Abstract
An understanding of the distribution of colored dissolved organic matter (CDOM) in the oceans and its role in the global carbon cycle requires a better knowledge of the colored materials produced and consumed by marine phytoplankton and bacteria. In this work, we examined the net uptake and release of CDOM by a natural bacterial community growing on DOM derived from four phytoplankton species cultured under axenic conditions. Fluorescent humic-like substances exuded by phytoplankton (excitation/emission [Ex/Em] wavelength, 310 nm/392 nm; Coble's peak M) were utilized by bacteria in different proportions depending on the phytoplankton species of origin. Furthermore, bacteria produced humic-like substances that fluoresce at an Ex/Em wavelength of 340 nm/440 nm (Coble's peak C). Differences were also observed in the Ex/Em wavelengths of the protein-like materials (Coble's peak T) produced by phytoplankton and bacteria. The induced fluorescent emission of CDOM produced by prokaryotes was an order of magnitude higher than that of CDOM produced by eukaryotes. We have also examined the final compositions of the bacterial communities growing on the exudates, which differed markedly depending on the phytoplankton species of origin. Alteromonas and Roseobacter were dominant during all the incubations on Chaetoceros sp. and Prorocentrum minimum exudates, respectively. Alteromonas was the dominant group growing on Skeletonema costatum exudates during the exponential growth phase, but it was replaced by Roseobacter afterwards. On Micromonas pusilla exudates, Roseobacter was replaced by Bacteroidetes after the exponential growth phase. Our work shows that fluorescence excitation-emission matrices of CDOM can be a helpful tool for the identification of microbial sources of DOM in the marine environment, but further studies are necessary to explore the association of particular bacterial groups with specific fluorophores.
INTRODUCTION
Colored dissolved organic matter (CDOM) is receiving increasing attention due to its important role in aquatic ecosystems. It regulates UV and visible light penetration in the water column, thus influencing primary productivity (2) and preventing cellular DNA damage (16, 17). CDOM can also form complexes with metals, reducing the concentrations of free ions in seawater (28). In addition, changes in the optical properties of CDOM are suitable to trace microbial and photochemical degradation processes (34, 45) and, more specifically, the in situ formation of biorefractory humic materials from bioavailable DOM (26). CDOM has also been the focus of remote sensing studies, given that the absorption spectrum of CDOM overlaps with that of chlorophyll a, affecting the satellite-derived estimates of phytoplankton biomass and activity in the oceans, especially in coastal areas (51).
The fraction of CDOM that emits induced fluorescent light is called fluorescent dissolved organic matter (FDOM). Two main groups of fluorophores have been differentiated (9): protein-like substances, which fluoresce around an excitation/emission wavelength (Ex/Em) of 280 nm/350 nm (peak T), and humic-like substances, which fluoresce at two pairs of wavelengths, an Ex/Em wavelength of 312 nm/380 to 420 nm (peak M) and an Ex/Em wavelength of 340 nm/440 nm (peak C). Both peaks M and C have been found in samples from freshwater and seawater environments. However, a shift to longer wavelengths of the humic-like peaks in freshwater samples has been reported (8), because terrestrial humic substances are more aromatic than marine humic substances (3). Additionally, and independently of their origins, humic substances also fluoresce in the UV-C region of the spectrum, at an Ex/Em wavelength of 250 nm/450 nm (8, 9).
Humic materials have been traditionally considered photodegradable but resistant to bacterial degradation. In fact, when humic-like FDOM is not exposed to natural UV radiation levels, it can accumulate in the ocean on centennial to millennial time scales within the global conveyor belt (7, 60). Therefore, the formation of bioresistant CDOM during the degradation of bioavailable DOM would allow the sequestration of anthropogenic CO2 in a dissolved organic form for hundreds to thousands of years as part of the recently coined “microbial carbon pump” sequestration mechanism (19).
Global carbon flux estimates require quantitative information about the degradation rates of biogenic organic matter. Although the bioavailability of phytoplankton exudates has been recurrently studied (36, 55, 56), few works have dealt with the exudation of CDOM by phytoplankton (44). In fact, marine FDOM has been considered mainly a by-product of bacterial metabolism (7, 34, 60), until some recent studies showed that it can be produced by zooplankton (59), krill (37), and also phytoplankton cells (44). Among these FDOM producers, phytoplankton is the most abundant organism in terms of biomass. Therefore, a quantitative and qualitative analysis of the FDOM produced by phytoplankton and its comparison with that produced by bacteria are essential to better understand the distributions of CDOM in the oceans as well as the role of CDOM in the global carbon cycle.
In addition, while the bacterioplankton structure is modulated by the amount and quality of the available substrates (e.g., see references 41 and 46), the activities of different groups of bacteria can shape organic matter compositions. However, to the best of our knowledge, there is a lack of studies examining the changes in FDOM and bacterial community composition under controlled conditions. Since fluorescence excitation-emission matrices (EEMs) are fingerprints of DOM structures, the study of these EEMs in relation to different bacterial groups could be a step further in the characterization of DOM produced by bacteria and in the identification of DOM sources in the environment.
The objective of the present work is to quantify the net production and consumption of CDOM by a natural bacterial community growing on exudates derived from four phytoplankton species (Chaetoceros sp., Skeletonema costatum, Prorocentrum minimum, and Micromonas pusilla) cultured under axenic conditions. To this end, the time course of the induced fluorescence excitation-emission properties of CDOM as well as its relationship with changes in the structure of the bacterial community were monitored. Moreover, we investigated whether the bacterial groups selected by the treatments can be associated with characteristic FDOM signals.
MATERIALS AND METHODS
Phytoplankton cultures.
Algal exudates were produced from axenic cultures of four phytoplankton species. The axenic species obtained from the Provasoli-Guillard National Center for Culture of Marine Phytoplankton (CCMP) (https://ccmp.bigelow.org/) were cultured under axenic conditions as described previously by Romera-Castillo et al. (44). The organisms used were the diatoms Chaetoceros sp. (CCMP199) and Skeletonema costatum (CCMP2092) (Greville) Cleve, the dinoflagellate Prorocentrum minimum (CCMP1329) (Pavillard) J. Schiller, and the prasinophyte Micromonas pusilla (R. W. Butcher) (CCMP1545) I. Manton and M. Parke. The four species produced significant amounts of FDOM (44).
The cultured volume (1.4 liters) of each phytoplankton species was filtered through a 0.2-μm Sterivex cartridge and inoculated with 150 ml of natural seawater filtered twice through a 0.6-μm polycarbonate filter to eliminate predators of bacteria, such as small flagellates. The inoculum was taken on 7 May 2008 from the Blanes Bay Microbial Observatory (http://www.icm.csic.es/bio/projects/icmicrobis/bbmo/). Each 1.55 liters of inoculated sample was then distributed into 36 polycarbonate bottles of 60 ml filled with 40 ml of the inoculated exudates.
