Abstract
Phages play a key role in the marine environment by regulating the transfer of energy between trophic levels and influencing global carbon and nutrient cycles. The diversity of marine phage communities remains difficult to characterize because of the lack of a signature gene common to all phages. Recent studies have demonstrated the presence of host-derived auxiliary metabolic genes in phage genomes, such as those belonging to the Pho regulon, which regulates phosphate uptake and metabolism under low-phosphate conditions. Among the completely sequenced phage genomes in GenBank, this study identified Pho regulon genes in nearly 40% of the marine phage genomes, while only 4% of nonmarine phage genomes contained these genes. While several Pho regulon genes were identified, phoH was the most prevalent, appearing in 42 out of 602 completely sequenced phage genomes. Phylogenetic analysis demonstrated that phage phoH sequences formed a cluster distinct from those of their bacterial hosts. PCR primers designed to amplify a region of the phoH gene were used to determine the diversity of phage phoH sequences throughout a depth profile in the Sargasso Sea and at six locations worldwide. phoH was present at all sites examined, and a high diversity of phoH sequences was recovered. Most phoH sequences belonged to clusters without any cultured representatives. Each depth and geographic location had a distinct phoH composition, although most phoH clusters were recovered from multiple sites. Overall, phoH is an effective signature gene for examining phage diversity in the marine environment.
INTRODUCTION
Marine viruses merit study not only because of their sheer abundance but also because of the critical roles they play in the Earth's biogeochemical cycles (11). The majority of these viruses are phages (viruses that infect bacteria). Because phages are host-specific predators that influence the composition of the bacterial community (9, 47), it is essential to understand the diversity of marine phages. Microscopy-based methods have only limited resolution for analyzing marine phage diversity, and therefore genetic methods are preferable. However, identification of phages in environmental samples is hampered by the lack of a single gene found in all phages (50). Nonetheless, some genes are shared within groups of phages, and these “signature genes” can be used as markers to examine the diversity of a phage group of interest (70). Several signature genes have been developed to examine the diversity of phages in the marine environment, including structural genes (61, 64, 86), replication genes (10, 33), and auxiliary metabolic genes (14, 54, 60, 68, 80).
Auxiliary metabolic genes (AMGs) are phage-borne metabolic genes that were typically thought to be restricted to cellular genomes yet have been identified in phage genomes through sequencing (11). Numerous AMGs involved in photosynthesis, carbon metabolism, and nucleotide metabolism have been identified in marine phages (14, 35, 36, 42, 43, 65, 68, 78, 80). In addition, marine phages carry AMGs involved in nutrient limitation (51, 65, 67, 78), such as those belonging to the Pho regulon, which regulates phosphate uptake and metabolism under low-phosphate conditions (24, 77). Here we examined the presence of genes belonging to the Pho regulon in completely sequenced phage genomes and demonstrated the utility of phoH as a new signature gene for the study of marine phage diversity. Newly described PCR primers were used to amplify phoH from viral samples collected throughout the world's oceans. A high diversity of phoH genes was found in marine viral communities, with the types of phoH identified varying with depth and location.
MATERIALS AND METHODS
Prevalence of Pho regulon genes in phages.
To determine the presence of Pho regulon genes in completely sequenced phage genomes, a pool of bacterial Pho regulon genes was collected from three bacterial strains. First, the nucleotide sequences of the 35 genes of the Pho regulon (amn, eda, phnCDEFGHIJKLMNOP, phoABEHRU, psiEF, pstABCS, ugpABCEQ, yibD, and ytfK) (24) from Escherichia coli strain K-12 (substrain MG1655; accession number U00096) were retrieved from GenBank. Next, potential Pho regulon genes from Prochlorococcus marinus strain NATL1A (accession number NC_008819) were collected by using the 35 E. coli Pho regulon genes as the query in a TBLASTX (3) search against the genome of NATL1A. Twelve of the 35 queries produced hits with E values of <0.001. Those hits in the genome of NATL1A (genes annotated as eda, phoB, phoH, phoR, pstA, pstB, pstC, salX, and potA and genes with the locus tags NATL1_02681, NATL1_11521, and NATL1_07881) were added to the Pho regulon genes from E. coli. One additional NATL1A gene, locus tag NATL1_20941, was included because although it was the second-best hit (when E. coli's phnL was used as the query), it is annotated as a phosphate transporter in the NATL1A genome. Finally, genes from P. marinus strain NATL2A (accession number CP000095) that were predicted to be part of that cyanobacterium's Pho regulon (63) were included in the pool. This step added 20 genes, with the locus tags PMN2A_0440, PMN2A_0439, PMN2A_0438, PMN2A_0249, PMN2A_0435, PMN2A_0436, PMN2A_0437, PMN2A_0549, PMN2A_0496, PMN2A_0959, PMN2A_0559, PMN2A_0742, PMN2A_1499, PMN2A_1369, PMN2A_0714, PMN2A_0311, PMN2A_0310, PMN2A_0309, PMN2A_0308, and PMN2A_0307. This combined pool of bacterial Pho regulon genes from E. coli and P. marinus contained 68 sequences. To identify Pho regulon genes in phage genomes, each sequence was compared by BLASTX (3) against the GenBank nonredundant (nr) database (using default parameters), limiting the subject organisms to viruses (taxonomy identification no. [taxid] 10239). All significant hits (E value < 0.001) were confirmed through reciprocal BLASTP analysis against the GenBank nr database.
Collection and processing of depth profile samples.
To examine the difference in phoH composition of the phage community present at different depths, small-scale samples were collected from throughout a depth profile (0, 200, 500, and 1,000 m) at the Bermuda Atlantic Time-series Study site (31°40′N, 64°10′W) in September 2008. Whole seawater samples (100 ml) were filtered through a 0.22-μm Sterivex filter (Millipore, Billerica, MA) and then onto a 0.02-μm Anotop filter (Whatman, Piscataway, NJ). Anotop filters were stored at −80°C until DNA was extracted with a MasterPure complete DNA and RNA purification kit (Epicentre Biotechnologies, Madison, WI) following the protocol of Culley and Steward (17). Briefly, filters were defrosted, and all liquid was purged from the filter by pushing air through with a sterile syringe. A flame-sealed pipette tip was used to temporarily seal the filter outlet, and a mixture of 400 μl of 2× T&C lysis buffer (from the MasterPure kit) and 50 μg proteinase K was forced onto the filter. The filter was then incubated for 10 min in the air at 65°C before the lysate was expelled into a microcentrifuge tube and immediately placed on ice. Then 150 μl of MPC protein precipitation reagent (from the MasterPure kit) was added to the lysate and vortexed vigorously for 10 s. The debris was pelleted by centrifugation at 10,000 × g for 10 min. Isopropanol was added to the recovered supernatant, and the tube was inverted 30 to 40 times. The DNA was then pelleted by centrifugation at 20,000 × g at 4°C for 10 min and washed twice with 75% ethanol. Extracted DNA was resuspended in sterile water and stored at −20°C.
