained in plasmids pSGM-2, pSGM-4, and pSGM-10 revealed the presence of DNA fragments of 9.5 kb, 8.7 kb, and 8.9 kb in length from chromosomes X, V, and II, respectively. In total, 11 complete and 3 partial open reading frames (ORFs) were identified. The former were PCR amplified from yeast genomic DNA, ligated into the YEplac195 multicopy expression vector, and retransformed into the S. cerevisiae CEN.PK2-1C wild-type strain. This led to the identification of ALY2 (ART3/YJL084C), CAJ1 (YER048C), and UBP13 (YBL0621) as responsible for the cold-tolerant phenotype found in pSGM-2, pSGM-4, and pSGM-10 transformants, respectively.
Endocytic adaptor Aly2p.
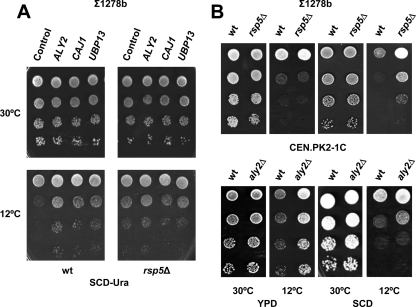
Aly2p/Art3p belongs to the ARTs (for arrestin-related trafficking adaptors), a protein family in yeast (32) that is composed of nine members (44). Like other ART proteins, Aly2p is predicted to contain N-terminal (pfam00339) and C-terminal (pfam02752) arrestin fold domains (structural elements also found in the mammalian α- and β-arrestin family; see reference 6) and two PY motifs, this being the conserved sequence recognized by the Rsp5p/Npi1p ubiquitin ligase (9). It has been proposed that members of the ART family function as adaptor proteins for Rsp5p, promoting endocytosis of plasma membrane-associated proteins (cargoes) in response to environmental cues or by targeting damaged or superfluous plasma membrane proteins for degradation (32). Recently, Aly2p has been shown to mediate endocytosis of the aspartic acid/glutamic acid transporter Dip5p by recruitment of Rsp5p (26). Consistent with all of this, the lack of Rsp5p (Fig. 2A and B) resulted in increased growth at 12°C, whereas Aly2p overexpression in this strain did not increase cold growth above the RSP5 deletion-mediated rate (Fig. 2A). Furthermore, the absence of a functional Aly2 protein had no major effects on cold growth (Fig. 2B). Similar results were found for aly2Δ mutants of either the CEN.PK2-1C (Fig. 2B) or the BY4741 (data not shown) background. Since ARTs have been proposed to confer specificity to endocytosis (44), the overproduction of Aly2p might improperly recruit Rsp5p to substrates that are not relevant for low temperature growth, decreasing endocytosis of essential permeases under this condition. In this respect, a recent study by O'Donnell et al. (43) has demonstrated that Aly2p stimulates the trafficking of Gap1p, the general amino acid permease, from endosomes to the trans-Golgi network, thus increasing the level of the transporter within cells and at the plasma membrane.
Fig. 2.
The effect of cold sensitivity suppressors Aly2p, Caj1p, and Ubp13p depends on the existence of a functional ubiquitin ligase, Rsp5p. (A) Cells of the rsp5Δ mutant strain and its corresponding parental (Σ1278b) were transformed with YEplac195 (URA3)-based plasmids containing the whole ORFs for ALY2, CAJ1, and UBP13 including its own promoter and terminator and tested for growth on SCD-Ura solid medium at 30 or 12°C. (B) Growth of rsp5Δ and aly2Δ mutant cells and their corresponding wild-type strains, Σ1278b and CEN.PK2-1C, respectively, was examined on rich YPD and minimal SCD medium. In all cases, cells were pregrown and spotted onto solid medium plates as described in Fig. 1. In all cases, a representative experiment is shown.
J protein Caj1.
