Abstract
Most activated sludge treatment plants suffer from the presence of foams on the surfaces of their aeration reactors. These are often stabilized by hydrophobic mycolic acid-synthesizing actinobacterial species. A polyvalent Siphoviridae phage, GTE7, which lysed several Gordonia and Nocardia species, is described here. Its genome has a modular structure similar to that described for Rhodococcus phage ReqiDocB7. In laboratory-scale experiments, we showed that GTE7 prevents stabilization of foams by these Gordonia and Nocardia species.
TEXT
The appearance of stable foams on the surfaces of aerobic reactors of activated sludge wastewater treatment plants is a common feature and an operational problem for which no reliable and universally applicable control strategy exists currently (3). Its cause is the overproliferation of hydrophobic bacterial populations, among which are the mycolic acid-producing Actinobacteria, the mycolata (3, 5, 11, 18, 19). This group includes the genera Corynebacterium, Dietzia, Gordonia, Skermania, Mycobacterium, Nocardia, Rhodococcus, and Tsukamurella (5). Lytic phages for these mycolata can be isolated readily from activated sludge mixed liquor, and the suggestion has been made that these phages may provide a targeted, environmentally friendly, and safe method for controlling foam formation by reducing the causative bacterial cell numbers below the threshold needed for foam stabilization (14, 16, 21, 23). Laboratory-scale studies suggest that these values are host cell specific (14), and so before such an application can be undertaken, suitable phages need to be purified for each of the foaming bacteria and characterized.
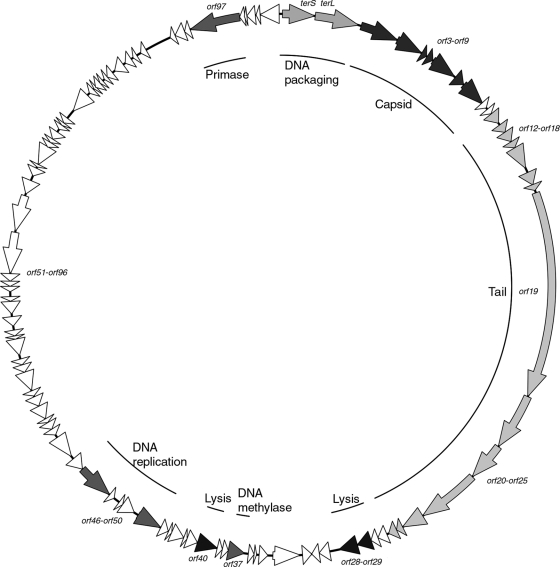
Consequently, as part of attempts to assess the feasibility of this control strategy, we describe a novel Gordonia phage, GTE7, that also targets some Nocardia species. Phage GTE7 was isolated from a wastewater treatment plant in Bendigo (Victoria, Australia) based on its ability to form small (<1-mm diameter) plaques on lawn plates of Gordonia terrae (Ben601) after incubation at 30°C for 2 days. Transmission electron microscopy (TEM) (15) revealed that GTE7 belongs to the Siphoviridae family, possessing a characteristic long noncontractile tail (∼438 nm) and isometric capsid head (∼70 nm) (Fig. 1). The burst size of GTE7 was determined (15) as 83 ± 3 PFU/cell, with a latency period of 2 h in peptone-yeast extract calcium (PYCa) broth at 30°C. When GTE7 was screened for its ability to lyse 65 different mycolata strains (see Table S1 in the supplemental material), it generated lytic plaques on lawn plates of five G. terrae strains (Ben601, Ben602, Ben603, Ben604, and Gter34), two of Gordonia malaquae (A554 and A448), and one of Gordonia australis (18F3M), Gordonia amicalis (Ben607), Nocardia nova (Nnov47), and Nocardia asteroides (Nast23). In comparison to Gordonia phage GTE2 (16), GTE7 lysed a wider range of mycolata species, making it potentially more attractive as a biocontrol agent for foaming. The molecular basis for this broad-host-range phenotype is unknown, but it does suggest that GTE7 is binding to a widespread and conserved receptor(s). GTE7 phage DNA was isolated and sequenced as described previously (15). This genome is organized as a circularly permuted double-stranded DNA molecule of 74,431 bp with a G+C content of 56.8 mol%. At the DNA level, only 6% of the sequence shares similarity with that of the phage ReqiDocB7 genome (20), and the remainder (94%) shares no sequence similarity to any other DNA sequence in GenBank. The GTE7 phage genome has 102 putative open reading frames (ORFs) and one tRNA. The first 31 ORFs are transcribed in one direction, and the following 71 ORFs are transcribed in the opposite direction (Fig. 2), an arrangement similar to that of the Rhodococcus phage ReqiDocB7 genome (20). Only 13 ORFs could be annotated functionally against known protein sequences using the BLASTP algorithm (see Table S2 in the supplemental material).