Collection and processing of geographic samples.
To examine the biogeography of phage phoH sequences, samples were collected from the Sargasso Sea, British Columbia coastal waters, the Gulf of Mexico, Raunefjorden, Kongsfjorden, and the Mediterranean Sea. Large-scale samples (approximately 250 liters) from 0 m and 100 m from the Sargasso Sea (31°40′N, 64°10′W) were concentrated by tangential flow filtration with 100-kDa filters (GE Healthcare, Piscataway, NJ) to a volume of approximately 50 ml. These viral concentrates were filtered through 0.22-μm Sterivex filters to remove bacteria and stored at 4°C until further processing. Viruses were further concentrated and purified from the Sargasso Sea concentrates by polyethylene glycol precipitation followed by cesium chloride density-dependent centrifugation. Solid polyethylene glycol 8000 (PEG 8000) was added to the concentrates at a final concentration of 10% (wt/vol), and the concentrates were stored at 4°C overnight. The concentrates were then centrifuged for 40 min at 11,000 × g and 4°C to pellet the viruses. The pelleted viruses were resuspended in 0.02-μm-filtered seawater and further purified through ultracentrifugation in a cesium chloride density gradient with layers of 1.2 g/ml, 1.5 g/ml, and 1.7 g/ml (22,000 rpm on a Beckman SW40 Ti rotor for 3 h at 4°C). The viral fractions were further concentrated with a Microcon centrifugal filter device (Millipore), and viral DNA was extracted using the formamide method as described by Sambrook et al. (53). The Raunefjorden (60°16.2′N, 5°12.5′E) and Kongsfjorden (79°00′N, 11°40′E) samples were prefiltered through 0.45-μm-pore-size low-protein-binding Durapore membrane filters 142 mm in diameter (Millipore) in order to remove cellular organisms. The filtrate was then concentrated to approximately 45 ml using a QuixStand benchtop system with 100-kDa hollow fiber cartridges (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). The samples from the Gulf of Mexico (pool of 41 samples collected between 1994 and 2001 from the surface to 164 m) and British Columbia coastal waters (pool of 85 samples collected between 1996 and 2004 from the surface to 245 m) were collected as described by Angly et al. (4) and processed as outlined by Suttle et al. (71). Briefly, the samples were prefiltered through 142-mm-diameter glass fiber filters with a 1.2-μm pore size (Advantec MFS, Dublin, CA) or a 0.7-μm pore size (Whatman, Clifton, NJ), followed by filtration through 0.45-μm or 0.2-μm-pore-size Durapore membrane filters (Millipore, Bedford, MA). Concentration of virus-sized particles from the filtrate was completed with 10-kDa or 30-kDa spiral-wound cartridges (Amicon/Millipore, Billerica, MA). Concentrates were stored in the dark at 4°C until further processing. Mediterranean samples (43°41′N, 7°19′E) were collected and concentrated as described by Bonilla-Findji et al. (8). The samples were prefiltered through 0.8-μm polycarbonate filters (142-mm diameter) (Osmonics, Inc., Minnetonka, MN), followed by tangential flow filtration through 0.2-μm Durapore polycarbonate filters (Millipore) and concentration on 100-kDa spiral polyethersulfone cartridges (Millipore). For all locations except the Sargasso Sea, viral DNA was obtained by incubating 500 μl of viral concentrate at 90°C twice for 2 min, placing the concentrate on ice between incubations. Then, 20 μl of 0.5 M EDTA (pH 8.0) and 5 μl of freshly made proteinase K (10 mg/ml) were added, and the mixture was incubated for 10 min at 55°C. After the addition of 25 μl of 10% sodium dodecyl sulfate, the mixture was further incubated for 1 h at 55°C. The DNA was cleaned with a DNA Clean and Concentrator kit (Zymo Research Corp., Irvine, CA) following the manufacturer's instructions and resuspended in 20 μl of sterile water.
Primer design and DNA amplification.
phoH primers were designed based on a CLUSTALX (73) alignment of the full-length phoH gene from Synechococcus phage S-PM2, Prochlorococcus phages P-SSM2 and P-SSM4, and Vibrio phage KVP40. PCR primers vPhoHf (5′-TGCRGGWACAGGTAARACAT-3′) and vPhoHr (5′-TCRCCRCAGAAAAYMATTTT-3′) were used to amplify a product of approximately 420 bp. The 50-μl reaction mixture for PCR amplification of the phoH gene contained 1 U Apex Taq DNA polymerase (Genesee Scientific, San Diego, CA), 1× Apex Taq reaction buffer, 1.5 mM Apex MgCl2, a 0.5 μM concentration of each primer, 0.2 mM deoxynucleoside triphosphates, and 0.04% bovine serum albumin. The reaction conditions were (i) 5 min of initial denaturation at 95°C; (ii) 35 cycles of 1 min of denaturation (95°C), 1 min of annealing (53°C), and 1 min of extension (72°C); and (iii) 10 min of final extension at 72°C. Before amplification of the phoH gene, DNA from the Sargasso Sea samples was amplified by the strand displacement method of the Illustra GenomiPhi V2 DNA amplification kit (GE Healthcare, Piscataway, NJ) according to the manufacturer's instructions.
Cloning and sequencing.
phoH PCR products were cloned into vectors and used to transform competent cells. After screening, the inserts in positive transformants were sequenced. PCR products from the Sargasso Sea were cloned using the TOPO TA cloning kit for sequencing (Invitrogen, Carlsbad, CA) and were sequenced by Beckman Coulter Genomics (Danvers, MA). PCR products from the remaining samples were cloned with the StrataClone PCR cloning kit (Stratagene, La Jolla, CA) and sequenced by LGC Genomics (Berlin, Germany). PCR products from the cyanophage isolates were directly sequenced (without cloning) by the University of Florida (Gainesville, FL).
Phylogenetic analysis.
Vector and low-quality sequences were trimmed with Sequencher 4.7 (Gene Codes, Ann Arbor, MI). The Sargasso Sea samples were dereplicated using FastGroup II at a level of 99% sequence identity with gaps (84). Reference sequences from cultured phages were obtained from GenBank and through amplification of phoH from cyanophages isolated from the Gulf of Mexico on Synechococcus WH7803 (41). All sequences were aligned at the amino acid level using CLUSTALW (using default parameters) as implemented in TranslatorX (1). The amino acid alignment (see Fig. 1) or back-translated nucleotide alignments (see Fig. 2 and 4) were then used to build maximum-likelihood phylogenetic trees with PhyML 3.0 (21). Protein-coding sequences such as phoH are more conserved at the amino acid level than they are at the nucleotide level (1), and thus alignments are more accurate when conducted at the amino acid level. The back-translated nucleotide sequences obtained from the amino acid alignments were used to build the trees in order to better reflect the diversity of the phoH sequences in the environment. Nonparametric branch supports were determined by an approximate likelihood ratio test (5). Nodes with branch support values of ≤50 were collapsed using Mesquite (version 2.74) (37). Phylogenetic trees were edited with MEGA 5 (31).