The bottles were incubated for 34 days and sampled at 0, 4, 12, 21, and 34 days. At each sampling time, 4 of the initial 36 bottles per treatment were sacrificed: 3 replicates were used for dissolved organic carbon (DOC) and FDOM analyses and for bacterial counts, and 1 was used for catalyzed reporter deposition fluorescence in situ hybridization (CARD-FISH).
Bacterial community biomass and structure.
Heterotrophic bacteria in the three bottles were counted with a FACSCalibur (Becton Dickinson) flow cytometer equipped with a 15-mW argon-ion laser (488-nm emission) as described previously by Gasol and Del Giorgio (15). For each sample, 4 ml of water was collected and fixed immediately with cold 10% glutaraldehyde (final concentration, 1%), left in the dark for 10 min at room temperature, and then stored at −80°C. Later, 400 μl of the sample received a diluted SYTO-13 (Molecular Probes Inc., Eugene, OR) stock (10:1) at a final concentration of 2.5 μmol liter−1, was left for about 10 min in the dark to complete the staining, and was run in the flow cytometer. At least 30,000 events were acquired for each subsample (usually 90,000 events). Fluorescent beads (Fluoresbrite carboxylate microspheres, 1 μm; Polysciences Inc., Warrington, PA) were added at a known density as internal standards. The bead standard concentration was determined by epifluorescence microscopy. Heterotrophic bacteria were detected by their signature in a plot of side scatter (SSC) versus FL1 (green fluorescence). Data analysis was performed with the Paint-A-Gate software (Becton Dickinson).
Bacterial cell size (V) (in μm3 cell−1) was estimated by using the relationship between the average bacterial cell size and the average fluorescence of the SYTO-13-stained sample relative to the beads (FL1 bacteria/FL1 beads), as reported previously by Gasol and Del Giorgio (15), as follows: V = 0.0075 + 0.11 × (FL1 bacteria/FL1 beads).
The bacterial biomass (BB) (in pg C cell−1) was calculated by using the carbon-to-volume (V) (in μm3 cell−1) relationship derived previously by Norland (35) from the data of Simon and Azam (52), as follows: BB = 0.12 × V0.7.
The bacterial community composition (BCC) was determined by CARD-FISH. Three- to five-milliliter samples were fixed with paraformaldehyde (1% final concentration), and bacterial cells were collected onto 0.2-μm polycarbonate filters (Whatman) and kept at −80°C until hybridization. Four horseradish peroxidase probes were used, according to a protocol described previously by Pernthaler et al. (39), ALT1413, CF319a, ROS537, and SAR11, targeting the Alteromonas, Bacteroidetes, Roseobacter, and SAR11 clades, respectively (H. Sarmento and J. M. Gasol, submitted for publication). The NON338 probe was used as a negative control. After permeabilization with lysozyme and achromopeptidase, several filter sections were cut and hybridized for 2 h at 35°C (39). Probe GAM42a was used with competitor oligonucleotides as described previously (27). Filter portions were then counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (1 μg ml−1) before enumeration with an epifluorescence microscope. A minimum of 10 fields per filter portion were counted, and at least 400 cells per group were counted. The selected groups were expressed as a percentage relative to the total number of DAPI-stained bacterial cells.
DOC and FDOM.
Triplicate DOC and FDOM samples were filtered through GF/F filters with an acid-cleaned syringe. Milli-Q water was filtered through the filtration system, and no significant enrichment was observed for DOC and FDOM concentrations, discarding contamination during filtration.
Approximately 10 ml of water was collected into precombusted (450°C for 12 h) glass ampoules for DOC analysis. H3PO4 was added to acidify the sample to pH <2, and the ampoules were heat sealed and stored in the dark at 4°C until analysis. DOC was measured with a Shimadzu TOC-V organic carbon analyzer. The system was standardized daily with potassium hydrogen phthalate. Each ampoule was injected 3 to 5 times, and the average area of the 3 replicates that yielded a standard deviation of <1% was chosen to calculate the average DOC concentration of each sample after the subtraction of the average area of the freshly produced UV-irradiated Milli-Q water used as a blank. The performance of the analyzer was tested with the DOC reference materials provided by D. Hansell (University of Miami). We obtained a concentration of 45.2 ± 0.3 μmol C liter−1 for the deep ocean reference (Sargasso Sea deep water, 2,600 m), minus blank reference materials, on the day when the samples were analyzed. The nominal DOC value provided by the reference laboratory (certified reference materials for DOC analysis, batch 04, 2004) is 45 μmol C liter−1.
Samples for FDOM analysis were measured immediately after collection. Single measurements at specific excitation/emission wavelengths as well as excitation-emission matrices (EEMs) of the aliquots were performed with a Perkin-Elmer LS 55 luminescence spectrometer, equipped with a xenon discharge lamp, equivalent to 20 kW for an 8-μs duration. The detector was a red-sensitive R928 photomultiplier, and a photodiode worked as a reference detector. Slit widths were 10.0 nm for the excitation and emission wavelengths, and the scan speed was 250 nm min−1. Measurements were performed at a constant room temperature of 20°C in a 1-cm quartz fluorescence cell. For comparison with other works, the Ex/Em wavelengths used for single measurements were those established previously by Coble (8): Ex/Em wavelength of 280 nm/350 nm (peak T) as an indicator of protein-like substances and Ex/Em wavelengths of 320 nm/410 nm (peak M) and Ex/Em wavelengths of 340 nm/440 nm (peak C) as indicators of humic-like substances. According to methods described previously by Coble (8), fluorescence measurements were expressed in quinine sulfate units (QSU) by calibrating the Perkin-Elmer LS 55 instrument at Ex/Em wavelengths of 350 nm/450 nm against a quinine sulfate dihydrate (QS) standard made up in 0.05 mol liter−1 sulfuric acid.
EEMs were performed to track possible changes in the positions of the protein- and humic-like fluorescence peaks. These matrices were generated by combining 21 synchronous Ex/Em fluorescence spectra of the sample, obtained for excitation wavelengths of 250 to 400 nm and an offset between the excitation and emission wavelengths of 50 nm for the 1st scan and 250 nm for the 21st scan. Rayleigh scatter does not need to be corrected when the EEMs are generated from synchronous spectra, and Raman scatter was corrected by subtracting the EEM of pure water (Milli-Q) from the EEM of the sample.
The paired Student t test was used to check for significant differences in the measured variables between the initial, intermediate, and final incubation times (53).
RESULTS
Time course of bacterial biomass.
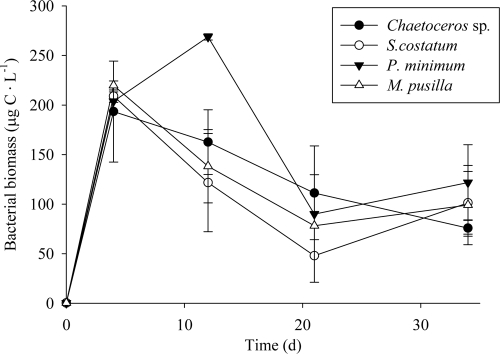
The bacterial biomass increased similarly in the four cultures throughout the initial 4 days of exponential growth (Fig. 1), reaching concentrations that ranged from 193 ± 51 to 220 ± 5 μg C liter−1. It kept increasing until day 12 only for the treatment with P. minimum exudates, reaching 269 ± 3 μg C liter−1. The bacterial biomass (BB) slightly decreased after day 4, except on P. minimum exudates, where it decreased after day 12. The growth rates of the natural bacterial community (ΔBB) were not significantly different (P > 0.05) among the four treatments amended with exudates of different phytoplankton species.