Fig. 1.
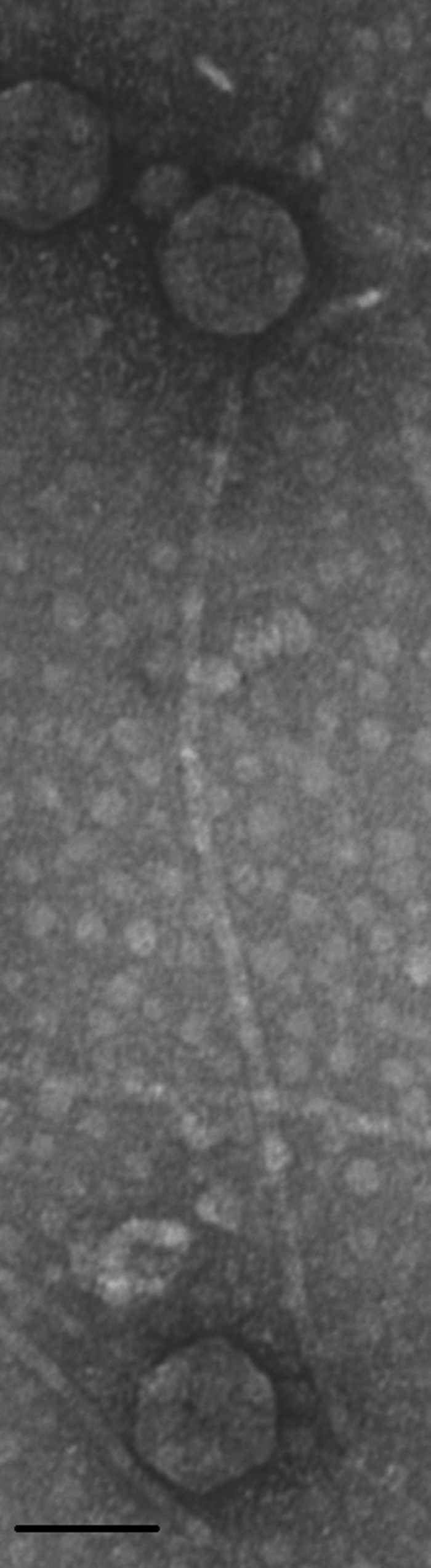
Electron micrograph of GTE7. Bar = 70 nm.
Fig. 2.
Circular map of the GTE7 genome. Arrows represent the putative genes and the direction in which they are transcribed. Modules are shaded in similar colors, and the inner circle indicates the genes encoded within the modules.
The Siphoviridae genomes sequenced so far share an organization of the packaging and structural proteins (1, 6). They encode either one or two DNA packaging proteins (17). In the GTE7 phage, the second ORF (terL) shares identity with the large terminase subunit of the ReqiDocB7 phage, which is required for DNA packaging into the phage head (17). No small terminase could be identified in the GTE7 phage based on sequence homologies. In other phage genomes, the small terminase is expressed from a gene upstream of the large terminase gene. The gene upstream of terL (i.e., terS) appears to be a fused version of two genes in phage ReqiDocB7. Neither has been assigned a putative function. Based on its location in phage GTE7, we hypothesize that this is the small terminase subunit.
