Abstract
Plant and microbial community composition in connection with soil chemistry determines soil nutrient cycling. The study aimed at demonstrating links between plant and microbial communities and soil chemistry occurring among and within four sites: two pine forests with contrasting soil pH and two grasslands of dissimilar soil chemistry and vegetation. Soil was characterized by C and N content, particle size, and profiles of low-molecular-weight compounds determined by high-performance liquid chromatography (HPLC) of soil extracts. Bacterial and actinobacterial community composition was assessed by terminal restriction fragment length polymorphism (T-RFLP) and cloning followed by sequencing. Abundances of bacteria, fungi, and actinobacteria were determined by quantitative PCR. In addition, a pool of secondary metabolites was estimated by erm resistance genes coding for rRNA methyltransferases. The sites were characterized by a stable proportion of C/N within each site, while on a larger scale, the grasslands had a significantly lower C/N ratio than the forests. A Spearman's test showed that soil pH was correlated with bacterial community composition not only among sites but also within each site. Bacterial, actinobacterial, and fungal abundances were related to carbon sources while T-RFLP-assessed microbial community composition was correlated with the chemical environment represented by HPLC profiles. Actinobacteria community composition was the only studied microbial characteristic correlated to all measured factors. It was concluded that the microbial communities of our sites were influenced primarily not only by soil abiotic characteristics but also by dominant litter quality, particularly, by percentage of recalcitrant compounds.
INTRODUCTION
Plant community composition induces changes in microbial community structure and soil nutrient cycling and vice versa (3, 27). Thus, the chemical quality of plant inputs affects the microbial community structure because litter character can select for particular groups of microorganisms possessing the respective catabolic pathways required for its decomposition (45). The microbial community reacts not only to the added plant substrate but also to the soil chemical conditions by changing its structure and biomass turnover rate (31). Soil pH may serve as an integrating variable in these processes (38). Variations in soil pH and nutrient content may influence decomposition by limiting the growth or activity of microbial decomposers (5), but it can also affect plant community structure, which, in turn, determines the character of the organic matter (3).
Concordant patterns between plant communities, soil organic matter, and microbial communities along major environmental gradients have been demonstrated for deciduous forests (44, 55) and tundra grasslands (19, 60) at a continental or landscape level (38, 59) or within one relatively homogeneous area (55, 19, 58). In these studies only several bacterial phyla (Alphaproteobacteria, Acidobacteria, Bacteriodetes, Chloroflexi, and Actinobacteria) varied in their abundance and diversity under site-specific conditions, which were mostly characterized by pH. Among these organisms, Actinobacteria have long been known as efficient degraders of organic matter, including recalcitrant biopolymers (2, 4, 13, 42), and they were also known to enter the decomposition process in the later stages, thus at the period or sites in which the more recalcitrant compounds predominate (45, 4). The distribution and relative abundance of actinobacteria were found associated with soil pH and moisture deficit (38); they increased with soil N content (47) and followed individual plant species (46). Consequently, the actinobacterial community might reflect site-specific characteristics related to locally occurring decomposition pathways.
The present study investigated the relationships between plant and microbial communities and the soil characteristics at two forest and two grassland sites. The sites were selected to possess both similar and contrasting characteristics. In both forests, pine needles formed the major portion of recalcitrant litter, but they had contrasting soil pH and species richness. The two grassland soils had similar pHs but differed in bedrock composition, vegetation richness, and soil organic carbon and nitrogen contents.
Terminal restriction fragment length polymorphism (T-RFLP) profiles were used to characterize community composition, and real-time PCR was used for quantity assessments. To obtain more information about the metabolic contributions of actinobacteria, their community was assessed not only by 16S rRNA genes but also by horizontally transferred erm genes coding for rRNA methyltransferases, which confer resistance to secondary metabolites inhibiting protein synthesis, including macrolides, lincosamides, and streptogramin B (24). Profiles of extractable low-molecular-weight compounds were determined using high-performance liquid chromatography (HPLC) for characterizing the soluble soil chemical environment and thus providing information about decomposition chemistry. The aim was to explore links between plant and microbial communities, together with the soil chemistry, occurring among and within selected sites, with close attention paid to participation of actinobacteria.
MATERIALS AND METHODS
Sites and soil sampling.
Three sites located in the Czech Republic and one in Austria were chosen. Soil pH, bedrock, and vegetation cover contrasted the sites that otherwise shared dry local conditions given by their exposure toward the south or west (Table 1). The two forest sites, Merkenstein (M) and Zakopana (Z), were covered with pine forest, Pinus nigra Arnold and P. sylvestris L., respectively; they differed in soil pHs. The M site forest is azonal and considered natural without human impact, with trees about 200 years old. The Z site is located in a nature protected area and has limited management; the trees are 50 to 200 years old. The two grasslands were both alkaline; Oblik (O) vegetation was dominated by forbs while Devin (D) was dominated by grasses. Both grassland sites are located in a nature protected area. They were originally pastures, but in the past several decades they have been managed by mowing and only partially by goat grazing. At each site, five soil samples were collected, approximately 1 m apart from each other in an area of 3 by 3 m during October 2007. A sample of 10 by 10 cm (approximately 1 kg) was cut from the upper horizon, homogenized, and divided into three subsamples for subsequent analyses. For microbial analysis, the soil was placed in 50-ml sterile plastic tubes, cooled during transportation, and stored at −20°C upon arrival at the laboratory. For chemical analyses, 100-ml glass bottles were filled with approximately 60 ml of loose soil and overlaid with a mixture of acetone-water-trifluoroacetic acid (80:19:1, vol/vol/vol) (7). The remaining soil was placed in a plastic bag for determination of soil pH, organic C, and total N contents.