Fig. 1.
Time course of the bacterial biomass (BB) during the incubation time.
Time course of dissolved organic carbon.
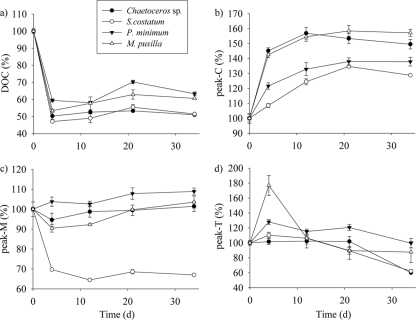
Two phases can be distinguished in the time course of DOC concentrations (Fig. 2a). A sharp decrease of the DOC concentration occurred during the initial 4 days of bacterial exponential growth, reaching concentrations that were 47.1% ± 0.3% to 59.5% ± 0.9% of the initial DOC concentration. Diatom exudates presented the most abrupt decrease in DOC concentrations, and P. minimum exudates presented the smallest one. In the second phase, from days 4 to 34, DOC concentrations tended to increase until final stagnation or a small decrease.
Fig. 2.
Time courses of DOC (a), peak C (b), peak M (c), and peak T (d) during the incubation time. Values are reported as percentages of their respective initial concentrations.
Time course of fluorescent CDOM.
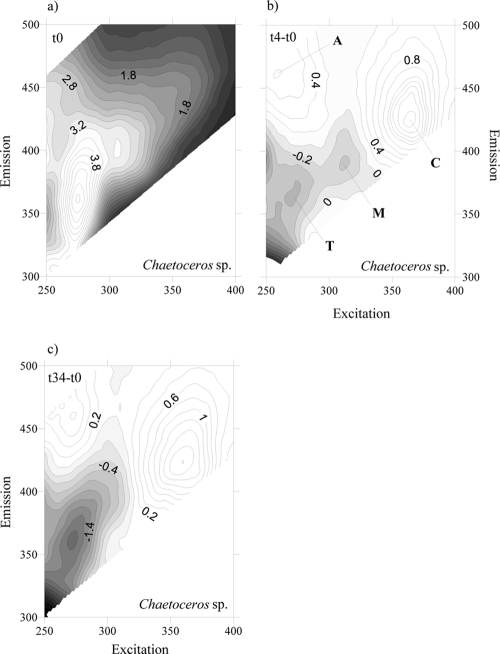
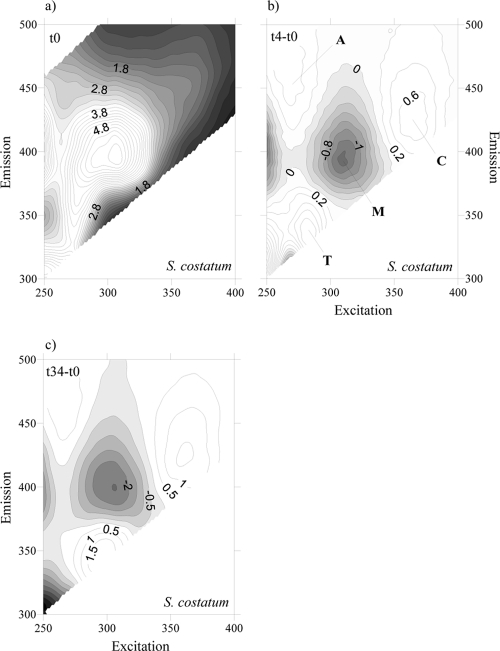
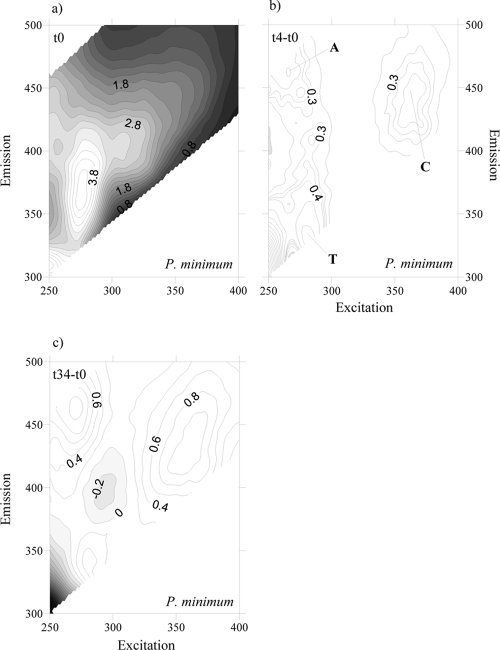
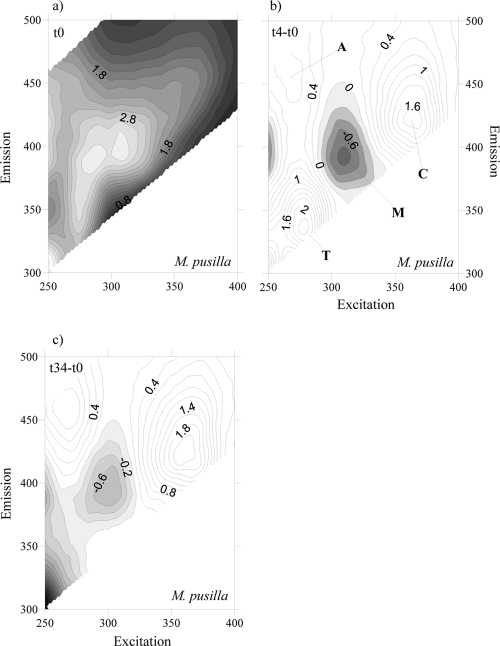
To account for the net production and consumption of any CDOM fluorophore after 4 days of exponential growth, the fluorescence excitation-emission matrices (EEMs) at the initial time were subtracted from the corresponding EMMs at day 4 for each culture. At day 4, all EEMs showed the production of a humic-like fluorophore that peaked around an Ex/Em wavelength of 354 nm/424 nm (Fig. 3b, 4b, 5b, and 6b), i.e., Coble's peak C (9). After 34 days of incubation, peak C was still present in the EEMs of all the cultures, but its fluorescence intensity had increased (Fig. 3c, 4c, 5c, and 6c). On the contrary, a second humic-like fluorophore that peaked around an Ex/Em wavelength of 310 nm/392 nm, Coble's peak M (9), decreased after 4 days of exponential growth in 3 of the 4 cultures (Fig. 3b, 4b, and 6b). In the P. minimum exudates, peak M took slightly longer to diminish (Fig. 5b and c).