The predicted amino acid sequences of orf3 to orf27 suggest that they encode the GTE7 phage structural proteins and many share a high level of identity with genes from phage ReqiDocB7 (20). The gene immediately downstream of terL, orf3, encodes the conserved motif DUF935, indicative of phage portal proteins. The major capsid protein is encoded by orf9 based on its similarity with other major capsid proteins and the characteristic conserved domain pfam03864. The putative genes orf4 to orf8 probably encode other capsid-related proteins since in all other Siphoviridae phage genomes, the structural genes are clustered into functional domains (1). Thus, genes encoding the phage head morphogenesis proteins are followed by tail-encoding genes (1). This arrangement is also observed in phage GTE7. Genes transcribed downstream of the major capsid protein, orf10 to orf15, encode proteins with no known function, but they share similarity to proteins in phage ReqiDocB7 (20). Again, it is hypothesized that these genes also encode phage structural proteins. The largest gene in the GTE7 phage genome is orf19, which shares identity to phage tape measure proteins (TMP) and contains the appropriate conserved domains found there (10). The genes upstream, orf17 and orf18, appear to be involved in tail assembly, where orf18 is translated via a conserved translational slippage mechanism, a common feature of phages (24). Siphoviridae genomes are typically arranged such that genes encoding the major tail protein precede the two tail assembly genes with translational slippage and are followed by TMP genes (1). Assuming the GTE7 phage fits this pattern, it seems reasonable to predict that its orf16 encodes the major tail protein.
The remainder of genes (orf20 to orf27) in this cluster may encode other GTE7 phage structural proteins, although their function is unknown. A conserved domain, PHA02579, identified in the predicted protein encoded by orf25, is often associated with phage proteins forming the virion baseplate (12).
Organization of the Siphoviridae phage lysis gene modules varies. All characterized phage lysis modules contain an endolysin and a holin gene (22). The GTE7 phage genome has three putative lysin genes downstream of the structural module, namely, orf28, orf29, and orf40 (Fig. 2). The predicted protein of orf28 has an amino acid sequence highly similar to that of an N-acetylmuramoyl-l-alanine amidase in a Micromonospora sp., with the pfam01510 motif characteristic of peptidoglycan recognition proteins (PGRP). The same motif is found in lysin proteins of other phages, including T7 and TPA2 (2, 9, 15). Immediately adjacent to orf28 is another suspected lysin-encoding gene, orf29. Orf29 is closely related to LysA proteins in Rhodococcus phages ReqiPine5 and ReqiDocB7 (20). It contains the pfam01551 diagnostic of M23 peptidase proteins. Orf40 is closely related to the LysB of Rhodococcus phage ReqiDocB7, with the pfam01083 motif reported in cutinase enzymes. While some phage genomes may encode two lysin genes (13), it is unusual for a phage to produce three lysins. No gene encoding a holin protein could be identified in the GTE7 genome.
The putative genes orf32 to orf102 are transcribed in the opposite direction of the first 31 genes. Most gene products in this cluster have no known function based on their predicted peptide sequences compared to those in public databases. orf37 encodes a DNA methylase, tentatively identified from the presence of a conserved pfam01555 motif and its encoded protein sequence similarity to the DNA methylase protein in Rhodococcus phage ReqiDocB7 (20). Orf47 is a predicted exonuclease from the presence of a pfam00929 domain (10). orf50 and orf97 are predicted to encode a putative helicase and primase, respectively. The predicted amino acid sequence of Orf50 possesses a conserved pfam00176 domain with no defined function, but which may be involved in DNA repair, recombination, and chromatin unwinding (4). Within the same cluster, orf46 encoding the beta subunit of DNA polymerase III was identified from its high level of similarity to the DnaN protein sequence in Rhodococcus phage ReqiDocB7 and from the presence of the conserved domain cd00140, which is characteristic (8).
The remainder of this gene string has no known function, although orf48, orf72, and orf102 encode proteins with conserved domains. Orf48 contains a pfam00176 domain characteristic of excisionase-type proteins. Genes orf71 and orf72 encode proteins with high similarities to those encoded by phage ReqiDocB7 genes (20). These were identified in ReqiDocB7 from the presence of conserved domains (20). In phage GTE7, orf72 encodes a conserved domain pfam07728 characteristic of an ATP-hydrolyzing domain. The Orf71 predicted sequence is significantly similar to the protein vWFA, encoded by Rhodococcus phage ReqiDocB7, but no conserved domain was seen in this protein in phage GTE7. In phage ReqiDocB7, a von Willebrand factor type A (vWFA) conserved domain was identified, suggesting that these genes evolved along a shared lineage. The gene orf102 encodes a COG4951 domain, commonly encountered in protein sequences of unknown function in bacteria.