Table 1.
Sites, soil characteristics, and diversity of vegetation and actinobacteria
| Parameter | Value for the parameter at the indicated sitea |
|||
|---|---|---|---|---|
| Devin | Oblik | Merkenstein | Zakopana | |
| Country, region | Czech Republic, South Moravia | Czech Republic, North Bohemia | Austria, Lower Austria | Czech Republic, Central Bohemia |
| Slope | Southwest | West | West | South |
| Coordinates | 48°51′55″N, 16°38′35″E | 50°24′40″N, 13°48′15″E | 47°57′58″N, 16°08′37″E | 50°31′00″N, 15°04′00″E |
| Type | Grassland | Grassland | Forest | Forest |
| pH ± SD | 7.8 ± 0.07 A | 7.9 ± 0.04 B | 8.1 ± 0.17 C | 3.7 ± 0.13 D |
| Bedrock | Limestone | Basalt | Dolomite | Sandstone |
| Silt (%) | 25 | 29 | 10 | 5 |
| Clay (%) | 3 | 6 | 0 | 0 |
| C ± SD (%) | 15.2 ± 2.3 A | 4.3 ± 0.1 B | 21.9 ± 1.1 C | 38.9 ± 1.1 D |
| N ± SD (%) | 1.3 ± 0.2 A | 0.3 ± 0.02 B | 0.7 ± 0.1 B | 1.3 ± 0.04 A |
| C/N ratio | 11.97 | 13.56 | 33.70 | 29.56 |
| Tree cover (%) | 0 | 0 | 50 | 70 |
| Shrub cover (%) | 0 | 0 | 10 | 0 |
| Herb cover (%) | 75 | 75 | 90 | 15 |
| Forb cover (% herbs) | 29 | 83 | 64 | 67 |
| Grass cover (% herbs) | 71 | 17 | 36 | 33 |
| Fisher's α (plants) | 5.48 | 18.46 | 20.82 | 0.7007 |
| H′ (log e; plants) | 1.925 | 3.032 | 2.957 | 1.05 |
Letters A to D indicate the statistical relations between sites (P < 0.05) based on ANOVA and Tukey posthoc tests.
Plant diversity.
Plant diversity was assessed at all sites using a seven-level quantity evaluation using the following scale: 1, 2, 3, 4, 5, r (rare), and + (present) for the least-numerous species (8).
Soil pH, total carbon, and nitrogen.
Soil particle size was determined by standard sieving method using about 1 kg of soil. Soil pH was measured by a Multi 350 WTW glass electrode in a soil-water extract for which 20 g of soil was extracted with 50 ml of distilled water and set at room temperature for 12 h. Organic C and total N were determined by high-temperature combustion followed by gas separation in a Vario Max CNS apparatus (Elementar Analysensysteme, Hanau, Germany).
HPLC analysis of soluble low-molecular-weight soil metabolites.
Soil samples were extracted with approximately 50 ml of acetone-water-trifluoroacetic acid (80:19:1, vol/vol/vol) at room temperature for 2 to 3 days. The extracts were filtered and vacuum concentrated. Portions of the extract (∼20 mg) were dissolved in water. The resulting aqueous suspension was chromatographed over Amberlite XAD 1180 (2 to 3 g of aqueous suspension; diameter of glass column, 1 cm). Fifty ml of water (MilliQ quality) was used as effluent for the first (hydrophilic) fraction; 75 ml of absolute ethanol was used for the second (lipophilic) fraction. The ethanolic fraction was evaporated and diluted in water at a concentration of 10 mg ml−1. The HPLC system was a Dionex Summit (Dionex, Sunnyvale, CA) equipped with a photodiode array detector and a Famos autosampler (LC Packings, San Francisco, CA). The column was a Phenomenex Synergi Max C12 (150 by 2 mm; 4-μm particle size). The column oven was adjusted to 40°C, and the flow rate was 0.2 ml min−1. Solvent A was water–methanol–o-phosphoric acid (9:1:0.5, vol/vol/vol); solvent B was pure MeOH. The gradient started with 100% of solvent A for 2 min and then increased linearly to 100% of solvent B within 98 min. The final concentration was held for a further 10 min. Five microliters of each sample was injected. UV spectra were recorded from 450 to 220 nm. The UV spectra were compared to an in-house library of reference spectra and to literature (35) for tentative assignments. The principal component analysis (PCA) was conducted using the HPLC signal obtained at 229 nm of data points collected at intervals of 0.65-s retention time from 5 to 107 min.
Extraction of soil DNA.
Soil DNA was extracted by the SK method described by Sagova-Mareckova et al. (51), which used bead beating and phenol-chloroform extraction, followed by purification with CaCl2 and a GeneClean minicolumn (Q-Biogene, Heidelberg, Germany).
PCR amplification of 16S rRNA and erm genes.