Fig. 3.
FDOM excitation-emission matrices of incubation on Chaetoceros sp. exudates at day 0 (a), day 4 minus day 0 (b), and day 34 minus day 0 (c). Different peaks are indicated.
Fig. 4.
FDOM excitation-emission matrices of incubation on S. costatum exudates at day 0 (a), day 4 minus day 0 (b), and day 34 minus day 0 (c). Different peaks are indicated.
Fig. 5.
FDOM excitation-emission matrices of incubation on P. minimum exudates at day 0 (a), day 4 minus day 0 (b), and day 34 minus day 0 (c). Different peaks are indicated.
Fig. 6.
FDOM excitation-emission matrices of incubation on M. pusilla exudates at day 0 (a), day 4 minus day 0 (b), and day 34 minus day 0 (c). Different peaks are indicated.
Coble's UV protein-like peak T (9), which peaks at around Ex/Em wavelengths of 275 nm/361 nm, was also present in the cultures grown on the four phytoplankton species (Fig. 3a, 4a, 5a, and 6a). After 4 days of incubation, the intensity of peak T increased in the cultures grown on S. costatum, M. pusilla, and P. minimum exudates (Fig. 4b, 5b, and 6b) but decreased slightly in the culture grown on Chaetoceros sp. exudates (Fig. 3b). In addition, all the EEMs showed a production of Coble's UV humic-like peak A (9) at around 261 nm/463 nm (Fig. 3b, 4b, 5b, and 6b).
Changes in the intensities of peaks C, M, and T, relative to values at the initial time, during the course of the incubations are presented in Fig. 2 for an easier comparison between the exudates of the different phytoplankton species. The initial values of these parameters are summarized in Table 1. The intensity of peak C increased significantly (P < 0.001) until day 21 in all cultures (Fig. 2b), with the highest variation during the first 4 days; the corresponding net production rates are summarized in Table 2. On the contrary, the intensity of peak M increased significantly (P < 0.05) only in the P. minimum exudate culture (Fig. 2c). For the other 3 species, a significant decrease (P < 0.05) occurred during the initial 4 days of incubation. The proportion of bioavailable FDOM excreted by phytoplankton was extremely different depending on the species: 30.3% ± 0.3% of the marine humic-like substances (peak M) excreted by S. costatum were bioavailable, while the proportion decreased to 5% ± 2% and 10% ± 2% in Chaetoceros sp. and M. pusilla, respectively. Since the incubations were performed in the dark, photobleaching of peak M can be discarded. During the stationary phase, the intensity of peak M experienced a significant (P < 0.05) increase only from days 12 to 21. Net bacterial production rates of peaks C and M per unit of carbon biomass and day were calculated for all treatments, during the exponential phase, to compare them with the net production rates of humic-like substances by marine phytoplankton during the exponential growth phase reported previously by Romera-Castillo et al. (44). We obtained net production rates of peak C of between 15 × 10−4 ± 2 × 10−4 and 75 × 10−4 ± 3 × 10−4 QSU μg C−1 liter day−1, and the highest net consumption rates of peak M, −117 × 10−4 ± 4 × 10−4 QSU μg C−1 liter day−1, corresponded to the bacteria grown on S. costatum exudates (Table 2).
Table 1.
Initial values of average bacterial biomass, dissolved organic carbon, and fluorescence of CDOM at peak M, peak C, and peak T
| Exudate | Avg BB (μg C liter−1) ± SD | Avg DOC (μmol C liter−1) ± SD | Avg peak M (QSU) ± SD | Avg peak C (QSU) ± SD | Avg peak T (QSU) ± SD |
|---|---|---|---|---|---|
| Chaetoceros sp. | 0.41 ± 0.02 | 287.8 ± 2.2 | 3.12 ± 0.06 | 2.07 ± 0.01 | 4.32 ± 0.04 |
| S. costatum | 0.23 ± 0.03 | 304.5 ± 0.4 | 4.73 ± 0.01 | 2.22 ± 0.00 | 4.05 ± 0.02 |
| P. minimum | 0.31 ± 0.05 | 304.9 ± 1.0 | 2.96 ± 0.02 | 1.97 ± 0.03 | 3.77 ± 0.06 |
| M. pusilla | 0.35 ± 0.02 | 242.1 ± 0.6 | 3.00 ± 0.01 | 2.03 ± 0.02 | 2.91 ± 0.02 |
Table 2.
Net production of dissolved organic carbon and fluorescent CDOM by bacteria versus phytoplankton during exponential phase in the four culturesa
| Phytoplankton species | Avg BB from days 0 to 4 (μg C liter−1) ± SD | ΔDOC/(B · t) (μg C μg C−1 day−1) | Δpeak T/(B · t) (QSU μg C−1 liter day−1) | Δpeak M/(B · t) (QSU μg C−1 liter day−1) | Δpeak/(B · t) (QSU μg C−1 liter day−1) |
|---|---|---|---|---|---|
| Bacteria grown on: | |||||
| Chaetoceros sp. | 31.3 ± 0.8 | −11.3 ± 0.4 | 6 × 10−4 ± 7 × 10−4 | −13 × 10−4 ± 4 × 10−4 | 75 × 10−4 ± 3 × 10−4 |
| S. costatum | 30.7 ± 1.0 | −15.77 ± 0.08 | 34 × 10−4 ± 7 × 10−4 | −117 × 10−4 ± 4 × 10−4 | 15 × 10−4 ± 2 × 10−4 |
| P. minimum | 31.4 ± 1.6 | −11.8 ± 0.2 | 84 × 10−4 ± 8 × 10−4 | 9 × 10−4 ± 3 × 10−4 | 34 × 10−4 ± 3 × 10−4 |
| M. pusilla | 34.1 ± 0.8 | −9.9 ± 0.1 | 164 × 10−4 ± 19 × 10−4 | −21 × 10−4 ± 3 × 10−4 | 63 × 10−4 ± 2 × 10−4 |
| Production by: | |||||
| Chaetoceros sp. | 116 × 10−3 ± 11 × 10−3 | 9.1 × 10−4 ± 0.8 × 10−4 | 4.8 × 10−4 ± 0.1 × 10−4 | 2.4 × 10−4 ± 0.05 × 10−4 | |
| S. costatum | 93 × 10−3 ± 13 × 10−3 | 5.8 × 10−4 ± 0.1 × 10−4 | 4.5 × 10−4 ± 0.1 × 10−4 | 1.3 × 10−4 ± 0.02 × 10−4 | |
| P. minimum | 97 × 10−3 ± 9 × 10−3 | 3.2 × 10−4 ± 0.2 × 10−4 | 1.3 × 10−4 ± 0.03 × 10−4 | 0.8 × 10−4 ± 0.04 × 10−4 | |
| M. pusilla | 86 × 10−3 ± 5 × 10−3 | 2.6 × 10−4 ± 0.02 × 10−4 | 3.0 × 10−4 ± 0.1 × 10−4 | 1.7 × 10−4 ± 0.02 × 10−4 |
B, average biomass for each kind of organism; t, time (in days) that the exponential phase lasted.