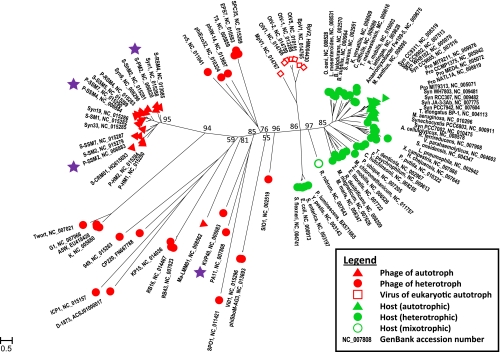
Fig. 1.
Phylogenetic tree (from an amino acid alignment) showing the relationship among the phoH genes of completely sequenced bacteria, phages, and eukaryotic viruses. The scale bar shows substitutions per site. phoH primers were designed from sequences marked with a star.
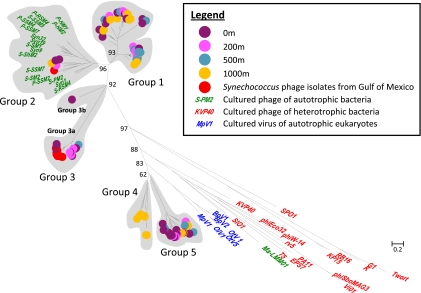
Fig. 2.
Phylogenetic tree (from a nucleotide alignment) showing the relationship among phoH sequences from environmental virus samples throughout a depth profile in the Sargasso Sea and phoH sequences from cultured phages and viruses. Group classifications for environmental sequences are indicated. The scale bar shows substitutions per site.
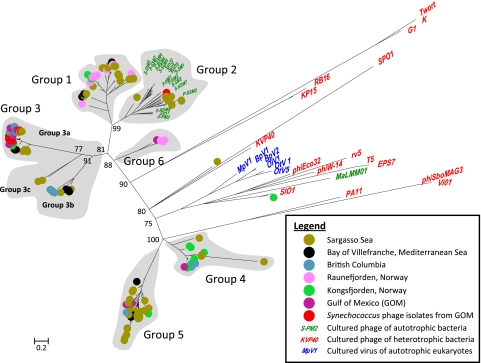
Fig. 4.
Phylogenetic tree (from a nucleotide alignment) showing the biogeography of phoH sequences from environmental virus samples from six locations. phoH sequences from cultured phages and viruses are also shown. Group classifications for environmental sequences are indicated; groups 1 through 5 are the same as groups 1 through 5 in Fig. 2. The scale bar shows substitutions per site.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to GenBank and assigned accession numbers JF963974 through JF964262.
RESULTS AND DISCUSSION
Pho regulon genes in phages.
The Pho regulon contains a group of genes whose products control the uptake and metabolism of phosphate by the cell in response to phosphate limitation (24, 77). Phosphorus is essential for cell survival due to its presence in membrane lipids and nucleic acids, as well as its roles in posttranslational protein modification and energy transfer (6, 79). In E. coli, expression of the Pho regulon is activated when phosphate is limited (77). There is direct evidence that at least 31 genes are part of the Pho regulon, and indirect evidence of several more (24).
Genes involved in phosphate limitation (i.e., phoH, pstS, and phoA) have been previously identified in the genomes of marine phages (15, 39, 43, 44, 51, 65–67, 69, 78), as well as in marine metagenomes (52, 58, 60, 80). To determine the prevalence of these and other Pho regulon genes in phage genomes, BLAST similarity searches (3) were performed using Pho regulon genes from the genomes of E. coli strain K-12 substrain MG1655, P. marinus strain NATL1A, and P. marinus strain NATL2A against the virus subset of the nr database. Of the 35 Pho regulon genes examined, only five (phoH, pstS, phoA, phoE, and ugpQ) were found in phage genomes (Table 1). phoH was the gene most commonly found in phages, occurring in 42 of the 602 completely sequenced phage genomes in the GenBank database (as of 26 May 2011). A phosphate transporter subunit gene, pstS, occurred in nine phages whose genomes are completely sequenced (66). These relative frequencies support prior analyses of the Global Ocean Sampling (GOS) metagenome showing that scaffolds containing phoH genes included a much higher percentage of viral open readings frames than scaffolds containing pstS genes (80). phoA, a gene of the Pho regulon that encodes bacterial alkaline phosphatase (24, 77), was found in two fully sequenced phages, located next to pstS in the genomes (S-SM1 and S-SM2 [66]). The metagenomic GOS data revealed that uncultured cyanophages contained phoA as well (26). A phage that contained neither phoH nor pstS nonetheless possessed a different Pho regulon gene; the enterobacterial phage P7 contained nmpC, a gene encoding an outer membrane porin precursor homologous to porins of the phoE family (77). In addition, three Staphylococcus phages (G1, K, and A5W) contained ugpQ, which encodes a glycerophosphoryl diester phosphodiesterase (74). Interestingly, the genomes of marine phages appeared to be enriched in Pho regulon genes compared to the genomes of phages from other environments. Forty-four percent of the phage genomes containing Pho regulon genes were isolated from the marine environment (19 out of 43), while marine phages comprised only a small proportion (8%) of the 602 completely sequenced phage genomes in GenBank. Among the completely sequenced phage genomes in GenBank, nearly 40% of the marine phages contained Pho regulon genes, while only 4% of nonmarine phage genomes contained these genes. Thus, the data from this study show that it is not equally likely for sequenced marine and nonmarine phages to contain Pho regulon genes, although this result could be biased by the representation of phage genomes in GenBank. These data support previous assertions that it may be advantageous for marine phages to encode genes involved in phosphate regulation because phosphate is often a limiting nutrient in the oceans (26, 36, 51, 65, 66).
Table 1.