The primers are listed in Table 2. All PCRs were performed on a TGRADIENT Thermocycler (Whatman Biometra, Göttingen, Germany). Fifty nanograms (in approximately 1 to 3 μl) of template DNA was preincubated with 3 μl of bovine serum albumin (10 mg ml−1) and 6 μl of Tris-EDTA (pH 8) at 90°C for 1 min before addition to the PCR mix (25). PCR amplification for the 16S rRNA gene consisted of an initial denaturation at 94°C for 2 min, followed by 35 cycles of 94°C for 45 s, annealing at 57°C for 45 s, and 1 min 30 s of extension at 68°C, with a final extension step at 68°C for 5 min. The PCR protocol for the erm gene amplification consisted of an initial denaturation at 94°C for 3 min, followed by 35 cycles of 94°C for 1 min, annealing at 56°C for 30 s, and 30 s of extension at 68°C, with a final extension step at 68°C for 5 min. LA polymerase (Top-Bio, Prague, Czech Republic) was used. Unlabeled PCR products were purified using a QIAquick PCR purification kit (Qiagen, Hilden, Germany), cloned to the pGEM-T Vector (Promega, Madison, WI), and sequenced from universal primers (Macrogen, South Korea), while labeled ones were subjected to the same purification procedure and subsequent T-RFLP analysis. Corrected clone sequences (Chromas; Technelysium, Tewantin, Queensland, Australia) were NAST aligned (15), checked for occurrence of chimeric sequences by Bellerophon, version 3 (29), and assigned to the existing taxa using the Ribosomal Database Project (RDP) classifier tool (56). Initial alignments of partial 16S rRNA gene (1,076 to 1,126 bp) and erm gene (322 to 409 bp) sequences were performed using Muscle, version 3.6 (17). Phylogeny was inferred by the neighbor-joining method based on the Jukes-Cantor distance matrix (1,000 bootstrap resamplings) in the Phylip, version 3.67, package (20).
Table 2.
Primers
| Experiment | Primer | Sequence (5′–3′) | Target | Label | Reference |
|---|---|---|---|---|---|
| T-RFLP + cloning | 16Seu27f | AGAGTTTGATCMTGGCKCAG | Bacteria, 16S rRNA | HEX | 9 |
| 783ra | CTACCAGGGTATCTAATCCTG | Bacteria, 16S rRNA | 52 | ||
| 783rb | CTACCGGGGTATCTAATCCCG | Bacteria, 16S rRNA | 52 | ||
| 783rc | CTACCCGGGTATCTAATCCGG | Bacteria, 16S rRNA | 52 | ||
| 1114rACT | GAGTTGACCCCGGCRGT | Actinobacteria, 16S rRNA | 36 | ||
| 16Seu27f | AGAGTTTGATCMTGGCKCAG | Actinobacteria, 16S rRNA | FAM | 9 | |
| 16Sact623r | ACACCAGGAATTCCAGTCTC | Actinobacteria, 16S rRNA | 51 | ||
| rB1f | ARCWCGGYCAGAAYTTYCT | erm | HEX | 9 | |
| rB1r | CGSGCSACYTCCCAYTG | erm | 9 | ||
| qPCR | ITS1F | CTTGGTCATTTAGAGGAAGTAA | Fungi, 16S rRNA | 22 | |
| ITS4 | TCCTCCGCTTATTGATATGC | Eukaryota, 23S rRNA | 57 | ||
| Eub338 | ACTCCTACGGGAGGCAGCAG | Bacteria, 16S rRNA | 37 | ||
| Actino235 | CGCGGCCTATCAGCTTGTTG | Actinobacteria, 16S rRNA | 54 | ||
| Eub518a | ATTACCGCGGCTGCTGG | Bacteria, 16S rRNA | 43 |
Eub518 was used as a reverse primer with both Eub338 and Actino235.
Terminal restriction fragment length polymorphism analysis.
Double-color T-RFLP was used for the bacterial community analysis (32). Briefly, PCR amplicons of the 16S rRNA gene region with primers labeled with the fluorescent dye hexachlorofluorescein (HEX) for bacteria and 6-carboxyfluorescein (FAM) for actinobacteria were mixed in ratio of 2:1 and cleaved by AluI. The erm gene PCR amplicon was cleaved by MspI. After inactivation of the restriction enzymes and purification with Sigma Spin Post-Reaction Clean-Up columns (Sigma-Aldrich, St. Louis, MO), 5 μl of cleaved PCR product was mixed with 15 μl of HiDi-Formamide (Applied Biosystems, Carlsbad, CA) and 0.3 μl of 500 ROXTM size standard (Applied Biosystems, Carlsbad, CA). The samples were subjected to fragment analysis on a 96-capillary sequencer (Applied Biosystems, Foster City, CA). T-RFLP analyses were performed in one run for all samples. Crude profiles were filtered to the largest T-RF peaks, representing 95% of the total of peak heights (areas) to eliminate the noise intermittently exceeding the cutoff limit (1).
qPCR.