CAJ1 encodes a member of the cytosolic class II J proteins (also known as Hsp40s), which are defined by the presence of an ∼65-amino-acid (aa) J domain (pfam00226), formerly identified in the bacterial heat shock gene DnaJ (45, 62). All of them are obligate cochaperones of 70-kDa heat shock proteins, Hsp70s, stimulating their ATPase activity and thus allowing them to function in multiple cellular processes (30, 49). Specifically, Caj1p was isolated from a yeast calmodulin-binding protein fraction (40). The Ca2+ regulatory protein calmodulin controls numerous targets (65), among them the serine/threonine protein phosphatase calcineurin (15), which has, in turn, been found to downregulate amino acid transporter endocytosis, triggered by several stress conditions (11). Accordingly, expression in a high copy number of CAJ1 might somehow increase calcineurin activity, thus reducing the cold-instigated turnover of transporters and membrane-anchored proteins. To test this hypothesis, CAJ1 transformants of cna1Δ cna2Δ, cnb1Δ, and crz1Δ mutant strains (16, 57) were examined for growth at 12°C. In its native form, calcineurin is present as a heterodimer containing a catalytic subunit encoded by the functionally redundant genes CNA1 and CNA2, complexed with a regulatory subunit, the gene product of CNB1. Active calcineurin dephosphorylates the transcriptional factor Crz1p (36, 57), which regulates the expression of most salt-responsive genes (66). However, the high copy number of CAJ1 increased cold growth independently of calcineurin-Crz1p pathway functioning (see Fig. S1 in the supplemental material). Furthermore, CAJ1 had no effect on yeast sensitivity to Mn2+ cations, a well-known stressor triggering this signaling route.
Then, the growth of YEpCAJ1 transformants of the rsp5Δ mutant strain was tested at 12°C. Overproduction of Caj1p has been reported to confer growth resistance to the drug FTY720, an immunosuppressive agent that inhibits amino acid transport in S. cerevisiae (63). Although the mechanism of Caj1p-mediated FTY720 toxicity suppression is unknown, it has been hypothesized that this J protein might influence amino acid uptake and/or protein degradation (63). In this respect, tryptophan transporters Tat1p and Tat2p, and ubiquitin proteases Ubp5p and Ubp11p, have also been isolated as multicopy suppressors of FTY720-induced growth inhibition (63). As can be seen in Fig. 2A, the effect of Caj1p at 12°C was completely masked by the absence of a functional Rsp5p ubiquitin ligase. Hence, our results suggest that Caj1p could play a specific role in modulating the activity of Hsp70s chaperones in the ubiquitin-mediated reprogramming of proteins at low temperature.
Ubiquitin C-terminal hydrolase UBP13.
The protease Ubp13p is a member of the family of deubiquitinating enzymes (Dubs), which comprises 17 potential genes in S. cerevisiae (7). Dubs are ubiquitin-specific proteases which release the peptide from ubiquitin-conjugated cargoes, thus reversing the ubiquitination process. As a consequence, Dubs are required for both ubiquitin homeostasis and proteasome-dependent proteolysis (31, 41). In particular, it has been shown that three Dubs, DOA4, UBP6, and UBP14, are involved in pressure-induced degradation of Tat2p since a lack of any of these proteins causes stabilization of the tryptophan transporter and allows yeast cells to grow at high pressure (38). In contrast, we found that overexpression of UBP13 was beneficial to cells at low temperature, whereas its absence had no effect on growth at 12°C (data not shown). Moreover, these effects were again dependent on the existence of a functional Rsp5 protein (Fig. 2A). In this context, it is possible that Ubp13p might have either specific or overlapping functions at low temperature, as previously suggested for other deubiquitinating enzymes (7).
Cold regulation of membrane transporters and functional role of Rsp5p.
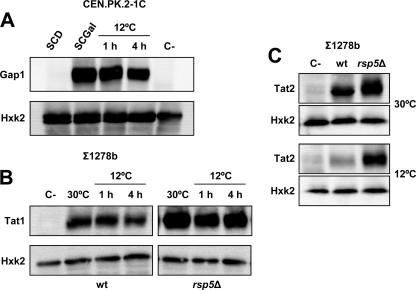
The above results would suggest that cold is perceived as a stress condition for membrane transporter degradation and that Rsp5p might be involved in this process. To test this, first the levels of general amino acid permease Gap1p were analyzed in cells transformed with plasmid YCpJ25, containing the GAL1 promoter-dependent GFP-tagged GAP1 gene (18). As can be seen, the amount of Gap1p decreased when cells of the wild-type strain CEN.PK2-1C were transferred from 30 to 12°C at 4 h (Fig. 3A).