The increasing availability of genome sequences of phages targeting the Actinobacteria is revealing novel insights into their evolution. Mycobacterium phages are the largest such group whose genomes have been sequenced (7). Of 70 complete sequences now available for Mycobacterium phages, several phylogenetically related groups are recognizable (7). Genome sequence data from Tsukamurella phage TPA2 (15) and Rhodococcus phage ReqiPine5 (20) showed that both can be assigned to the same group based on similarity, despite their lack of DNA sequence identity (7). However, Gordonia phage GTE2 (16) could not be assigned, and it has been grouped with phages called singletons, sharing no relationship with any other known phage (7). While phage GTE7 also fails to cluster with other groups, its genome has a high level of similarity to Rhodococcus phage ReqiDocB7. This suggests that the ReqiDocB7 and GTE7 phages may share an evolutionary lineage, despite clear differences in their host ranges. Phages GTE7 and ReqiDocB7 form a novel group, and perhaps the Mycobacterium phage grouping should be extended to include it.
The hosts that GTE7 infects all have hydrophobic mycolic acid-containing cell surfaces and stabilize foaming in laboratory-scale experiments (14). The ability to control a bacterium responsible for foam stabilization targeted by the GTE7 phage was pursued. Bacterial hosts producing scum (14) were excluded. The optical density at 600 nm (OD600) of cultures were all adjusted to ∼1.0 prior to addition of phage GTE7 (multiplicity of infection [MOI]= 0.3).
The OD600 of cultures exposed to phage GTE7 after 24 h of incubation at 30°C fell to an OD600 of ∼0.7, and the mycolata CFU/ml was reduced by at least 1,000-fold (data not shown). The phage had no impact on foam stabilization by Gordonia aichiensis (Raic22), which is not a host for the GTE7 phage. The reduction in OD600 was lower than expected, probably because considerable amounts of cell debris remained after lysis. Unlike similar GTE2 phage data (16), these results (Table 1) suggest that all GTE7 phage host bacteria retained some capability for foaming, but foam stability was markedly reduced upon phage exposure (Table 1). This difference in response is not surprising since the GTE7 host bacteria produce surfactants that markedly reduce foaming threshold values (14). Thus, any surfactant produced by these bacteria in culture would persist over into the foaming assay. Unstable foam formation is still a desirable operational outcome since it will dissipate rapidly and hence not accumulate in activated sludge reactors.
Table 1.
Influence of GTE7 on production of stable foams by selected mycolata strains under laboratory conditions
| Culture | Foaming scorea |
|
|---|---|---|
| Without phage | In the presence of GTE7a | |
| Gordonia aichiensis (Raic22)b | Stable foam (>15 cm) | Stable foam (>15 cm) |
| Gordonia terrae (Gter34/Ben601/Ben602) | Intermittent stable films | Fragile bubbles (1 cm) |
| Gordonia amicalis (Ben607) | Fragile bubbles (1 cm) | Fragile bubbles (1 cm) |
| Nocardia asteroides (Nast23) | Stable films (>8 cm) | Fragile bubbles (1 cm) |
Foaming scores are in accordance with the scale from Petrovski et al. (14).
This culture was used as a negative control. GTE7 does not lyse this host.
Nucleotide sequence accession number.
The genome sequence for Gordonia phage GTE7 has been deposited in GenBank under accession number JN035618.