Primers used in quantitative PCR (qPCR) are listed in Table 2. Quantification of the total bacteria and actinobacteria by the 16S rRNA gene copy number and of fungi by the internal transcribed spacer (ITS) I and II copy number in the extracted soil DNA was performed using an iCycler iQ Multicolor Real Time PCR Detection System (Bio-Rad Laboratories, Veenendaal, The Netherlands). SYBR green (Bio-Rad Laboratories, The Netherlands) was used as a double-stranded DNA (dsDNA) binding dye. Baseline and threshold calculations were performed with the I-Cycler software (version 3.1). Amplification was done with SYBR green Super Mix (Bio-Rad Laboratories, Veenendaal, The Netherlands). The amplification consisted of 40 cycles including denaturation (30 s at 95°C), annealing (35 s at 54°C), and elongation (45 s at 72°C). 16S rRNA gene and ITS region sequences cloned to the StrataClone cloning vector (Agilent Technologies, Santa Clara, CA) were used for calibration. The inhibition was tested by serial DNA dilution from each site. Melting curves were recorded to ensure qPCR specificity. The qPCR measurements were done in duplicate.
Statistical analyses.
Differences between sites in soil pH, C and N contents, T-RF numbers, and qPCR were tested using analysis of variance (ANOVA) and a Tukey posthoc test. The HPLC absorption signals were baseline corrected and normalized by division through the median of the absorption values. A principle component analysis (PCA) was carried out using Simca-P, version 11, software (Umetrics, Umea, Sweden). Similarly, the T-RFLP projection was done using Sammon's multidimensional scaling from the R computing environment.
For the T-RFLP and HPLC profiles, the Manhattan metric (sum of absolute differences) was used to calculate the distance matrices. For the T-RFLP, HPLC profiles, and testing the phylogeny of 16S rRNA and erm genes, analysis of similarities (ANOSIM) and its modifications were used (34, 39). All the computations were conducted with R (http://www.r-project.org).
The correlations between HPLC profiles, T-RFLP profiles, abundances of bacteria, actinobacteria, and fungi (qPCR), and soil content of organic C and total N were tested using Spearman's rank correlation coefficient on the distance matrices as a measure of similarity because the scales of the analyses were generally different. A Mantel test (or null correlation of the Spearman's correlation coefficient) was used to make a formal test of null correlation against positive correlation. The site-specific results were further tested to see if the positive correlation can be explained purely by the specificity of the sites, using a stratified Mantel test with the sites as strata.
Nucleotide sequence accession numbers.
The sequences obtained in this study were deposited in GenBank under accession numbers HM480603 to HM480705 for 16S rRNA genes of actinobacteria and HM480498 to HM480602 for erm genes.
RESULTS
Soil pH, carbon, and nitrogen content.
The sites differed in carbon (C) and nitrogen (N) contents. The O site, a basalt grassland, had the lowest content of C and N. The C/N ratio was low for the O site and the limestone grassland D site, but it was high for pine forests in the limestone M site and the sandstone Z site. Also, the grassland sites were higher in their small particle contents, silt, and clay than the forest sites. The pHs of soils from sites O, M, and D were alkaline, and the soil from site Z was acidic (Table 1).
Plant diversity.
The 9-m2 plot at the O site contained 35 plant species from 15 plant families, the plot at the M site had 34 plant species of 19 plant families, and the plot at the D site had 15 plant species from 8 families. In contrast, the plot at the Z site had only 3 plant species from 2 families. Consequently, the calculated diversity indices were highest at sites O (Fisher's α = 18.46, Shannon's H′ = 3.032) and M (α = 20.82, H′ = 2.957); D was less diverse (α = 5.48, H′ = 1.925), and the lowest diversity of the investigated sites was at Z (α = 0.70, H′ = 1.055) (Table 1). The O site was dominated by forbs, which covered an area four times as large as that covered by grasses. The D site was dominated by Poaceae grasses and Cyperaceae. The diversity of herbs at the M site was comparable to that of the O site, whereas the acidic Z site was poor. Here, shrubs belonging to the family of Ericaceae dominated (see Table S1 in the supplemental material).
HPLC analyses of extractable low-molecular-weight compounds.
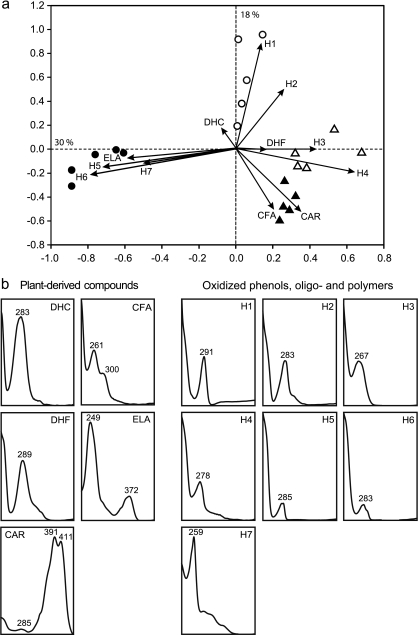
The HPLC profiles of soil extracts differed significantly between sites (P < 0.001). The O site profiles clustered separately from the other sites by the first PCA axis, which explained 30% of variation (Fig. 1a). The D site profiles were again separated from those of the sites M and Z by the second PCA axis, which explained 22% of variation. In a closer evaluation, the chromatograms from sites D, M, and Z showed rather similar peak patterns due to the absence of prominent peaks, while the chromatograms from the O site showed a series of notable peaks. The chromatograms of all sites contained peaks with UV spectra characteristics for both phenolic and nonphenolic compounds, but only peaks with UV spectra of phenolic compounds showed vectors supporting the obtained ordination of the sites (Fig. 1b). Among these vectors, plant-derived compounds, such as a dihydroflavonoid (DHF), a dihydrochalcone (DHC), coniferyl alcohol (CFA), and an ellagic acid derivative (ELA), could be tentatively identified. The only aliphatic but highly unsaturated compound among these was a carotenoid (CAR) found in the Z site. In addition to the above, a number of site-specific peaks (Fig. 1a, H1 to H7) were assigned as oxidized phenols that originated during decomposition. Their retention time suggested that they constituted oxidized phenolic oligomers.