Fig. 3.
The ubiquitin ligase Rsp5 influences the steady-state levels of different plasma membrane permeases, which are downregulated in response to low temperature. (A) CEN.PK2-1C cells transformed with plasmid YCpJ25 (18) were pregrown overnight in SCD and then refreshed (OD600 ∼0.25) in SCG (90 min), containing galactose as the sole carbon source. Then, 2% glucose was added and the culture was transferred to a 12°C chilled water bath for the times indicated. Preparation of protein extracts, SDS-PAGE separation, and visualization of Gap1-GFP and Hxk2p (loading control) were performed as described in Materials and Methods. A protein extract from untransformed cells was used as the negative control (C−). (B) Cells of the rsp5Δ mutant strain and its corresponding parental (Σ1278b) carrying a 3HA-TAT1c plasmid (2) were grown in SCD-Ura, refreshed in the same medium, and incubated at 30°C until the culture reached an OD600 of ∼0.5 or at 12°C for the periods indicated. Cells were processed and protein extracts analyzed as described above, except that Tat1p was detected using an anti-HA rabbit polyclonal antibody. (C) The same strains as in B were transformed with plasmid 3HA-TAT2c (2), and protein extracts were analyzed as above, except that the cold treatment was extended to a 24-h period. In all cases, a representative experiment is shown.
Then, the levels of Tat1p and Tat2p were examined in cells of the wild-type and rsp5Δ mutant cells transformed with plasmid p3HA-TAT1c or p3HA-TAT2c, each encoding an N-terminally hemagglutinin (HA)-tagged, fully functional Tat1 or Tat2 protein (1, 2). As expected, the level of both tryptophan permeases was higher in cells of the rsp5Δ mutant grown at 30°C than in the corresponding parental strain (Fig. 3B and C). Exposure to low temperature caused a reduction in Tat1p in both wild-type and mutant cells, suggesting that another ubiquitin ligase may control the regulation of Tat1p in response to low temperature, as reported for yeast cells subjected to high hydrostatic pressure (2). Nevertheless, cold-instigated changes in the level of Tat1p (Fig. 3B), such as those shown in Gap1p (Fig. 3A), were scarce after 4 h of exposure to low temperature. To examine whether a longer incubation period at 12°C can cause stronger effects, 3HA-Tat2p cells were analyzed after 24 h of cold treatment (OD600 of ∼0.4 to 0.5). As shown, long-term exposure to low temperature strongly reduced the level of Tat2p in wild-type cells (Fig. 3C). However, this change was less pronounced in rsp5Δ mutant cells, which still showed an intense Tat2p band after 24 h at 12°C (Fig. 3C). Hence, cold exposure of yeast cells triggers the downregulation of various membrane permeases, and Rsp5p plays a role in this mechanism.
Overexpression of ALY2 increases Tat2p abundance, which is essential in the cold growth of yeast cells.
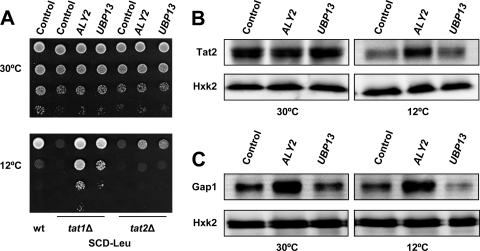
We tried to further clarify the involvement of tryptophan permeases in the cold sensitivity suppression mechanism. The study was restricted to the effects mediated by ALY2 and UBP13 because LEU2-based plasmids containing CAJ1 could not be obtained. First, we analyzed whether the improved cold growth was dependent on the presence of a functional Tat1p or Tat2p permease. CEN.PK2-1C mutant strains lacking TAT1 or TAT2 were constructed and transformed with plasmids for ALY2 or UBP13. As shown, deletion of either of these tryptophan permeases prevented cell proliferation at 12°C under conditions of limited tryptophan availability, SCD medium (Fig. 4A). A high expression of ALY2 or UBP13 conferred cold growth, but this effect was much more pronounced in a strain lacking TAT1 than in the tat2Δ mutant.