Genes of the Pho regulon found in the genomes of fully sequenced phages
| Phage | Presence of: |
Host | Host trophic status | Marine origin | ||||
|---|---|---|---|---|---|---|---|---|
| phoH | pstS | phoA | phoE (nmpC) | ugpQ | ||||
| SPO1 | X | Bacillus subtilis | Heterotroph | |||||
| CP220 | X | Campylobacter | Heterotroph | |||||
| D-1873 | X | Clostridium botulinum | Heterotroph | |||||
| phiW-14 | X | Delftia acidovorans | Heterotroph | |||||
| P7 | X | Enterobacteria | Heterotroph | |||||
| RB43 | X | Enterobacteria | Heterotroph | |||||
| phiEco32 | X | Escherichia coli | Heterotroph | |||||
| RB16 | X | Escherichia coli | Heterotroph | |||||
| rv5 | X | Escherichia coli | Heterotroph | |||||
| T5 | X | Escherichia coli | Heterotroph | |||||
| KP15 | X | Klebsiella pneumoniae | Heterotroph | |||||
| 949 | X | Lactococcus lactis | Heterotroph | |||||
| Ma-LMM01 | X | Microcystis aeruginosa | Autotroph | |||||
| P-HM1 | X | Prochlorococcus | Autotroph | X | ||||
| P-HM2 | X | Prochlorococcus | Autotroph | X | ||||
| P-RSM4 | X | X | Prochlorococcus | Autotroph | X | |||
| P-SSM2 | X | X | Prochlorococcus | Autotroph | X | |||
| P-SSM4 | X | X | Prochlorococcus | Autotroph | X | |||
| P-SSM7 | X | X | Prochlorococcus | Autotroph | X | |||
| PA11 | X | Pseudomonas aeruginosa | Heterotroph | |||||
| SIO1 | X | Roseobacter SIO67 | Heterotroph | X | ||||
| SPC35 | X | Salmonella enterica and Escherichia coli | Heterotroph | |||||
| EPS7 | X | Salmonella enterica serovar Typhimurium | Heterotroph | |||||
| Vi01 | X | Salmonella enterica serovar Typhi Vi | Heterotroph | |||||
| phiSboM-AG3 | X | Shigella boydii | Heterotroph | |||||
| A5W | X | X | Staphylococcus aureus | Heterotroph | ||||
| G1 | X | X | Staphylococcus aureus | Heterotroph | ||||
| K | X | X | Staphylococcus aureus | Heterotroph | ||||
| Twort | X | Staphylococcus aureus | Heterotroph | |||||
| S-CRM01 | X | Synechococcus | Autotroph | |||||
| S-PM2 | X | Synechococcus | Autotroph | X | ||||
| S-RSM4 | X | Synechococcus | Autotroph | X | ||||
| S-SM1 | X | X | X | Synechococcus | Autotroph | X | ||
| S-SM2 | X | X | X | Synechococcus | Autotroph | X | ||
| S-SSM5 | X | X | Synechococcus | Autotroph | X | |||
| S-SSM7 | X | X | Synechococcus | Autotroph | X | |||
| Syn1 | X | Synechococcus | Autotroph | X | ||||
| S-ShM2 | X | Synechococcus and Prochlorococcus | Autotroph | X | ||||
| Syn19 | X | X | Synechococcus and Prochlorococcus | Autotroph | X | |||
| Syn33 | X | Synechococcus and Prochlorococcus | Autotroph | X | ||||
| Syn9 | X | Synechococcus and Prochlorococcus | Autotroph | X | ||||
| ICP1 | X | Vibrio cholerae | Heterotroph | |||||
| KVP40 | X | Vibrio parahaemolyticus | Heterotroph | X | ||||
Given that phoH is much more abundant in phage genomes than any of the other Pho regulon genes, it is possible that PhoH in phages serves a role unrelated to phosphate uptake. The study that first identified and characterized the phoH gene noted that PhoH could bind ATP, and that it was probably a cytoplasmic protein involved in the uptake of phosphate under conditions of phosphate starvation (29, 38). However, despite the well-studied nature of the Pho regulon and the presence of phoH genes in a wide array of phage genomes, the function of PhoH remains unknown. The gene product is likely an ATPase, given its conserved nucleoside triphosphate hydrolase domain (24, 30, 38, 66). If PhoH hydrolyzes ATP, the resulting reaction would release energy to drive another reaction, presumably to assist in the uptake of phosphate by the cell. Kazakov et al. (27) examined homologs of the phoH genes from E. coli and Bacillus subtilis; they found that the homologs clustered into three groups. The positions of the homologs and their presence in two different clusters of orthologous groups suggested several different potential functions for PhoH, including fatty acid beta oxidation, phospholipid metabolism, and metal-dependent RNA modification (27).
Not only does the role of PhoH remain unclear, but the expression of phoH under conditions of phosphate stress has also been shown to vary among species. In E. coli, phoH is upregulated when the cell is subjected to phosphate stress (66, 77). Similarly, levels of phoH mRNA transcripts in Corynebacterium glutamicum are 4.6 times higher when phosphate is limited than when the cell has sufficient phosphate (25). In contrast, phoH is downregulated in Synechococcus sp. WH8102 under conditions of phosphate limitation (72). In two strains of Prochlorococcus (MED4 and MIT9313), there is no change in the expression of phoH when phosphate is limited (40). The only study to examine the link between phage infection and phoH expression demonstrated an increase in the phoH transcript level in Prochlorococcus MED4 upon infection with phage P-SSP7 (34). At 4 h postinfection, phoH is upregulated by a factor of 1.8. It has been hypothesized that the increased expression of the gene could represent a response by the host to the stress of phage infection (34). Upregulation of phosphate-uptake genes, whether carried by the host or by the phage, may work to the advantage of the phage, since phosphorus is a key limiting nutrient in the marine environment. Thus, the existence of Pho regulon genes in phage genomes may constitute a selective advantage to the phage, enabling phosphate uptake during infection and allowing further phage replication despite phosphate limitation (34, 36, 55, 65, 79).
phoH as a signature gene for phage identification.
Several signature genes are currently being used to study phage diversity, but each of these marker genes has limitations. For example, primers available for amplifying the DNA polymerase gene of T7-like podophages are restricted to only a subset of that phage group (10, 33). Structural genes such as g20, which encodes a portal protein (14, 64, 86), and g23, which encodes a major capsid protein (20), are also commonly used as genetic markers in phages. However, the available primers for these genes are restricted to myophages, with the g20 primers specifically targeting cyanomyophages (20, 47, 86). Although primers for genes homologous to psbA and psbD (encoding photosystem II reaction center proteins D1 and D2) have proven useful for phage identification (14, 35, 36, 68), the ability of the psb primers to characterize phage diversity is limited to cyanophages.