Fig. 1.
(a) HPLC-UV profiles of extractable low-molecular-weight compounds at four sites: Z (▴), M (▵), D (○), and O (●). PCA was based on a normalized signal at 229 nm (for details, see Materials and Methods). (b) UV spectra of peaks (compounds) that supported the ordination obtained. The compounds were classified either as plant-derived or oxidized phenolic oligomers. DHF, dihydroflavonoid; DHC, dihydrochalcone; CFA, coniferyl alcohol; ELA, ellagic acid derivative; CAR, carotenoid; H1 to H7, site-specific oxidized phenols.
Taxonomic diversity by T-RFLP.
The number of terminal restriction fragments (T-RF) for total bacteria and actinobacteria differed between sites. The highest average number of T-RFs for total bacteria was found in the Z site (80 T-RFs) while the lowest bacterial T-RF number was obtained in the D site (64 T-RFs). For actinobacteria, the highest numbers of T-RFs were found in the M (57 T-RFs) and O (56 T-RFs) sites, while they were again lowest in the D site. Differences were found for the number of T-RFs common to both bacterial and actinobacterial profiles, which was significantly lower at site D than at the M site (P = 0.0013) and also than at the O site (P = 0.0031) (see Table S2 in the supplemental material). Furthermore, using the double-color analysis of the T-RFLP profiles, the percentage of actinobacterial T-RFs present in the universal profiles was 31% in D, 32% in Z, 40% in M, and 44% in O.
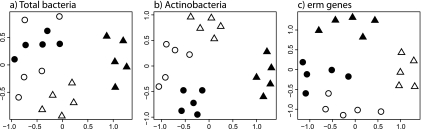
Bacterial (Fig. 2a) and actinobacterial (Fig. 2b) communities differed significantly between sites (both, P < 0.001 by ANOSIM) by the T-RFLP analysis. The first axis distinguished the communities by soil pH; the second axis separated the high-pH forest site from the high-pH grassland sites, the individual samples of which appeared mixed and close together. Discrimination of the two grassland sites was more pronounced in the actinobacterial communities.
Fig. 2.
T-RFLP profiles of 16S rRNA genes of bacteria (a) and actinobacteria (b) and of erm genes (c) in soil communities sampled at four sites: Z (▴), M (▵), D (○), and O (●). Sammon's multidimensional scaling projection based on Manhattan distance matrices was calculated for all profiles.
Identification of actinobacteria by cloning sequencing.
By cloned sequences, Micromonosporaceae and Pseudonocardiaceae were typical for the neutral to alkaline pH sites, and Nocardioidaceae and Propionibacteriaceae were typical for the grassland sites. At the low-pH site Z, the sequences showed the most distinct actinomycete community because of a large site-specific actinobacterial clade determined there. The O and M sites showed only small site-specific sequence clusters while all actinobacterial sequences of the D site always clustered together with those of the other sites (see Fig. S1 in the supplemental material). Similar to the T-RFLP results, the highest number of actinobacterial operational taxonomic units (OTUs) defined at the 90% similarity level was at the O (16) and M (12) sites, and lowest was at the Z site (5) (see Fig. S3b in the supplemental material). The Libshuff test showed marginally significant differences of actinobacterial sequences between sites Z and D and between Z and O.
Erm genes by T-RFLP and by cloning and sequencing.
The highest number of T-RFs of the resistance genes (92 T-RFs) was determined for D, and the lowest (53 T-RFs) was for site M (see Table S2 in the supplemental material), quite in contrast to the number of peaks representing actinobacterial richness of the 16S rRNA genes. The T-RFLP analysis of the erm genes showed that their variability was significantly related to a particular site (P < 0.001). Yet the separation of the sites was different than that for the 16S rRNA genes because the high-pH forest site M was separated along the first axis from all the other sites (Fig. 2c). The second axis separated the low-pH forest site from the grassland sites. Cloning and sequencing of the erm genes confirmed the site specificity of the resistance gene pool; in particular, the sequences of the forests Z and M formed distinctly separated clusters (see Fig. S2 in the supplemental material). Comparisons of the acquired sequences with the database revealed that those coming from site M were mostly of the producer type (71%), while this proportion was only 32% at site O, 18% at D, and 1% at the Z site. The erm gene genotype clusters defined at the 70% similarity level showed the highest number occurring at the O site. The highest local specificity of these genotypes occurred at the O and Z sites (see Fig. S3c). The Libshuff test showed significant differences in the erm gene sequences between the sites D and O.
Quantitative analyses of soil microorganisms.
Abundances of total bacteria, actinobacteria, and fungi differed significantly between sites. The lowest abundances of all microorganism groups were found at the O site, showing significant differences for bacteria and actinobacteria from levels at the D and M sites. Fungi were more abundant in the forest sites, with a significant difference occurring between the O and Z sites (Fig. 3).
Fig. 3.