Fig. 4.
A high copy number of ALY2 reduces the cold-induced downregulation of the tryptophan transporter Tat2p, which is important for cold growth. (A) YEplac181 (control, LEU2), YEp181ALY2 (ALY2), and YEp181UBP13 (UBP13) transformants of tat1Δ and tat2Δ mutants of the S. cerevisiae CEN.PK2-1C background were examined for growth at 30 or 12°C. YEplac181 transformants (control) of the wild-type (wt) strain were also spotted as a reference. Cells were pregrown in liquid SCD-Leu, and then the cultures were diluted and spotted onto solid medium as described for Fig. 1. (B) Cell lysates were subjected to SDS-PAGE and immunoblotted with anti-HA or anti-HxK2 antibody. Analysis was performed of CEN.PK2-1C derivative strains cotransformed with plasmids 3HA-TAT2c (URA3) and YEplac181 (control, LEU2), YEp181ALY2 (ALY2), or YEp181UBP13 (UBP13). Cells were pregrown at 30°C and protein extracts were treated as described for Fig. 3C. (C) The levels of the general amino acid permease Gap1p were analyzed in cells cotransformed with plasmid YCpJ25, which contains the GAL1-promoter dependent GFP-tagged GAP1 gene (18), and YEplac181 (Control, LEU2), YEp181ALY2 (ALY2), or YEp181UBP13 (UBP13). Cells were grown as described for Fig. 3A, except that the cold treatment was extended to a 24-h period. Preparation of protein extracts, SDS-PAGE separation, and visualization of Gap1-GFP and Hxk2p (loading control) were performed as described in Materials and Methods. In all cases, a representative experiment is shown.
Then, we examined whether the level of Tat2p was affected by overexpression of ALY2 or UBP13. As a control, the levels of Gap1p were also checked. LEU2-based plasmids carrying the multicopy suppressors were used to transform p3HA-TAT2c and YCpJ25 derivatives of the CEN.PK2-1C strain. As can be seen in Fig. 4B, neither ALY2 nor UBP13 appeared to affect the level of Tat2p in cells grown at 30°C. Overexpression of UBP13 had limited effects, if any, in the abundance of Tat2p at 12°C. However, a high expression of ALY2 seemed to inhibit cold-stimulated Tat2p degradation, since Tat2p levels remained stable after 24 h at 12°C (Fig. 4B). ALY2 overexpression also increased Gap1p levels, although its effects were already evident at 30°C (Fig. 4B), whereas UBP13 caused no changes or even appeared to reduce the abundance of the permease in cells grown at 12°C (Fig. 4B). Overall, our results suggests that downregulation of Tat2p is the main cause for yeast growth inhibition at low temperature. Overexpression of ALY2 counteracts the cold-instigated downregulation of Tat2p and increases the relative content of Gap1p, although other plasma membrane transporters must also be considered to fully explain the ability of ALY2 and in particular UBP13 transformants to grow under low-temperature conditions.
Enhanced growth of industrial strains at low temperature.
We were interested in investigating whether increasing the gene dosage of cold sensitivity suppressors might be a useful approach to improve growth at low temperature of industrial strains. To address this, first Ura− and Ura− Trp− auxotrophic derivatives of the QA23 wine strain were selected. Single and double mutants were transformed with URA3-based plasmids YEpALY2, YEpCAJ1, and YEpUBP13 and tested for growth at low temperature. As expected, enhanced growth at 12°C was observed only for Trp− transformants and not for prototroph strains (data not shown). Thus, the ability of these three genes to increase yeast growth at 12°C appeared to be closely related to their capacity to interfere in the cold-instigated regulation of amino acid transporters. At this point, the question arose as to whether this property may be extended to other nutrient permeases and, in that case, under which conditions the overexpression of ALY2, CAJ1, or UBP13 could confer cold growth advantages in prototroph industrial strains.