The presence of phoH in phages that infect both heterotrophic and autotrophic hosts suggests that it could potentially capture a broad range of phages and therefore be used to analyze phage diversity. phoH genes have been found in many phages infecting autotrophic bacteria (Table 1), such as the cyanophages P-SSM2 and P-SSM4, which infect Prochlorococcus (65), cyanophage Syn9, which infects Synechococcus (78), and cyanophage Ma-LMM01, which infects Microcystis aeruginosa (82). In addition, phoH genes have been detected in a range of phages infecting heterotrophic bacteria, such as roseophage SIO1, a phage of Roseobacter (51), PA11, a phage of Pseudomonas aeruginosa (32), and KVP40, a broad-host-range vibriophage (44). Another advantage of phoH as a signature gene for examining phage diversity is that this gene is not restricted to one morphological type of phage. The phoH gene has been found in the genomes of podophages, such as the enterobacterial phage phiEco32 (57), in siphophages, such as enterobacterial phage EPS7 (23) and enterobacterial phage T5 (76), and also in myophages, such as Bacillus phage SPO1 (62). Among heterotrophic marine phages, phoH has been detected in both podophages (such as Roseobacter phage SIO1 [51]) and myophages (such as vibriophage KVP40 [44]); however, among sequenced cyanophages, phoH has so far been identified only in myophages (65–67). Finally, phoH genes are not restricted to phages and have also been detected in viruses that infect autotrophic eukaryotes. For example, several viruses of unicellular photosynthetic marine green algae of the Ostreococcus genus, as well as viruses infecting Micromonas and Bathycoccus, have been shown to contain phoH (18, 45, 79).
phoH has been found in phages and viruses isolated from a wide variety of geographic areas, including the coast of Japan (KVP40 [44]), coastal lagoons in the northwestern Mediterranean Sea (OlV1 and MpV1 [45]), the coast of southern California (SIO1 [51]), the Red Sea (S-RSM4 [43]), the Sargasso Sea (P-SSM4 [69]), the English Channel (S-PM2 [39, 43, 81]), the Pacific Ocean near Hawaii (P-HM1 and P-HM2 [66]), and the coast of Massachusetts by Woods Hole (Syn9 [43, 78]). While most of the cultured phages containing the phoH gene originated from marine waters, some were isolated from other habitats. For example, SPO1 was isolated from soil in Japan (62); EPS7 was isolated from Korean sewage samples (23); phiEco32 was found in a river in Tbilisi, Georgia (57); Ma-LMM01 was isolated from a lake in Japan (83); Vi01 came from human stool samples from Canadian patients with typhoid fever (49); and phage KP15 was obtained from sewage samples from Warsaw, Poland (48).
Comparison of phage and host phoH.
Comparison of other AMGs in phages and the hosts they infect has demonstrated that many of these genes are evolving differently from their host counterparts. Phylogenetic analysis reveals that phage and host versions of the photosynthesis gene psbA tend to cluster separately, though not completely (14, 54). However, the phage genes group next to the genes from their hosts: psbA from phages that infect Synechococcus form a sister clade to Synechococcus psbA genes, and psbA genes from phages that infect Prochlorococcus form a sister clade to Prochlorococcus psbA genes (22, 36, 68, 80, 85). A similar pattern exists for psbD genes, which are involved in photosynthesis (36, 54, 68, 80), and PTOX genes (encoding plastoquinol terminal oxidase) (43). Recent research reveals that phage-borne PSI genes are also evolving separately from the host versions of those genes (2, 59). Finally, analysis of GOS data shows that NAD(P)H dehydrogenase genes in phages mainly cluster separately from bacterial versions (59), and mazG genes from cyanophages cluster separately from host Prochlorococcus and Synechococcus versions of the gene (12). These phylogenetic patterns suggest that after the host genes have been incorporated into phage genomes, the selective pressure on those genes changes in such a way that it becomes possible to distinguish between host and phage versions.
Phylogenetic analysis of the phoH gene from the genomes of fully sequenced phages and bacteria revealed that phages clustered separately from hosts (Fig. 1), thereby demonstrating that phoH can be used as a signature gene to discriminate between host and phages when phage diversity is being investigated. Within that primary division, there was further resolution by trophic strategy. Cyanobacteria formed their own well-supported clade, while the heterotrophic bacteria formed several separate phoH clusters. Similarly, phages clustered according to the nutrition mode of their hosts; there was a well-supported clade of cyanophages, while the heterotrophic phages fell into other groups. phoH of phages infecting heterotrophs displayed a greater diversity than phoH of those infecting autotrophs. Viruses of eukaryotes also formed their own well-supported cluster.
Phage phoH diversity throughout the water column in the Sargasso Sea.
The diversity of phoH throughout a depth profile at the Bermuda Atlantic Time-series Study site in the Sargasso Sea was examined to determine whether distinct phage types were present throughout the water column. A significant diversity of phoH sequences was identified along the depth profile containing samples from 0, 200, 500, and 1,000 m. The depth profile phoH sequences formed five distinct clusters, identified as groups 1 through 5, with the majority of the Sargasso Sea sequences belonging to clusters without any cultured isolates (Fig. 2). This is similar to the situation observed for several other signature genes in the marine environment (10, 20, 33) and demonstrates that environmental phages are not well represented by the phage isolates currently available in culture. Since many of the environmental phoH groups do not contain cultured isolates, it is possible that some of the sequences obtained in this study are not viral in origin. However, several steps were taken during sample processing to ensure removal of host DNA, including filtration of all samples and density-dependent centrifugation of some samples. Phylogenetic trees containing environmental phoH sequences alongside cultured phages, viruses, and hosts revealed that none of the environmental sequences clustered with those of hosts (data not shown), which is not surprising given that the primers were designed specifically based on phage sequences. Nonetheless, although it is extremely likely that the environmental phoH sequences are viral in origin, the possibility that the samples contain host-derived DNA, such as that contained within gene transfer agents or transducing particles, cannot be excluded.
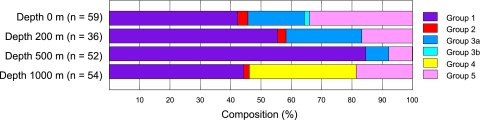
The phoH composition of the phage community varied throughout the water column (Fig. 3). Changes in the composition of the phage community are likely driven by differences in the composition of the host community, which has been studied in great detail at this site (13, 19, 46, 75). All depths were dominated by sequences belonging to group 1, which did not contain any cultured representatives. Group 2, which contained most of the phoH sequences of cultured cyanophages that have been fully sequenced, comprised only a minor component of the sequences recovered at any depth. Each depth contained sequences from multiple groups, and the proportion of sequences represented by each group varied among depths (Fig. 3). For instance, although group 1 sequences were found at all four depths, over 80% of the 500-m sequences belonged to group 1, while just over 40% of the 0-m and 1,000-m sequences belonged to group 1. Group 3 sequences were more abundant in the photic zone, decreasing with depth and not detected in the 1,000-m sample. In contrast, group 4 comprised 35% of the phoH sequences recovered from 1,000 m and was not detected at any of the other depths. It is not surprising that the 1,000-m sequences were distinct from those of the other depths, because the 1,000-m phage community would not be expected to contain the cyanophages that populate the photic zone.
Fig. 3.
Composition of phoH sequences found at each depth in the Sargasso Sea, based on the groups defined in the phylogenetic tree in Fig. 2.
Biogeography of phage phoH sequences.