Quantitative real-time PCR of total bacteria, actinobacteria, and fungi determined at four sites with respective standard deviations. Significant differences by ANOVA and a Tukey posthoc test for bacteria were between O and D (P = 0.008) and O and M (P = 0.04); for actinobacteria they were between O and D (P = 0.001), O and M (P = 0.002), Z and D (P = 0.005), and Z and M (P = 0.01); for fungi they were between O and Z (P = 0.02).
Relationships among chemical environment and microbial communities.
Two types of relationships were assessed (Table 3): the first one (before adjustments) demonstrated a comparison among variables of all sites while the second one (after adjustments using the stratified Mantel test) demonstrated a comparison within each site. The relationships of the first group showed a significant correlation between HPLC, T-RFLP profiles, and bacterial, actinobacterial, and fungal abundances. Also, T-RFLP profiles of erm genes were significantly correlated with T-RFLP profiles of bacteria and actinobacteria and also with abundances of actinobacteria and fungi. Significant correlations of all T-RFLP profiles and fungal abundances were also found with organic C content while only actinobacterial T-RFLP profiles were correlated with N content. Finally, soil pH correlated positively with the HPLC and T-RFLP profiles of bacteria, actinobacteria, and fungi while it was negatively correlated with fungal qPCR and N and C content.
Table 3.
Relationships between microbial and chemical characteristics: values of Spearman's correlation coefficient
| Experiment type and target or parameter | Spearman's correlation coefficient fora: |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T-RFLP |
qPCR |
N content | Corg contentb | pH | HPLC | |||||
| Actinobacteria | Bacteria | erm genes | Actinobacteria | Bacteria | Fungi | |||||
| T-RFLP | ||||||||||
| Actinobacteria | 0.93 AA,B | 0.69 AA | 0.27 A | 0.19 A | 0.39 A | 0.27 A | 0.88 AA | 0.79 AA | 0.31 A | |
| Bacteria | 0.93 AA, B | 0.67 AA | 0.16 A | 0.15 | 0.26 A | 0.18 | 0.84 AA | 0.78 AA,B | 0.27 A | |
| erm genes | 0.69 AA | 0.67 AA | 0.15 A | 0.03 | 0.17 A | 0.14 | 0.5 AA | 0.54 AA | 0.32 AA | |
| qPCR | ||||||||||
| Actinobacteria | 0.27 A | 0.16 A | 0.15 A | 0.9 AA,B | 0.56 A,B | 0.45 | 0.31 | −0.25 | 0.39 AA | |
| Bacteria | 0.19 A | 0.15 | 0.03 | 0.9 AA,B | 0.6 A,BB | 0.31 | 0.29 | −0.27 | 0.48 AA | |
| Fungi | 0.39 A | 0.26 A | 0.17 A | 0.56 A,B | 0.6 A,BB | 0.53 | 0.87 AA | −0.59 A | 0.62 AA | |
| N content | 0.27 A | 0.18 | 0.14 | 0.45 | 0.31 | 0.53 | 0.69 A,BB | −0.89 AA | 0.31 A | |
| Corg contentb | 0.88 AA | 0.84 AA | 0.5 AA | 0.31 | 0.29 | 0.87 AA | 0.69 A,BB | −0.78 A | 0.45 A | |
| pH | 0.79 AA | 0.78 AA,B | 0.54 AA | −0.25 | −0.27 | −0.59 A | −0.89 AA | −0.78 A | 0.19 A | |
| HPLC | 0.31 A | 0.27 A | 0.32 AA | 0.39 AA | 0.48 AA | 0.62 AA | 0.31 A | 0.45 A | 0.19 A | |
For the values in boldface, the Spearman's correlation coefficient was used to test the null correlation. The other relationships were tested using a Mantel test to test the null correlation against positive correlation. P values are indicated as follows: A, 0.01 < P < 0.05; AA, 0.01 < P < 0.001. Adjusted P values (stratified Mantel test) are indicated as follows: B, 0.01 < P < 0.05; BB, 0.01 < P < 0.001.
Corg, organic carbon.
The relationships of the second group (within sites) showed a significant correlation between C and N content. Abundances of bacteria, actinobacteria, and fungi were significantly correlated with each other, and a significant correlation was determined between pH and the bacterial T-RFLP profile (Table 3).
Comparison of plant diversity with richness of actinobacterial 16S rRNA and erm genes showed parallels with the above-described links among the microbial communities' structure, C and N content, and pH. Sites which differed in plant diversity tended to differ in the same way also in their actinobacterial 16S rRNA and erm gene richness (established at 90% and 70% sequence similarity, respectively.) In particular, the M and D sites were similar while the Z site differed from the other three sites in vegetation cover, actinobacteria, and erm genes (see Fig. S3 in the supplemental material).