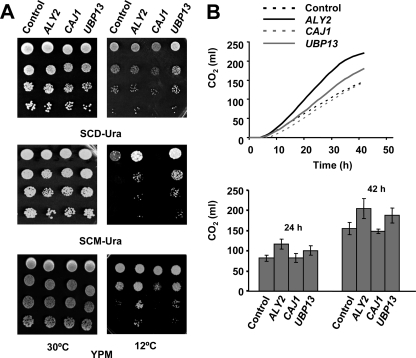
An important feature of the S. cerevisiae genome is the presence of many glucose transporters. Indeed, at least 17 genes, HXT1 to HXT17, need to be deleted to ensure that Saccharomyces is unable to take up and to grow on glucose as the sole carbon source (64). Conversely, maltose is internalized in most S. cerevisiae strains by single genes, MAL31/MAL61. Furthermore, yeast maltose transport is regulated by catabolite inactivation through Rsp5p- and Doa4p-mediated proteolysis (34). As shown in Fig. 5A, overexpression of ALY2 and UBP13 conferred enhanced growth at 12°C to prototroph cells of the wine strain QA23 cultivated on SC with maltose as the sole carbon source (SCM), whereas no differences could be found at 30°C. Similar results were observed in rich medium YP supplemented with maltose (YPM) (Fig. 5A). No effects were observed in YEpCAJ1 transformants, suggesting that this J protein alters targets other than maltose permease.
Fig. 5.
Engineering of ALY2 or UBP13 improves the cold growth and fermentative activity of the prototroph industrial QA23 strain on maltose. (A) Ura− derivatives of the wine strain QA23 ho were transformed with YEplac195-based plasmids (URA3) containing ALY2, CAJ1, or UBP13 and assayed for growth at 30 or 12°C on SC medium containing glucose (SCD-Ura) or maltose (SCM-Ura) as the sole carbon source, or on rich YP-maltose (YPM). Transformants carrying the empty plasmid were also tested (control). In all cases, cells were pregrown and treated as described in the Fig. 1. In all cases, a representative experiment is shown. (B) The fermentative performance of the mentioned strains was tested on liquid YPM at 12°C by measuring the CO2 production in a Fermograph apparatus. Cell biomass was prepared and CO2 was measured as described in Materials and Methods. Values are expressed as milliliters of CO2 and represent the temporal change graph of total gas (top) or the total volume of gas produced after 24 or 42 h of fermentation (bottom). At least three independent experiments were conducted for each yeast strain. The error (bottom) was calculated using the following formula: (1.96 × SD)/√n, where n is the number of measurements.
Thus, the gassing power of the recombinant strains was inspected. As can be seen in Fig. 5B (top), the rate of CO2 production by UBP13, and especially ALY2 transformants, was clearly higher than that recorded for YEpCAJ1 or control cells in liquid YPM at 12°C. Thus, CO2 production attained by overexpression of ALY2 after 42 h of fermentation at low temperature was about 30% higher than that observed with the control strain under these conditions (Fig. 5B, bottom).
Concluding remarks.
Our work has identified low-temperature sensitivity suppressors in S. cerevisiae for the first time. Since nutrient permeases are essential for cold growth, it was expected that this approach would identify, among others, genes involved in regulating the membrane protein repertoire and activity. Indeed, three poorly characterized genes (ALY2, CAJ1, and UBP13) were found to provide enhanced growth at 12°C. Of these, only ALY2 has been studied in greater depth recently (26, 43). According to these reports, the α-arrestin Aly2p mediates the endocytosis of Dip5 (26) and regulates the trafficking of Gap1p in response to nutrient signaling (43). Indeed, based on our analysis, not only Aly2p but also Caj1p and Ubp13p comprise or are involved in the activity of the ubiquitin machinery. This machinery controls membrane protein trafficking in yeast, with the ubiquitin ligase Rsp5p as a major player in this regulatory system. Our analysis demonstrated that tryptophan permeases, in particular Tat2p, but also other transporters like Gap1p, are downregulated at low temperature and that their steady-state level is controlled by Rsp5p. Furthermore, we also showed that Aly2p inhibits the cold-instigated downregulation of Tat2p and increases the abundance of Gap1p. Nevertheless, further studies are required to understand and clarify the exact suppressor mechanism for Aly2p, Caj1p, and Ubp13p. Since many other genes have been identified as being involved in the plasma membrane transporter regulation at different steps, it is to be expected that more cold suppressors may be identified. This would help to elucidate the exact machinery operating at low temperatures and identify new candidates providing increased growth under this restrictive condition. In light of our results, this indeed has potential biotechnological applications to improve the growth and activity of industrial strains under nutrient-limiting conditions.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Léon, F. Abe, B. André, M. Cyert, J. M. Guillamón, and A. Colosio for providing us with plasmids and strains and L. Perales for technical assistance.