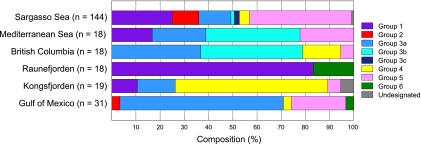
In addition to the depth profile of the phoH gene, the biogeography of phoH was studied in viral concentrates from six locations around the world (the Gulf of Mexico, the Arctic Ocean, British Columbia coastal waters, the Mediterranean Sea, the Sargasso Sea, and a site near the coast of Norway). Along with the five groups identified in the depth profile phylogenetic tree, the global study also revealed a sixth cluster, group 6, which did not appear in the sequences from the Sargasso Sea (Fig. 4). phoH composition differed for the phage community from each location, with no single group found at all sites (Fig. 5). Different phoH groups dominated at different locations. For example, group 1 represented over 80% of the sequences from Raunefjorden but only approximately 10% of the sequences from Kongsfjorden. Group 3 represented less than 20% of the sequences from Kongsfjorden but nearly 80% of the British Columbia sequences and was not present at all in the Raunefjorden sequences. Group 5 was also absent from the Raunefjorden profile and varied from approximately 5% in Kongsfjorden to over 40% in the Sargasso Sea. While the Raunefjorden, Kongsfjorden, and Mediterranean samples were all drawn from the surface, samples from the other three locations were pooled from the surface down to 100 m (Sargasso Sea), 164 m (Gulf of Mexico), and 245 m (British Columbia). Given that the depth profile drawn from the Sargasso Sea (Fig. 3) showed that each depth exhibited a distinct phoH composition, further work is required in order to better resolve biogeographical differences in phoH sequences.
Fig. 5.
Composition of phoH sequences detected at each location from the biogeographical survey, based on the groups defined in the phylogenetic tree in Fig. 4.
In light of these different profiles, it is apparent that the phoH gene can distinguish phage communities from different locations and serve as a useful biogeographical marker. However, this analysis also points out gaps in our knowledge. It is somewhat surprising that group 2, which contained almost all of the completely sequenced cyanophage isolates in GenBank, was found at only two of the studied locations: the Sargasso Sea and the Gulf of Mexico. In contrast, the cultured cyanophages from the Gulf of Mexico whose phoH genes were sequenced in this study belonged almost entirely to group 3a, which was represented in every studied location except one. Consistent with group 3a originating from cyanophages, in the Sargasso Sea depth profile, group 3a was most abundant in the photic zone and was not present at 1,000 m. This supports the idea that the sequenced cyanophages currently in the database do not adequately represent total cyanophage diversity and that more cyanophages need to be cultured and sequenced. In addition, although a great deal of phoH diversity was elucidated using these primers, it is notable that only two of the 248 environmental phoH sequences in the global study appeared in the group with the cultured heterotrophic phages. This suggests that the cultured heterotrophic phages are not well represented in the marine environment, or that the phoH primers used in this study do not amplify the phoH gene of many of the cultured heterotrophic phages. Designing additional phoH primers to capture more of the cultured heterotrophic phages, as well as the viruses infecting photosynthetic eukaryotes, will enable a broader analysis of phoH diversity in the marine viral community. Since many of the major phoH groups identified in the environmental samples did not contain cultured representatives, it is unknown whether these groups consisted of cyanophages or heterotrophic phages. As additional phage-host systems are isolated, insight into the identity of the phages in the phoH environmental clusters will be gained.
Despite the different phoH compositions identified at the disparate locations, most of the phoH groups were found at multiple sites. This supports previous research suggesting that phages are not limited by geography. Sano et al. (56) examined phages from four different environments (soil, marine sediment, freshwater, and seawater) and discovered that soil, freshwater, and sediment phages can propagate on hosts from the marine environment. That study also showed that marine phages from one location can infect hosts from a different marine location (56). Signature gene studies using both DNA polymerase and structural genes have detected identical phage sequences from widely separated geographical locations, as well as from different habitats (10, 33, 61). Studies of phages infecting Vibrio species also demonstrated that genetically related vibriophages can be found throughout the water column, as well as in marine locations separated by up to 4,500 miles (16, 28). Metagenomic sequencing of viral communities from throughout the world's oceans confirmed that the majority of the viral genotypes are shared between locations, with differences between sites being driven by changes in the relative abundance of specific viruses (4). A more recent analysis of the GOS expedition supported these results, finding differential distribution of myophages, podophages, and siphophages by location while further establishing that many AMGs occur in phages worldwide (80). These studies, in combination with the phoH data presented here, support the idea that “everything is everywhere, but the environment selects” (7) and suggest that the selection may be driven not only by the composition of the host community but also by auxiliary metabolic genes present in the phage genomes.
ACKNOWLEDGMENTS
Thanks go to Hilde Kristiansen, Rachel Parsons, Craig Carlson, and many past and present members of the Suttle lab for collecting and processing the virus communities, as well as the crews of the R/V Atlantic Explorer, R/V Longhorn and CCGS Vector for logistical support.
This research was funded by grants from the National Science Foundation to MB (Microbial Interactions and Processes MCB-0701984 and Division of Biological Infrastructure DBI-0850206). The University of Bergen received financial support from the Norwegian Research Council for research program “Viral lysis and programmed cell death in marine phytoplankton” (VIPMAP, 186142/V40) and the project ERC Advanced Grant “MIcrobial Network OrganiSation” (MINOS). Collection of the virus communities from British Columbia and the Gulf of Mexico was supported by grants to C.A.S. from the Natural Science and Engineering Council of Canada, the National Science Foundation and the Office of Naval Research. D.B.G. was supported by a Presidential Doctoral Fellowship from the University of South Florida and the Von Rosenstiel Endowed Fellowship. A.V. was supported by an Erskine Fellowship (University of Canterbury, New Zealand).