DISCUSSION
The HPLC profiles of soil extracts reflected the soil chemical environment mostly corresponding to soil pH but also reflecting plant community composition. The low-pH site Z showed no characteristic peaks of oxidized phenolic oligomers but only those resembling unmodified plant-derived compounds, suggesting a low rate of oxidative decomposition typical for low-pH soils (6). In the alkaline sites, the HPLC profiles of M and D sites were closer to the Z site, reflecting a significant proportion of recalcitrant litter, while at the O site soil extracts contained prominent peaks that might be attributed to high occurrence of oxidized, lower-molecular-weight phenol oligomers (12). That is consistent with poorly decomposable pine needles influencing decomposition at the M site (50), with relatively recalcitrant litter of grasses dominating the plant community at the D site (11), and with readily decomposable litter consisting mostly of forbs at the O site. The relationships were further supported by the highest C/N ratio and C content occurring at the Z site, where recalcitrant litter accumulated, and the lowest C and N content at the O site, where the high quality of plant litter and nutrient-rich bedrock suggested a fast decomposition rate. The HPLC profiles of low-molecular-weight compounds were correlated with all measured microbial and soil characteristics, thus supporting the observed mutual interactions between soil organic matter chemistry, soil characteristics, and long-term decomposition processes (23). The characteristic peaks of HPLC profiles separated the sites by soil pH and litter quality, therefore specifying their dominant decomposition pathways as a locally specific combination of decomposition catalyzed by microbially produced exoenzymes or abiotically by transition metals (30, 10, 18).
Soil pH has been found to have a strong influence on composition of soil bacterial communities in many different environments by using various methods over different scales (38, 48, 58). In our study, the communities of bacteria and actinobacteria were primarily distinguished by pH according to both T-RFLP and cloning combined with sequencing. The effect of pH on the composition of bacterial community was significant even within each site, reflecting very small pH changes (the order of 0.1) occurring within distances of a meter, similar to the observations of Yergeau et al. (58) over larger distances. However, the erm genes showed locally specific patterns by T-RFLP and by cloning and sequencing, regardless of the soil pH, proving their horizontal transfer under local selection pressures (14).
The microbial community structure was also strongly responding to the recalcitrant litter. At the Z site, the most diverse bacterial communities (based on the number of T-RFs) and nonproducer type erm genes suggested that the diversification of bacteria was conditioned by large amounts of recalcitrant litter (16). The Z site conditions selected for a small but very specific actinobacterial community (lowest richness and lowest percentage of actinobacteria in the community by quantity), which consisted of Acidimicrobidae and sequences related to the recently described Trebon clade occurring in low-pH soil (33). At the M and D sites, the recalcitrant litter led to relatively high C content and corresponded with high abundance of both bacteria and actinobacteria (41) and also with the highest proportion of actinobacteria in the bacterial community. In addition, the M and D sites showed the highest richness of the producer and nonproducer types of erm genes, respectively, suggesting high secondary metabolic activity and preferred conditions for actinobacteria. The high number of T-RF peaks and distinct clusters of erm genes occurring in soils with a high pH and recalcitrant carbon also agree with observations demonstrating that secondary metabolite production changed with the type of carbon substrate and increased with the addition of the recalcitrant lignin (53). The highly diverse and decomposable litter at the O site supported the highest percentage of actinobacterial T-RFs in double-color profiles and also the highest richness of erm gene sequences. The data agreed with a previous finding that nutrient-poor soils sustain a lower quantity but larger diversity of decomposers (40).
The parallel increase in abundances of all the microbial groups would suggest limitation of resources (46, 58). The strong correlation between C and N occurring within each site indicated that local nutrient conditions were relatively homogeneous and most likely dependent on soil chemical conditions rather than on the litter of the nearby plants (30, 55). The quality of C appeared to be more important than its quantity because the HPLC profiles of extractable compounds were correlated with abundances of bacteria, fungi, and actinobacteria, while only in alkaline pH, correlation between C content and abundances was observed, similar to the report of Fierer et al. (21). For fungi, however, it seemed that the quantity of recalcitrant litter predicted their abundance regardless of the soil pH (49).
Finally, it is important to consider that different combinations of PCR primers/DNA extraction techniques recover different species as well as higher taxa (28). The presented results were based on one DNA extraction method; however, the method was developed to get a higher percentage of actinobacteria (51). Also, a limited set of primers was employed, but our approach can benefit from complementary use of primers for bacterial and actinobacterial 16S rRNA and additional use of the erm genes to access microbial community composition. T-RFLP can also introduce various biases; however, the relative comparisons of communities can be assessed reliably (26).
In conclusion, despite the diverseness of the sites, the microbial communities could be considered to be linked because their composition and abundances were often correlated. Similarly, the observed microbial community parameters were correlated to the chemical environment described by the extractable low-molecular-weight compounds. Actinobacterial community composition was the only studied microbial community characteristic correlated to all measured environmental factors.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by Academic Cooperation and Mobility grants (Aktion MEB 060817 and MEB 061111) awarded by the Ministry of Education, Youth and Sports of the Czech Republic and a Czech Science Foundation grant (P201/11/P290). Part of this research was supported also by Institutional Research Concepts MZE0002700604 and MSM002162839. Zdenek Kamenik was supported by an Ernst Mach grant from the Austrian Ministry for Science and Research.