This research was funded by CICYT projects (AGL2007-65498-C02-01) from the Ministry of Science and Innovation (MICINN, Spain). S.G.-M. was supported by a CSIC-JAE predoc fellowship, and M.J.H.-L. was the recipient of a contract within the AGL2007-65498-C02-01 project.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Abe F., Horikoshi K. 2000. Tryptophan permease gene TAT2 confers high-pressure growth in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:8093–8102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abe F., Iida H. 2003. Pressure-induced differential regulation of the two tryptophan permeases Tat1 and Tat2 by ubiquitin ligase Rsp5 and its binding proteins, Bul1 and Bul2. Mol. Cell. Biol. 23:7566–7584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Abe F., Minegishi H. 2008. Global screening of genes essential for growth in high-pressure and cold environments: searching for basic adaptive strategies using a yeast deletion library. Genetics 178:851–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aguilera J., Randez-Gil F., Prieto J. A. 2007. Cold response in Saccharomyces cerevisiae: new functions for old mechanisms. FEMS Microbiol. Rev. 31:327–341 [DOI] [PubMed] [Google Scholar]
- 5. Altschul S. F., et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alvarez C. E. 2008. On the origins of arrestin and rhodopsin. BMC Evol. Biol. 8:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Amerik A. Y., Li S. J., Hochstrasser M. 2000. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381:981–992 [DOI] [PubMed] [Google Scholar]
- 8. Beck T., Schmidt A., Hall M. N. 1999. Starvation induces vacuolar targeting and degradation of the tryptophan permease in yeast. J. Cell Biol. 146:1227–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belgareh-Touzé N., et al. 2008. Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking. Biochem. Soc. Trans. 36:791–796 [DOI] [PubMed] [Google Scholar]
- 10. Beltran G., Novo M., Guillamón J. M., Mas A., Rozès N. 2008. Effect of fermentation temperature and culture media on the yeast lipid composition and wine volatile compounds. Int. J. Food Microbiol. 121:169–177 [DOI] [PubMed] [Google Scholar]
- 11. Bultynck G., et al. 2006. Slm1 and Slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell. Biol. 26:4729–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen X., Sullivan D. S., Huffaker T. C. 1994. Two yeast genes with similarity to TCP-1 are required for microtubule and actin function in vivo. Proc. Natl. Acad. Sci. U. S. A. 91:9111–9115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chung N., Jenkins G., Hannun Y. A., Heitman J., Obeid L. M. 2000. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J. Biol. Chem. 275:17229–17232 [DOI] [PubMed] [Google Scholar]
- 14. Cross F. R. 1997. “Marker Swap” plasmids: convenient tools for budding yeast molecular genetics. Yeast 13:647–653 [DOI] [PubMed] [Google Scholar]
- 15. Cyert M. S. 2003. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311:1143–1150 [DOI] [PubMed] [Google Scholar]
- 16. Cyert M. S., Thorner J. 1992. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol. Cell. Biol. 12:3460–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cyert M. S., Kunisawa R., Kaim D., Thorner J. 1991. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc. Natl. Acad. Sci. U. S. A. 88:7376–7380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. De Craene J. O., Soetens O., Andre B. 2001. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J. Biol. Chem. 276:43939–43948 [DOI] [PubMed] [Google Scholar]
- 19. Dickson R. C., et al. 1997. Sphingolipids are potential heat stress signals in Saccharomyces. J. Biol. Chem. 272:30196–30200 [DOI] [PubMed] [Google Scholar]
- 20. Donalies U. E., Nguyen H. T., Stahl U., Nevoigt E. 2008. Improvement of Saccharomyces yeast strains used in brewing, wine making and baking. Adv. Biochem. Eng. Biotechnol. 111:67–98 [DOI] [PubMed] [Google Scholar]