In general, the production rate of peak T increased significantly in all cultures during the exponential growth phase (4 days) and then decreased up to the end of the experiment (Fig. 2d). The bacterial culture on M. pusilla exudates underwent the largest increase (77%). However, Chaetoceros sp. did not show a significant variation of the peak T production rate until the end of the incubation time (day 34), when it sharply decreased by 41%. The net production rate of peak T per carbon biomass and day ranged between 6 × 10−4 ± 7 × 10−4 and 164 × 10−4 ± 19 × 10−4 QSU μg C−1 liter day−1.
Time courses of bacterial community structure.
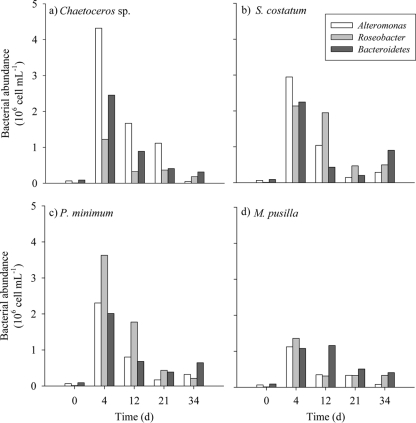
The most abundant groups of bacteria were identified and counted by CARD-FISH to examine their responses to the different substrates generated by the four phytoplankton species tested in this work (Fig. 7). Initial numbers of these bacterial groups were 2.49 × 104 cells ml−1, 6.42 × 104 cells ml−1, 1.21 × 106 cells ml−1, and 2.37 × 106 cells ml−1 for SAR11, Alteromonas, Roseobacter, and Bacteroidetes, respectively. In Chaetoceros sp. exudates, Alteromonas was the dominant bacterial group during the course of the incubation. Among the studied groups on S. costatum exudates, Alteromonas was dominant during the first 4 days, but it was replaced by Roseobacter thereafter. Roseobacter was also the dominant group in P. minimum exudates at all sampling times. In M. pusilla exudates, Roseobacter was the most abundant group during the exponential phase, but Bacteroidetes prevailed after day 12. Subtracting the cell abundance of each group at the initial time from that at day 4, we observed that Alteromonas was the group that developed better on diatom exudates (1.1 × 106 cells ml−1 day−1 and 7.3 × 105 cells ml−1 day−1 for Chaetoceros sp. and S. costatum, respectively), while it was Roseobacter in the case of the P. minimum (9.1 × 105 cells ml−1 day−1) and M. pusilla (3.4 × 105 cells ml−1 day−1) exudates. The abundance of SAR11, which was present at the initial time, decreased throughout the incubations in all exudates.
Fig. 7.
Time course of the abundances of the main bacterial groups grown on Chaetoceros sp. (a), S. costatum (b), P. minimum (c), and M. pusilla (d) exudates during the incubation time.
DISCUSSION
Net production and consumption of DOC and CDOM by marine bacteria.
The proportions of bacterial carbon consumption during the exponential growth phase (between 41 and 53% of the initial DOC) were similar to the 48% of labile DOM found previously by Puddu et al. (42) in phytoplankton exudates obtained by growing the diatom Cylindrotheca closterium in a nutrient-balanced medium. Previous works demonstrated that bacteria can take up humic substances retained by XAD resins (4, 18, 29). Indeed, during periods of low primary production, bacteria seem to use humic substances as a substrate (48, 56). Shimotori et al. (50) also observed a decrease of the FDOM measured at an Ex/Em wavelength of 315 nm/410 nm (equivalent to peak M) in their bacterial cultures but only from incubation times of 20 to 30 days, when the bacterial number started to decay. Their incubations consisted of artificial seawater amended with inorganic nutrients and glucose and inoculated with a natural bacterial community. Those authors attributed the FDOM uptake by bacteria in the stationary phase to the depletion of the amended glucose, the only carbon source for bacterial growth in their cultures. In our experiment, we used phytoplankton exudates that are known to be rich in carbohydrates (32), and the FDOM decrease that we observed occurred during the exponential growth phase of the experiments. Given the lability of the fresh materials exuded by phytoplankton, a full consumption of the exuded carbohydrates and the subsequent utilization of marine humic-like substances could have occurred in the short time that elapsed between the initial time and day 4. It was shown previously that humic substances retained by XAD resins can be adsorbed onto bacterial surfaces, although this adsorption is negligible at pHs typical of marine waters for the number of bacteria that our incubation reached (13). The peak M fluorophore consumed by bacteria in our experiment had been previously produced by phytoplankton (44). Therefore, phytoplankton cells exude fluorescent humic-like substances that are readily bioavailable to microbial degradation.
The different behaviors of peak M and peak C during the exponential phase of our incubations suggest different origins and fates for both fluorophores. Peak C was produced continuously in all cultures, while peak M was consumed during the exponential phase and produced during the senescence phase of the bacterial cultures. Since the peak M/peak C ratio did not change significantly from day 12 onwards, it is likely that the fluorescence intensity observed at the Ex/Em wavelengths of peak M during the senescence phase was due mostly to peak C tailing.
Regarding peak T, the release of this fluorophore by marine bacteria was previously reported (5). A pattern similar to that observed in our experiment, where the amount of peak T increased until the bacterial population reached the stationary growth phase and decreased from then on, was shown previously by Cammack et al. (5) in incubation experiments performed with lake waters of different trophic statuses. Those authors suggested that peak T reflects the balance between bacterial consumption and the production of a small fraction of the DOM pool and that it could be a by-product of bacterial metabolism. The significant positive linear correlation that we found between the fluorescence of peak T and the bacterial biomass (R2 = 0.41; P < 0.01; n = 16) supports this statement. Kawasaki and Benner (20) also found an increase in levels of total dissolved amino acids (TDAAs) matching a peak in bacterial biomass. Since we have used GF/F filters that could have not retained 100% of the BB, a partial contribution of the aromatic amino acid contained in cells to the peak T signal cannot be discarded.
Wavelength shifts in EEMs as indicators of changes in the chemical structure of CDOM.
EEMs provide complementary information about the chemical structure, especially the aromaticity, of the FDOM produced or consumed by marine phytoplankton and bacteria. The peak T produced by bacteria in our experiments was shifted to slightly longer excitation and shorter emission wavelengths than those for the peak T reported previously by Coble et al. (9). In this sense, tryptophan residues that are exposed to water have maximum fluorescence at emission wavelengths of about 340 to 350 nm, whereas totally buried residues (e.g., being part of peptides) fluoresce at about 330 nm (25). We found maximum intensities in the peak T region at an Ex/Em wavelength of 280 nm/335 nm, which is an indication of the release of aromatic amino acids by bacteria in a more buried, combined form rather than in a free form. That maximum matches with the fluorescence maximum of the enzyme nuclease, at an Ex/Em wavelength of 280 nm/334 nm (25). This exoenzyme could have been excreted by bacteria to hydrolyze DNA and consume their products (38). It differs from the protein-like substances excreted by phytoplankton, since the maximum peak produced by three species of phytoplankton in that region was at an Ex/Em wavelength of 275 nm/358 nm (44).