Footnotes
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Abascal F., Zardoya R., Telford M. J. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–W13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alperovitch-Lavy A., et al. 2011. Reconstructing a puzzle: existence of cyanophages containing both photosystem-I and photosystem-II gene suites inferred from oceanic metagenomic datasets. Environ. Microbiol. 13:24–32 [DOI] [PubMed] [Google Scholar]
- 3. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 4. Angly F. E., et al. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4:2121–2131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anisimova M., Gascuel O. 2006. Approximate likelihood-ratio test for branches: a fast, accurate, and powerful alternative. Syst. Biol. 55:539–552 [DOI] [PubMed] [Google Scholar]
- 6. Baek J. H., Lee S. Y. 2006. Novel gene members in the Pho regulon of Escherichia coli. FEMS Microbiol. Lett. 264:104–109 [DOI] [PubMed] [Google Scholar]
- 7. Becking L. G. M. B. 1934. Geobiologie of inleiding tot de milieukunde, vol. 18 WP Van Stockum & Zoon NV, The Hague, The Netherlands [Google Scholar]
- 8. Bonilla-Findji O., et al. 2008. Viral effects on bacterial respiration, production and growth efficiency: consistent trends in the Southern Ocean and the Mediterranean Sea. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 55:790–800 [Google Scholar]
- 9. Bratbak G., Thingstad F., Heldal M. 1994. Viruses and the microbial loop. Microb. Ecol. 28:209–221 [DOI] [PubMed] [Google Scholar]
- 10. Breitbart M., Miyake J. H., Rohwer F. 2004. Global distribution of nearly identical phage-encoded DNA sequences. FEMS Microbiol. Lett. 236:249–256 [DOI] [PubMed] [Google Scholar]
- 11. Breitbart M., Thompson L. R., Suttle C. A., Sullivan M. B. 2007. Exploring the vast diversity of marine viruses. Oceanography (Wash. D C) 20:135–139 [Google Scholar]
- 12. Bryan M. J., et al. 2008. Evidence for the intense exchange of MazG in marine cyanophages by horizontal gene transfer. PLoS One 3:17–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carlson C. A., et al. 2009. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 3:283–295 [DOI] [PubMed] [Google Scholar]
- 14. Chénard C., Suttle C. A. 2008. Phylogenetic diversity of sequences of cyanophage photosynthetic gene psbA in marine and freshwaters. Appl. Environ. Microbiol. 74:5317–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coleman M. L., et al. 2006. Genomic islands and the ecology and evolution of Prochlorococcus. Science 311:1768–1770 [DOI] [PubMed] [Google Scholar]
- 16. Comeau A. M., Chan A. M., Suttle C. A. 2006. Genetic richness of vibriophages isolated in a coastal environment. Environ. Microbiol. 8:1164–1176 [DOI] [PubMed] [Google Scholar]
- 17. Culley A. I., Steward G. F. 2007. New genera of RNA viruses in subtropical seawater, inferred from polymerase gene sequences. Appl. Environ. Microbiol. 73:5937–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Derelle E., et al. 2008. Life-cycle and genome of OtV5, a large DNA virus of the pelagic marine unicellular green alga Ostreococcus tauri. PLoS One 3:e2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. DuRand M. D., Olson R. J., Chisholm S. W. 2001. Phytoplankton population dynamics at the Bermuda Atlantic Time-series station in the Sargasso Sea. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 48:1983–2003 [Google Scholar]
- 20. Filée J., Tétart F., Suttle C. A., Krisch H. M. 2005. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc. Natl. Acad. Sci. U. S. A. 102:12471–12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 22. Hambly E., Suttle C. A. 2005. The viriosphere, diversity, and genetic exchange within phage communities. Curr. Opin. Microbiol. 8:444–450 [DOI] [PubMed] [Google Scholar]
- 23. Hong J., et al. 2008. Identification of host receptor and receptor binding module of a newly sequenced T5-like phage EPS7. FEMS Microbiol. Lett. 289:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hsieh Y.-J., Wanner B. L. 2010. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 13:198–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishige T., Krause M., Bott M., Wendisch V. F., Sahm H. 2003. The phosphate starvation stimulon of Corynebacterium glutamicum determined by DNA microarray analyses. J. Bacteriol. 185:4519–4529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kathuria S., Martiny A. C. 2011. Prevalence of a calcium-based alkaline phosphatase associated with the marine cyanobacterium Prochlorococcus and other ocean bacteria. Environ. Microbiol. 13:74–83 [DOI] [PubMed] [Google Scholar]
- 27. Kazakov A. E., Vassieva O., Gelfand M. S., Osterman A., Overbeek R. 2003. Bioinformatics classification and functional analysis of PhoH homologs. In Silico Biol. 3:3–15 [PubMed] [Google Scholar]
- 28. Kellogg C. A., Rose J. B., Jiang S. C., Thurmond J. M., Paul J. H. 1995. Genetic diversity of related vibriophages isolated from marine environments around Florida and Hawaii, U. S. A. Mar. Ecol. Prog. Ser. 120:89–98 [Google Scholar]
- 29. Kim S.-K., Makino K., Amemura M., Shinagawa H., Nakata A. 1993. Molecular analysis of the phoH gene, belonging to the phosphate regulon in Escherichia coli. J. Bacteriol. 175:1316–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Koonin E. V., Rudd K. E. 1996. Two domains of superfamily I helicases may exist as separate proteins. Protein Sci. 5:178–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar S., Nei M., Dudley J., Tamura K. 2008. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief. Bioinform. 9:299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kwan T., Liu J., DuBow M., Gros P., Pelletier J. 2006. Comparative genomic analysis of 18 Pseudomonas aeruginosa bacteriophages. J. Bacteriol. 188:1184–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Labonté J. M., Reid K. E., Suttle C. A. 2009. Phylogenetic analysis indicates evolutionary diversity and environmental segregation of marine podovirus DNA polymerase gene sequences. Appl. Environ. Microbiol. 75:3634–3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindell D., et al. 2007. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature 449:83–86 [DOI] [PubMed] [Google Scholar]
- 35. Lindell D., Jaffe J. D., Johnson Z. I., Church G. M., Chisholm S. W. 2005. Photosynthesis genes in marine viruses yield proteins during host infection. Nature 438:86–89 [DOI] [PubMed] [Google Scholar]
- 36. Lindell D., et al. 2004. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. U. S. A. 101:11013–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Maddison W. P., Maddison D. R. 2010. Mesquite: a modular system for evolutionary analysis. http://mesquiteproject.org
- 38. Makino K., Amemura M., Kim S.-K., Yokoyama K., Kimura S. 1998. Mechanism of transcriptional activation of the phosphate regulon in Escherichia coli. J. Microbiol. 36:231–238 [Google Scholar]
- 39. Mann N. H., et al. 2005. The genome of S-PM2, a “photosynthetic” T4-type bacteriophage that infects marine Synechococcus strains. J. Bacteriol. 187:3188–3200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martiny A. C., Coleman M. L., Chisholm S. W. 2006. Phosphate acquisition genes in Prochlorococcus ecotypes: evidence for genome-wide adaptation. Proc. Natl. Acad. Sci. U. S. A. 103:12552–12557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. McDaniel L. D., Delarosa M., Paul J. H. 2006. Temperate and lytic cyanophages from the Gulf of Mexico. J. Mar. Biol. Assoc. 86:517–527 [Google Scholar]
- 42. Millard A. D., Clokie M. R. J., Shub D. A., Mann N. H. 2004. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc. Natl. Acad. Sci. U. S. A. 101:11007–11012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Millard A. D., Zwirglmaier K., Downey M. J., Mann N. H., Scanlan D. J. 2009. Comparative genomics of marine cyanomyoviruses reveals the widespread occurrence of Synechococcus host genes localized to a hyperplastic region: implications for mechanisms of cyanophage evolution. Environ. Microbiol. 11:2370–2387 [DOI] [PubMed] [Google Scholar]