- 21. Entian K. D., Kötter P. 1998. Yeast mutant and plasmid collections. Methods Microbiol. 26:431–449 [Google Scholar]
- 22. Fleet G. H. 2008. Wine yeasts for the future. FEMS Yeast Res. 8:979–995 [DOI] [PubMed] [Google Scholar]
- 23. Galan J. M., Moreau V., Andre B., Volland C., Haguenauer-Tsapis R. 1996. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J. Biol. Chem. 271:10946–10952 [DOI] [PubMed] [Google Scholar]
- 24. Gietz R. D., Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534 [DOI] [PubMed] [Google Scholar]
- 25. Goldstein A. L., McCusker J. H. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- 26. Hatakeyama R., Kamiya M., Takahar T., Maeda T. 2010. Endocytosis of the aspartic acid/glutamic acid transporter Dip5 is triggered by substrate-dependent recruitment of the Rsp5 ubiquitin ligase via the arrestin-like protein Aly2. Mol. Cell. Biol. 30:5598–5607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huxley C., Green E. D., Dunham I. 1990. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 6:236. [DOI] [PubMed] [Google Scholar]
- 28. Inouye M. 1999. Cold-shock response and adaptation. J. Mol. Microbiol. Biotechnol. 1:191. [PubMed] [Google Scholar]
- 29. Ito H., Jukuda K., Murata K., Kimura A. A. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kampinga H. H., Craig E. A. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell. Biol. 11:579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kimura Y., Tanaka K. 2010. Regulatory mechanisms involved in the control of ubiquitin homeostasis. J. Biochem. 147:793–798 [DOI] [PubMed] [Google Scholar]
- 32. Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D. 2008. Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135:714–725 [DOI] [PubMed] [Google Scholar]
- 33. Los D. A., Murata N. 2004. Membrane fluidity and its roles in the perception of environmental signals. Biochim. Biophys. Acta 1666:142–157 [DOI] [PubMed] [Google Scholar]
- 34. Lucero P., Lagunas R. 1997. Catabolite inactivation of the yeast maltose transporter requires ubiquitin-ligase npi1/rsp5 and ubiquitin-hydrolase npi2/doa4. FEMS Microbiol. Lett. 147:273–277 [DOI] [PubMed] [Google Scholar]
- 35. Marchler-Bauer A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Matheos D. P., Kingsbury T. J., Ahsan U. S., Cunningham K. W. 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes Dev. 11:3445–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McCusker J. H., Davis R. W. 1991. The use of proline as a nitrogen source causes hypersensitivity to, and allows more economical use of 5FOA in Saccharomyces cerevisiae. Yeast 7:607–608 [DOI] [PubMed] [Google Scholar]
- 38. Miura T., Abe F. 2004. Multiple ubiquitin-specific protease genes are involved in degradation of yeast tryptophan permease Tat2 at high pressure. FEMS Microbiol. Lett. 239:171–179 [DOI] [PubMed] [Google Scholar]
- 39. Morsomme P., Boutry M. 2000. The plant plasma membrane H+-ATPase: structure, function and regulation. Biochim. Biophys. Acta 1465:1–16 [DOI] [PubMed] [Google Scholar]
- 40. Mukai H., et al. 1994. Isolation and characterization of CAJ1, a novel yeast homolog of dnaJ. Gene 145:125–127 [DOI] [PubMed] [Google Scholar]
- 41. Mukhopadhyay D., Riezman H. 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315:201–205 [DOI] [PubMed] [Google Scholar]
- 42. Nagayama A., Kato C., Abe F. 2004. The N- and C-terminal mutations in tryptophan permease Tat2 confer cell growth in Saccharomyces cerevisiae under high-pressure and low-temperature conditions. Extremophiles 8:143–149 [DOI] [PubMed] [Google Scholar]
- 43. O'Donnell A. F., Apffel A., Gardner R. G., Cyert M. S. 2010. Alpha-arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol. Biol. Cell 21:3552–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Polo S., Di Fiore P. P. 2008. Finding the right partner: science or ART? Cell 135:590–592 [DOI] [PubMed] [Google Scholar]