Polypeptides are part of the dissolved combined amino acid (DCAA) pool, and some authors have reported that protein and DCAAs are relatively more important as bacterial substrates than dissolved free amino acids (DFAAs) (10, 22, 24, 47). Using radiolabeled proteins to examine the relative significance of proteins versus DFAAs as bacterial substrates, Keil and Kirchman (21) found previously that bacteria preferred proteins in the oligotrophic Sargasso Sea (22) but DFAAs in the eutrophic Delaware Bay (21). Since our treatments with phytoplankton exudates were performed under nutrient-rich conditions, bacteria could have used DFAAs and released peptides.
Production of humic-like substances: marine phytoplankton versus marine bacteria.
Peak M has traditionally been associated with the humic materials produced in situ in marine ecosystems, since the EEMs of natural seawater samples usually present a fluorescence maximum in that region. In contrast, the EEMs of natural samples with a predominant terrestrial origin present a fluorescence maximum at peak C, shifted to significantly longer excitation and emission wavelengths than those of peak M (9). Therefore, in our culture experiments with marine bacteria growing on marine phytoplankton exudates, the production of peak M fluorescence should be expected. However, our study demonstrates that the main humic-like fluorophore produced by marine bacteria grown on marine phytoplankton exudates was peak C.
Furthermore, our results show that marine phytoplankton is able to produce substances fluorescing at peak M, which are consumed or transformed by marine bacteria that in turn produce other humic-like substances that fluoresce at peak C. Any shift of a humic-like fluorescent emission maximum to longer wavelengths is an unequivocal indication of increased aromaticity and polycondensation of humic materials (6). Therefore, attending to the position of both humic-like peak maxima, humic materials exuded by phytoplankton were more aliphatic (blue shifted) than the more aromatic humic materials produced by bacteria (red shifted). The preferential consumption of peak M by marine bacteria concurs with data from previous studies that reported higher bioavailabilities with increasing aliphatic carbon moieties in a compound (30, 54).
Considering our results, we could hypothesize that the more aliphatic humic-like substances that fluoresce at peak M are produced mostly as a by-product of eukaryote metabolism. Conversely, the more aromatic substances fluorescing at peak C could be associated with prokaryote by-products. This hypothesis is also supported by evidence that copepods exude humic-like substances that fluoresce at peak M (59) and that a strong signal in the peak C region was found in cultures of Synechococcus and Prochlorococcus (data not shown). Furthermore, it was reported previously that the FDOM intensity at peak M is higher in the euphotic zone (8), where phytoplankton is abundant, and decreases with depth. On the contrary, the intensity of peak C increases with depth (8), and it is known that organic matter transformations in the dark ocean are dominated by bacterial activity, which could generate higher fluorescence at this peak.
In general, the level of production of peaks T and C normalized to the biomass during the exponential growth phase was about one order of magnitude higher for marine bacteria than for marine phytoplankton (Table 2). This fact could explain the lack of a correlation between chlorophyll and CDOM that some authors reported previously (33, 43), which they attributed to the in situ production of CDOM by bacteria, concluding that phytoplankton was not a direct source of CDOM. Indeed, the consumption of the peak M fluorophore by bacteria also contributes to reduce the expected correlation between phytoplankton biomass and CDOM.
Selection of bacterial groups growing on different phytoplankton exudates.
Different bacterial groups were selected depending on the phytoplankton exudates on which they grew. This agrees with previous studies that concluded that the bacterioplankton structure is determined by the amount and quality of the substrates available in the ecosystem (e.g., see references 41 and 46). It is well known that both SAR11 and Roseobacter are specialized in processing low-molecular-weight organic substrates (1, 12, 31), but the former prevails in oligotrophic environments, and the latter prevails in meso- and eutrophic environments (14). On the other hand, the Bacteroidetes group is specialized in processing high-molecular-weight organic substrates (23, 58).
With these general considerations in mind, it can be straightforwardly explained why SAR11, which was present in the natural bacterial community of the (oligotrophic) Blanes Bay Observatory water that we used as an inoculum, was not selected in any of our (eutrophic) cultures. The fact that the highest cell numbers corresponded to Alteromonas grown on Chaetoceros sp. exudates after 4 days of incubation could be the result of the production, by this diatom species, of a specific substrate in which Alteromonas is specialized. Even if Schäfer et al. (49) did not find Gammaproteobacteria in 6 monoalgal diatom nonaxenic cultures, our work indicates that this class can also be associated with diatoms.
Suzuki et al. (57) found previously that the growth of members of the Flavobacteriaceae (subgroup of the Bacteroidetes) was not affected when inorganic nitrogen was excluded from the medium. On this basis, those authors suggested that these bacteria could utilize amino acids as a sole nitrogen source. Other studies reported that this group of bacteria notably grows when abundant amounts of dissolved proteins are available (11, 40). In our experiments, the negative correlation found between the variation in peak T (Δpeak T) and the variation in Bacteroidetes (ΔBacteroidetes) during the exponential phase (R2 = 0.98; P < 0.01; n = 4) supports this observation. Moreover, in the M. pusilla exudates, the largest amount of protein-like substances was produced during the exponential bacterial growth phase (Table 2 and Fig. 2d). These protein-like substances were likely in the form of proteins rather than free amino acids according to the shift of peak T observed in the EEM (Fig. 5b). This could also explain the dominance of Bacteroidetes after day 12 in the sample grown on M. pusilla exudates.
Although our results show that phytoplankton exudates influence bacterial community composition in each treatment, the association of each bacterial group with a particular fluorescent humic-like peak is not clear. More studies are needed to asses this, but from our results, it could be suggested that humic-like fluorescent material is a by-product of bacterial metabolism in general and not due to the activity of a particular bacterial group(s).
Conclusions.
Marine phytoplankton exudates contain fluorescent CDOM that is bioavailable to bacterial degradation; blue-shifted fluorescent humic-like substances (Coble's peak M) are readily taken up by marine bacteria that in turn exude red-shifted humic-like substances (Coble's peak C) during both the exponential and stationary growth phases. Based on these and previous results reported by other authors, we hypothesize that peak M could be a by-product preferentially associated with the catabolism of marine eukaryotic cells, whereas peak C could be associated with catabolism of marine prokaryotes.
Marine bacteria produced humic- and protein-like substances at rates that were an order of magnitude higher than those for phytoplankton when normalized to their respective biomasses. This fact, together with the photodegradability of aromatic compounds, is likely the reason behind the lack of correlation observed between phytoplankton biomass and CDOM in both in situ measurements and satellite-derived estimates.
Although bacterial growth was independent of the phytoplankton exudates in which they were grown, the exudates influenced bacterial community structure by preferentially selecting bacterial groups with contrasting substrate preferences.