- 44. Miller E. S., et al. 2003. Complete genome sequence of the broad-host-range vibriophage KVP40: comparative genomics of a T4-related bacteriophage. J. Bacteriol. 185:5220–5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moreau H., et al. 2010. Marine prasinovirus genomes show low evolutionary divergence and acquisition of protein metabolism genes by horizontal gene transfer. J. Virol. 84:12555–12563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morris R. M., et al. 2005. Temporal and spatial response of bacterioplankton lineages to annual convective overturn at the Bermuda Atlantic Time-series Study site. Limnol. Oceanogr. 50:1687–1696 [Google Scholar]
- 47. Mühling M., et al. 2005. Genetic diversity of marine Synechococcus and co-occurring cyanophage communities: evidence for viral control of phytoplankton. Environ. Microbiol. 7:499–508 [DOI] [PubMed] [Google Scholar]
- 48. Petrov V. M., Ratnayaka S., Nolan J. M., Miller E. S., Karam J. D. 2010. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol. J. 7:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pickard D., et al. 2010. A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar Typhi. J. Bacteriol. 192:5746–5754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rohwer F., Edwards R. 2002. The phage proteomic tree: a genome-based taxonomy for phage. J. Bacteriol. 184:4529–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rohwer F., et al. 2000. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with nonmarine phages. Limnol. Oceanogr. 45:408–418 [Google Scholar]
- 52. Rusch D. B., et al. 2007. The Sorcerer II global ocean sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed., vol. 1 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 54. Sandaa R.-A., Clokie M., Mann N. H. 2008. Photosynthetic genes in viral populations with a large genomic size range from Norwegian coastal waters. FEMS Microbiol. Ecol. 63:2–11 [DOI] [PubMed] [Google Scholar]
- 55. Sandaa R.-A. 2008. Burden or benefit? Virus-host interactions in the marine environment. Res. Microbiol. 159:374–381 [DOI] [PubMed] [Google Scholar]
- 56. Sano E., Carlson S., Wegley L., Rohwer F. 2004. Movement of viruses between biomes. Appl. Environ. Microbiol. 70:5842–5846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Savalia D., et al. 2008. Genomic and proteomic analysis of phiEco32, a novel Escherichia coli bacteriophage. J. Mol. Biol. 377:774–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sebastian M., Ammerman J. W. 2009. The alkaline phosphatase PhoX is more widely distributed in marine bacteria than the classical PhoA. ISME J. 3:563–572 [DOI] [PubMed] [Google Scholar]
- 59. Sharon I., et al. 2011. Comparative metagenomics of microbial traits within oceanic viral communities. ISME J. 5:1178–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sharon I., et al. 2007. Viral photosynthetic reaction center genes and transcripts in the marine environment. ISME J. 1:492–501 [DOI] [PubMed] [Google Scholar]
- 61. Short C. M., Suttle C. A. 2005. Nearly identical bacteriophage structural gene sequences are widely distributed in both marine and freshwater environments. Appl. Environ. Microbiol. 71:480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stewart C. R., et al. 2009. The genome of Bacillus subtilis bacteriophage SPO1. J. Mol. Biol. 388:48–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Su Z., Olman V., Xu Y. 2007. Computational prediction of Pho regulons in cyanobacteria. BMC Genomics 8:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sullivan M. B., et al. 2008. Portal protein diversity and phage ecology. Environ. Microbiol. 10:2810–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sullivan M. B., Coleman M. L., Weigele P., Rohwer F., Chisholm S. W. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sullivan M. B., et al. 2010. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ. Microbiol. 12:3035–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sullivan M. B., et al. 2009. The genome and structural proteome of an ocean siphovirus: a new window into the cyanobacterial ‘mobilome.’ Environ. Microbiol. 11:2935–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sullivan M. B., et al. 2006. Prevalence and evolution of core photosystem II genes in marine cyanobacterial viruses and their hosts. PLoS Biol. 4:e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sullivan M. B., Waterbury J. B., Chisholm S. W. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047–1051 [DOI] [PubMed] [Google Scholar]
- 70. Suttle C. A. 2005. Viruses in the sea. Nature 437:356–361 [DOI] [PubMed] [Google Scholar]
- 71. Suttle C. A., Chan A. M., Cottrell M. T. 1991. Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton. Appl. Environ. Microbiol. 57:721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tetu S. G., et al. 2009. Microarray analysis of phosphate regulation in the marine cyanobacterium Synechococcus sp. WH8102. ISME J. 3:835–849 [DOI] [PubMed] [Google Scholar]
- 73. Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tommassen J., et al. 1991. Characterization of two genes, glpQ and ugpQ, encoding glycerophosphoryl diester phosphodiesterases of Escherichia coli. Mol. Gen. Genet. 226:321–327 [DOI] [PubMed] [Google Scholar]
- 75. Treusch A. H., et al. 2009. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 3:1148–1163 [DOI] [PubMed] [Google Scholar]
- 76. Wang J., et al. 2005. Complete genome sequence of bacteriophage T5. Virology 332:45–65 [DOI] [PubMed] [Google Scholar]
- 77. Wanner B. L. 1996. Phosphorus assimilation and control of the phosphate regulon, p. 1357–1381 In Neidhardt F. C., Curtiss R. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed ASM Press, Washington, DC [Google Scholar]
- 78. Weigele P. R., et al. 2007. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ. Microbiol. 9:1675–1695 [DOI] [PubMed] [Google Scholar]
- 79. Weynberg K. D., Allen M. J., Ashelford K., Scanlan D. J., Wilson W. H. 2009. From small hosts come big viruses: the complete genome of a second Ostreococcus tauri virus, OtV-1. Environ. Microbiol. 11:2821–2839 [DOI] [PubMed] [Google Scholar]
- 80. Williamson S. J., et al. 2008. The Sorcerer II global ocean sampling expedition: metagenomic characterization of viruses within aquatic microbial samples. PLoS One 3:e1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wilson W. H., Joint I. R., Carr N. G., Mann N. H. 1993. Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus sp. strain WH7803. Appl. Environ. Microbiol. 59:3736–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yoshida T., et al. 2008. Ma-LMM01 infecting toxic Microcystis aeruginosa illuminates diverse cyanophage genome strategies. J. Bacteriol. 190:1762–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yoshida T., et al. 2006. Isolation and characterization of a cyanophage infecting the toxic cyanobacterium Microcystis aeruginosa. Appl. Environ. Microbiol. 72:1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yu Y., Breitbart M., McNairnie P., Rohwer F. 2006. FastGroupII: a web-based bioinformatics platform for analyses of large 16S rDNA libraries. BMC Bioinformatics 7:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zeidner G., et al. 2005. Potential photosynthesis gene recombination between Prochlorococcus and Synechococcus via viral intermediates. Environ. Microbiol. 7:1505–1513 [DOI] [PubMed] [Google Scholar]
- 86. Zhong Y., Chen F., Wilhelm S. W., Poorvin L., Hodson R. E. 2002. Phylogenetic diversity of marine cyanophage isolates and natural virus communities as revealed by sequences of viral capsid assembly protein gene g20. Appl. Environ. Microbiol. 68:1576–1584 [DOI] [PMC free article] [PubMed] [Google Scholar]