- 45. Qiu X. B., Shao Y. M., Miao S., Wang L. 2006. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 63:2560–2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Randez-Gil F., Sanz P., Entian K. D., Prieto J. A. 1998. Carbon source-dependent phosphorylation of hexokinase PII and its role in the glucose-signaling response in yeast. Mol. Cell. Biol. 18:2940–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Randez-Gil F., et al. 2003. Baker's yeast: challenges and future prospects, p. 57–97 In de Winde J. H. (ed.), Functional genetics of industrial yeasts. Springer-Verlag, Heidelberg, Germany [Google Scholar]
- 48. Rodríguez-Vargas S., Sánchez-García A., Martínez-Rivas J. M., Prieto J. A., Randez-Gil F. 2007. Fluidization of membrane lipids enhances the tolerance of Saccharomyces cerevisiae to freezing and salt stress. Appl. Environ. Microbiol. 73:110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sahi C., Craig E. A. 2007. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. U. S. A. 104:7163–7168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 51. Sanger F., Nicklen S., Coulson A. R. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463–5467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schmidt A., Hall M. N., Koller A. 1994. Two FK506 resistance-conferring genes in Saccharomyces cerevisiae, TAT1 and TAT2, encode amino acid permeases mediating tyrosine and tryptophan uptake. Mol. Cell. Biol. 14:6597–6606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Serrano R., Montesinos C., Sanchez J. 1988. Lipid requirements of the plasma membrane ATPases from oat roots and yeast. Plant Sci. 56:117–122 [Google Scholar]
- 54. Sigrist C. J., et al. 2002. PROSITE: a documented database using patterns and profiles as motif descriptors. Brief. Bioinform. 3:265–274 [DOI] [PubMed] [Google Scholar]
- 55. Sikorski R. S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Skrzypek M. S., Nagiec M. M., Lester R. L., Dickson R. C. 1998. Inhibition of amino acid transport by sphingoid long chain bases in Saccharomyces cerevisiae. J. Biol. Chem. 273:2829–2834 [DOI] [PubMed] [Google Scholar]
- 57. Stathopoulos A. M., Cyert M. S. 1997. Calcineurin acts through the CRZ1/TCN1-encoded transcription factor to regulate gene expression in yeast. Genes Dev. 11:3432–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thieringer H. A., Jones P. G., Inouye M. 1998. Cold shock and adaptation. BioEssays 20:49–57 [DOI] [PubMed] [Google Scholar]
- 59. Toyn J. H., Gunyuzlu P. L., White W. H., Thompson L. A., Hollis G. F. 2000. A counterselection for the tryptophan pathway in yeast: 5-fluoroanthranilic acid resistance. Yeast 16:553–560 [DOI] [PubMed] [Google Scholar]
- 60. Trivedi A., Singhal G. S., Prasad R. 1983. Effect of phosphatidylserine enrichment on amino acid transport in yeast. Biochim. Biophys. Acta 729:85–89 [DOI] [PubMed] [Google Scholar]
- 61. Wach A., Brachat A., Pöhlmann R., Philippsen P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793–1808 [DOI] [PubMed] [Google Scholar]
- 62. Walsh P., Bursać D., Law Y. C., Cyr D., Lithgow T. 2004. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 5:567–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Welsch C. A., Hagiwara S., Goetschy J. F., Movva N. R. 2003. Ubiquitin pathway proteins influence the mechanism of action of the novel immunosuppressive drug FTY720 in Saccharomyces cerevisiae. J. Biol. Chem. 278:26976–26982 [DOI] [PubMed] [Google Scholar]
- 64. Wieczorke R., et al. 1999. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464:123–128 [DOI] [PubMed] [Google Scholar]
- 65. Yamniuk A. P., Vogel H. J. 2004. Calmodulin's flexibility allows for promiscuity in its interactions with target proteins and peptides. Mol. Biotechnol. 27:33–57 [DOI] [PubMed] [Google Scholar]
- 66. Yoshimoto H., et al. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277:31079–31088 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.