ACKNOWLEDGMENTS
This work was supported by project SUMMER, grant number CTM2008-03309/MAR; C.R.-C. was funded by an I3P-CSIC predoctoral fellowship within the project MODIVUS, CTM2005-04795/MAR; and H.S. benefited from fellowships from the Spanish Ministerio de Educación y Ciencia (SB2006-0060 and JCI-2008-2727) and the Portuguese Fundação para a Ciência e a Tecnologia (FRH/BPD/34041/2006).
Footnotes
Published ahead of print on 8 July 2011.
REFERENCES
- 1. Alonso C., Pernthaler J. 2006. Roseobacter and SAR11 dominate microbial glucose uptake in coastal North Sea waters. Environ. Microbiol. 8:2022–2030 [DOI] [PubMed] [Google Scholar]
- 2. Arrigo K. R., Brown C. W. 1996. Impact of chromophoric dissolved organic matter on UV inhibition of primary productivity in the sea. Mar. Ecol. Prog. Ser. 140:207–216 [Google Scholar]
- 3. Benner R. 2003. Molecular indicators of the bioavailability of dissolved organic matter, p. 121–137 In Findlay S., Sinsabaugh R. (ed.), Aquatic ecosystems: interactivity of dissolved organic matter. Academic Press, New York, NY [Google Scholar]
- 4. Bussmann I. 1999. Bacterial utilization of humic substances from the Arctic Ocean. Aquat. Microb. Ecol. 19:37–45 [Google Scholar]
- 5. Cammack W. K. L., Kalff J., Prairie Y. T., Smith E. M. 2004. Fluorescent dissolved organic matter in lakes: relationships with heterotrophic metabolism. Limnol. Oceanogr. 49:2034–2045 [Google Scholar]
- 6. Chen J., LeBoeuf E. J., Dai S., Gu B. 2003. Fluorescence spectroscopic studies of natural organic matter fractions. Chemosphere 50:639–647 [DOI] [PubMed] [Google Scholar]
- 7. Chen R. F., Bada J. L. 1992. The fluorescence of dissolved organic matter in seawater. Mar. Chem. 37:191–221 [Google Scholar]
- 8. Coble P. G. 1996. Characterization of marine and terrestrial DOM in seawater using excitation-emission matrix spectroscopy. Mar. Chem. 51:325–346 [Google Scholar]
- 9. Coble P. G., Del Castillo C. E., Avril B. 1998. Distribution and optical properties of CDOM in the Arabian Sea during the 1995 Southwest Monsoon. Deep Sea Res. II 45:2195–2223 [Google Scholar]
- 10. Coffin R. B. 1989. Bacterial uptake of dissolved free and combined amino acids in estuarine waters. Limnol. Oceanogr. 34:531–542 [Google Scholar]
- 11. Cottrell M. T., Kirchman D. L. 2000. Natural assemblages of marine proteobacteria and members of the Cytophaga-Flavobacter cluster consuming low- and high-molecular-weight dissolved organic matter. Appl. Environ. Microbiol. 66:1692–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Del Giorgio P. A., Gasol J. M. 2008. Physiological structure and single-cell activity in marine bacterioplankton, p. 243–285 In Kirchman D. L. (ed.), Microbial ecology of the oceans, 2nd ed Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- 13. Fein J. B., Boily J.-F., Güçlü K., Kaulbach E. 1999. Experimental study of humic acid adsorption onto bacteria and Al-oxide mineral surfaces. Chem. Geol. 162:33–45 [Google Scholar]
- 14. Fuhrman J. A., Hagström Å. 2008. Bacterial and archaeal community structure and its patterns, p. 45–90 In Kirchman D. L. (ed.), Microbial ecology of the oceans, 2nd ed Wiley-Blackwell, Hoboken, NJ [Google Scholar]
- 15. Gasol J. M., Del Giorgio P. A. 2000. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 64:197 [Google Scholar]
- 16. Häder D.-P., Sinha R. P. 2005. Solar UV radiation-induced DNA damage in aquatic organisms: potential environmental impact. Mutat. Res. 571:221–233 [DOI] [PubMed] [Google Scholar]
- 17. Herndl G. J., Muller-Niklas G., Frick J. 1993. Major role of UV-B in controlling bacterioplankton growth in the surface layer of the ocean. Nature 361:717–719 [Google Scholar]
- 18. Hunt A. P., Parry J. D., Hamilton-Taylor J. 2000. Further evidence of elemental composition as an indicator of the bioavailability of humic substances to bacteria. Limnol. Oceanogr. 45:237–241 [Google Scholar]
- 19. Jiao N., et al. 2010. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat. Rev. Microbiol. 8:593–599 [DOI] [PubMed] [Google Scholar]
- 20. Kawasaki N., Benner R. 2006. Bacterial release of dissolved organic matter during cell growth and decline: molecular origin and composition. Limnol. Oceanogr. 51:2170–2180 [Google Scholar]
- 21. Keil R. G., Kirchman D. L. 1993. Dissolved combined amino acids: chemical form and utilization by marine bacteria. Limnol. Oceanogr. 38:1256–1270 [Google Scholar]
- 22. Keil R. G., Kirchman D. L. 1999. Utilization of dissolved protein and amino acids in the northern Sargasso Sea. Aquat. Microb. Ecol. 18:293–300 [Google Scholar]
- 23. Kirchman D. L. 2004. A primer on dissolved organic material and heterotrophic prokaryotes in the oceans, p. 31–66 In Follows M., Oguz T. (ed.), The ocean carbon cycle and climate. NATO science series. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 24. Kroer N. S., Jorgensen N. O. G., Coffin R. B. 1994. Utilization of dissolved nitrogen by heterotrophic bacterioplankton: a comparison of three ecosystems. Appl. Environ. Microbiol. 60:4116–4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lakowicz J. R. 1983. Principles of fluorescence spectroscopy. Plenum Press, New York, NY [Google Scholar]
- 26. Lønborg C., Alvarez-Salgado X. A., Martinez-Garcia S., Miller A. E. J., Teira E. 2010. Stoichiometry of dissolved organic matter and the kinetics of its microbial degradation in a coastal upwelling system. Aquat. Microb. Ecol. 58:117–126 [Google Scholar]
- 27. Manz W., Amann R., Ludwig W., Wagner M., Schleifer K. H. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria—problems and solutions. Syst. Appl. Microbiol. 15:593–600 [Google Scholar]
- 28. Midorikawa T., Tanoue E. 1998. Molecular masses and chromophoric properties of dissolved organic ligands for copper(II) in oceanic water. Mar. Chem. 62:219–239 [Google Scholar]
- 29. Moran M. A., Hodson R. E. 1990. Bacterial production on humic and nonhumic components of dissolved organic carbon. Limnol. Oceanogr. 35:1744–1756 [Google Scholar]
- 30. Moran M. A., Sheldon W. M., Zepp R. G. 2000. Carbon loss and optical property changes during long-term photochemical and biological degradation of estuarine dissolved organic matter. Limnol. Oceanogr. 45:1254–1264 [Google Scholar]