Supplementary Material
Acknowledgments
The research was supported by the Australian Research Council (ARC) Linkage grant (LP0774913) together with Melbourne Water (David Gregory) and South East Water (Graham Short), who are thanked for their financial support. S. Petrovski was funded by the ARC Linkage and La Trobe University grants.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Brüssow H., Desiere F. 2001. Comparative phage genomics and the evolution of Siphovirdae: insights from dairy phages. Mol. Microbiol. 39:213–222 [DOI] [PubMed] [Google Scholar]
- 2. Cheng X., Zhang X., Pflugrath J. W., Studier F. W. 1994. The structure of bacteriophage T7 lysozyme, a zinc amidase and an inhibitor of T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 91:4034–4038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de los Reyes F. L. 2010. Foaming, p. 215–259 In Seviour R. J., Nielsen P. H. (ed.), Microbial ecology of activated sludge. IWA Publishing, London, United Kingdom [Google Scholar]
- 4. Eisen J. A., Kevis S., Hanawait P. C. 1995. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 23:2715–2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goodfellow M., Maldonado L. A. 2006. The families Dietziaceae, Gordoniaceae, Nocardiaceae and Tsukamurellaceae, p. 843–888 In Dworkin M., Flakow S., Rosenberg E., Schleifer K.-H., Stackebrandt E. (ed.), The prokaryotes: archaea. Bacteria: firmicutes, actinomycetes. Springer, New York, NY [Google Scholar]
- 6. Hatfull G. F., Cresawn S. G., Hendrix R. W. 2008. Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution. Res. Microbiol. 159:332–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hatfull G. F., et al. 2010. Comparative genomic analysis of 60 Mycobacteriophage genomes: genome clustering, gene acquisition and gene size. J. Mol. Biol. 397:119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Herendeen D. R., Kelly T. J. 1996. DNA polymerase III: running rings around the fork. Cell 12:5–8 [DOI] [PubMed] [Google Scholar]
- 9. Inouye M., Arnheim N., Sternglanz R. 1973. Bacteriophage T7 lysozyme is a N-acetylmuramyl-l-alanine amidase. J. Biol. Chem. 248:7247–7252 [PubMed] [Google Scholar]
- 10. Koonin E. V., Rudd K. E. 1994. A conserved domain in putative bacterial and bacteriophage transglycosylase. Trends Biochem. Sci. 19:106–107 [DOI] [PubMed] [Google Scholar]
- 11. Kragelund C., et al. 2007. Ecophysiology of mycolic acid-containing Actinobacteria (Mycolata) in activated sludge foams. FEMS Microbiol. Ecol. 61:174–184 [DOI] [PubMed] [Google Scholar]
- 12. Miller E. S., et al. 2003. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 67:86–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Payne K., Sun Q., Sacchettini J., Hatfull G. F. 2009. Mycobacteriophage lysin B is a novel mycolylarabinoglactan esterase. Mol. Microbiol. 73:367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Petrovski S., et al. 2011. An examination of the mechanisms for stable foam formation in activated sludge systems. Water Res. 45:2146–2154 [DOI] [PubMed] [Google Scholar]
- 15. Petrovski S., Seviour R. J., Tillett D. 2011. Genome sequence and characterization of the Tsukamurella phage TPA2. Appl. Environ. Microbiol. 77:1389–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Petrovski S., Seviour R. J., Tillett D. 2011. Characterization of the genome of the polyvalent lytic bacteriophage GTE2, which has potential for biocontrol of Gordonia-, Rhodococcus-, and Nocardia-stabilized foams in activated sludge plants. Appl. Environ. Microbiol. 77:3923–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rao V. B., Feiss M. 2008. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 42:647–681 [DOI] [PubMed] [Google Scholar]
- 18. Soddell J. A. 1999. Foaming, p. 161–202 In Seviour R. J., Blackall L. L. (ed.), Microbiology of activated sludge. Kluwer Academic Publishers, Dordrecht, Netherlands [Google Scholar]
- 19. Soddell J. A., Seviour R. J. 1990. Microbiology of foaming in activated sludge plants—a review. J. Appl. Bacteriol. 69:145–176 [Google Scholar]
- 20. Summers E. J., et al. 2011. Genomic and functional analysis of Rhodococcus equi phages ReqiPepy6, ReqiPoco6, ReqiPine5, and ReqiDocB7. Appl. Environ. Microbiol. 77:669–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thomas J. A., Soddell J. A., Kurtböke D. I. 2002. Fighting foam with phages. Water Sci. Technol. 46:511–664 [PubMed] [Google Scholar]
- 22. Wang I., Smith D. L., Young R. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799–825 [DOI] [PubMed] [Google Scholar]
- 23. Withey S. E., Cartmell E., Avery L. M., Stephenson T. 2005. Bacteriophages potential for application in wastewater treatment processes. Sci. Total Environ. 339:1–18 [DOI] [PubMed] [Google Scholar]
- 24. Xu J., Hendrix R. W., Duda R. L. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11–21 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.