- 31. Moran M. A., González J. M., Kiene R. P. 2003. Linking a bacterial taxon to sulfur cycling in the sea: studies of the marine Roseobacter group. Geomicrobiol. J. 20:375–388 [Google Scholar]
- 32. Myklestad S. M. 2000. Dissolved organic carbon from phytoplankton, p. 111–148 In Wangersky P. (ed.), The handbook of environmental chemistry, vol. 5, part D Marine chemistry. Springer-Verlag, Berlin, Germany [Google Scholar]
- 33. Nelson N. B., Siegel D. A., Michaels A. F. 1998. Seasonal dynamics of colored dissolved material in the Sargasso Sea. Deep Sea Res. I 45:931–957 [Google Scholar]
- 34. Nieto-Cid M., Álvarez-Salgado X. A., Pérez F. F. 2006. Microbial and photochemical reactivity of fluorescent dissolved organic matter in a coastal upwelling system. Limnol. Oceanogr. 51:1391–1400 [Google Scholar]
- 35. Norland S. 1993. The relationship between biomass and volume of bacteria, p. 303–307 In Kemp P., Sherr B. F., Sherr E. B., Cole J. J. (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishing, Boca Raton, FL [Google Scholar]
- 36. Obernosterer I., Herndl G. J. 1995. Phytoplankton extracellular release and bacterial growth: dependence on the inorganic N:P ratio. Mar. Ecol. Prog. Ser. 116:247–257 [Google Scholar]
- 37. Ortega-Retuerta E., et al. 2009. Biogeneration of chromophoric dissolved organic matter by bacteria and krill in the Southern Ocean. Limnol. Oceanogr. 54:1941–1950 [Google Scholar]
- 38. Paul J. H., DeFlaun M. F., Jeffrey W. H. 1988. Mechanisms of DNA utilization by estuarine microbial populations. Appl. Environ. Microbiol. 54:1682–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pernthaler A., Pernthaler J., Amann R. 2004. Sensitive multi-color fluorescence in situ hybridization for the identification of environmental microorganisms, p. 3.11.711–3.11.726 In Kowalchuk G. A., de Bruijn F. J., Head I. M., Akkermans A. D., van Elsas J. D. (ed.), Molecular microbial ecology manual, 2nd ed., vol. 2 Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 40. Pinhassi J., et al. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13–26 [Google Scholar]
- 41. Pinhassi J., et al. 2004. Changes in bacterioplankton composition under different phytoplankton regimens. Appl. Environ. Microbiol. 70:6753–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Puddu A., et al. 2003. Bacterial uptake of DOM released from P-limited phytoplankton. FEMS Microbiol. Ecol. 46:257–268 [DOI] [PubMed] [Google Scholar]
- 43. Rochelle-Newall E. J., Fisher T. R. 2002. Production of chromophoric dissolved organic matter fluorescence in marine and estuarine environments: an investigation into the role of phytoplankton. Mar. Chem. 77:7–21 [Google Scholar]
- 44. Romera-Castillo C., Sarmento H., Álvarez-Salgado A. X., Gasol J. M., Marrasé C. 2010. Production of chromophoric dissolved organic matter by marine phytoplankton. Limnol. Oceanogr. 55:446–454 [Google Scholar]
- 45. Romera-Castillo C., et al. 2011. Fluorescence: absorption coefficient ratio—tracing photochemical and microbial degradation processes affecting coloured dissolved organic matter in a coastal system. Mar. Chem. 125:26–38 [Google Scholar]
- 46. Rooney-Varga J. N., et al. 2005. Links between phytoplankton and bacterial community dynamics in a coastal marine environment. Microb. Ecol. 49:163–175 [DOI] [PubMed] [Google Scholar]
- 47. Rosenstock B., Simon M. 1993. Use of dissolved combined and free amino acids by planktonic bacteria in Lake Constance. Limnol. Oceanogr. 38:1521–1531 [Google Scholar]
- 48. Rosenstock B., Zwisler W., Simon M. 2005. Bacterial consumption of humic and non-humic low and high molecular weight DOM and the effect of solar irradiation on the turnover of labile DOM in the Southern Ocean. Microb. Ecol. 50:90–101 [DOI] [PubMed] [Google Scholar]
- 49. Schäfer H., Abbas B., Witte H., Muyzer G. 2002. Genetic diversity of ‘satellite’ bacteria present in cultures of marine diatoms. FEMS Microbiol. Ecol. 42:25–35 [DOI] [PubMed] [Google Scholar]
- 50. Shimotori K., Omori Y., Hama T. 2009. Bacterial production of marine humic-like fluorescent dissolved organic matter and its biogeochemical importance. Aquat. Microb. Ecol. 58:55–66 [Google Scholar]
- 51. Siegel D. A., Maritorena S., Nelson N. B. 2002. Global distribution and dynamics of colored dissolved and detrital organic materials. J. Geophys. Res. 107:3228 [Google Scholar]
- 52. Simon M., Azam F. 1989. Protein content and protein sintesis rates of planktonic marine bacteria. Mar. Ecol. Prog. Ser. 51:201–213 [Google Scholar]
- 53. Sokal F. F., Rohlf F. J. 1984. Introduction to biostatistics. W. H. Freeman, New York, NY [Google Scholar]
- 54. Sun L., Perdue E. M., Meyer J. L., Weis J. 1997. Use of elemental composition to predict bioavailability of dissolved organic matter in a Georgia river. Limnol. Oceanogr. 42:714–721 [Google Scholar]
- 55. Sundh I. 1992. Biochemical composition of dissolved organic carbon derived from phytoplankton and used by heterotrophic bacteria. Appl. Environ. Microbiol. 58:2938–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sundh I., Bell R. T. 1992. Extracellular dissolved organic carbon released from phytoplankton as a source of carbon for heterotrophic bacteria in lakes of different humic content. Hydrobiologia 229:93–106 [Google Scholar]
- 57. Suzuki M. T., Preston C. M., Chavez F. P., DeLong E. F. 2001. Quantitative mapping of bacterioplankton populations in seawater: field tests across an upwelling plume in Monterey Bay. Aquat. Microb. Ecol. 24:117–127 [Google Scholar]
- 58. Teira E., et al. 2008. Linkages between bacterioplankton community composition, heterotrophic carbon cycling and environmental conditions in a highly dynamic coastal ecosystem. Environ. Microbiol. 10:906–917 [DOI] [PubMed] [Google Scholar]
- 59. Urban-Rich J., McCarty J. T., Fernández D., Acuña J. L. 2006. Larvaceans and copepods excrete fluorescent dissolved organic matter (FDOM). J. Exp. Mar. Biol. Ecol. 332:96–105 [Google Scholar]
- 60. Yamashita Y., Tanoue E. 2008. Production of bio-refractory fluorescent dissolved organic matter in the ocean interior. Nat. Geosci. 1:579–582 [Google Scholar]