We thank Katrin Glaser for her kind help with processing qPCR data, Jan Hubert for helpful discussions, and Steve Ridgill for English revisions.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Abdo Z., et al. 2006. Statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 8:929–938 [DOI] [PubMed] [Google Scholar]
- 2. Ball A. S., Godden B., Helvenstein P., Penninck M. J., McCarthy A. J. 1990. Lignocarbohydrate solubilization from straw by actinomycetes. Appl. Environ. Microbiol. 56:3017–3022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bardgett R. 2005. The biology of soil: a community and ecosystem approach. Oxford University Press, Oxford, United Kingdom [Google Scholar]
- 4. Bastian F., Bouziri L., Nicolardot B., Ranjard L. 2009. Impact of wheat straw decomposition on successional patterns of soil microbial community structure. Soil Biol. Biochem. 41:262–275 [Google Scholar]
- 5. Berg B. 2000. Litter decomposition and organic matter turnover in northern forest soils. Forest Ecol. Manag. 133:13–22 [Google Scholar]
- 6. Berg B., Laskowski R. 2006. Litter decomposition: a guide to carbon and nutrient turnover. Academic Press, Burlington, MA [Google Scholar]
- 7. Bonsall R. F., Weller D. M., Thomashow L. S. 1997. Quantification of 2,4-diacetylphloroglucinol produced by fluorescent Pseudomonas spp. in vitro and in the rhizosphere of wheat. Appl. Environ. Microbiol. 63:951–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Braun-Blanquet R. 1964. Pflanzensoziologie. Grundzüge der Vegetationskunde. Springer, Vienna, Austria [Google Scholar]
- 9. Čermák L., et al. 2008. Bacterial communities of two contrasting soils reacted differently to lincomycin treatment. Appl. Soil Ecol. 40:348–358 [Google Scholar]
- 10. Conrad R., Seiler W. 1985. Characteristics of abiological CO formation from soil organic matter, humic acids, and phenolic compounds. Environ. Sci. Technol. 19:1165–1169 [DOI] [PubMed] [Google Scholar]
- 11. Cornelissen J. H. C., Thompson K. 1997. Functional leaf attributes predict litter decomposition rate in herbaceous plants. New Phytol. 135:109–114 [DOI] [PubMed] [Google Scholar]
- 12. Cornwell W. K., et al. 2008. Plant species traits are the predominant control on litter decomposition rates within biomes worldwide. Ecol. Lett. 11:1065–1071 [DOI] [PubMed] [Google Scholar]
- 13. Crawford D. L. 1978. Lignocellulose decomposition by selected Streptomyces strains. Appl. Environ. Microbiol. 35:1041–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davelos Baines A. L., Xiao K., Kinkel L. L. 2007. Lack of correspondence between genetic and phenotypic groups amongst soil-borne streptomycetes. FEMS Microbiol. Ecol. 59:564–575 [DOI] [PubMed] [Google Scholar]
- 15. DeSantis T. Z., et al. 2006. NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res. 34:W394–W399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dilly O., Bloem J., Vos A., Munch J. C. 2004. Bacterial diversity in agricultural soils during litter decomposition. Appl. Environ. Microbiol. 70:468–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Edgar R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ekschmitt K., Liu M., Vetter S., Fox O., Wolters V. 2005. Strategies used by soil biota to overcome soil organic matter stability—why is dead organic matter left over in the soil? Geoderma 128:167–176 [Google Scholar]
- 19. Eskelinen A., Stark S., Männistö M. 2009. Links between plant community composition, soil organic matter quality and microbial communities in contrasting tundra habitats. Oecologia 161:113–123 [DOI] [PubMed] [Google Scholar]
- 20. Felsenstein J. 1989. PHYLIP: phylogeny inference package (version 3.2). Cladistics 5:164–166 [Google Scholar]
- 21. Fierer N., Grandy A. S., Six J., Paul E. A. 2009. Searching for unifying principles in soil ecology. Soil Biol. Biochem. 41:2249–2256 [Google Scholar]
- 22. Gardes M., Bruns T. D. 1993. ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113–118 [DOI] [PubMed] [Google Scholar]
- 23. Grandy A. S., Neff J. C., Weintraub M. N. 2007. Carbon structure and enzyme activities in alpine and forest ecosystems. Soil Biol. Biochem. 39:2701–2711 [Google Scholar]
- 24. Hachler H., Berger-Bächi B., Kayser F. H. 1987. Genetic characterization of a Clostridium difficile erythromycin-clindamycin resistance determinant that is transferable to Staphylococcus aureus. Antimicrob. Agents Chemother. 31:1039–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hartmann M., Frey B., Kflliker R., Widmer F. 2005. Semi-automated genetic analyses of soil microbial communities: comparison of T-RFLP and RISA based on descriptive and discriminative statistical approaches. J. Microbiol. Methods 61:349–360 [DOI] [PubMed] [Google Scholar]
- 26. Hartmann M., Widmer F. 2008. Reliability for detecting composition and changes of microbial communities by T-RFLP genetic profiling. FEMS Microbiol. Ecol. 63:249–260 [DOI] [PubMed] [Google Scholar]
- 27. Hobbie S. E. 1996. Temperature and plant species control over litter decomposition in Alaskan tundra. Ecol. Monogr. 66:503–522 [Google Scholar]
- 28. Hong S.-H., Bunge J., Leslin C., Jeon S., Epstein S. S. 2009. Polymerase chain reaction primers miss half of rRNA microbial diversity. ISME J. 3:1365–1373 [DOI] [PubMed] [Google Scholar]
- 29. Huber T., Faulkner G., Hugenholtz P. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319 [DOI] [PubMed] [Google Scholar]
- 30. Kemmitt S. J., et al. 2008. Soil mineralization of native soil organic matter is not regulated by the size, activity or composition of the soil microbial biomass—a new perspective. Soil Biol. Biochem. 40:61–73 [Google Scholar]
- 31. Kogel-Knabner I. 2002. The macromolecular organic composition of plant and microbial residues as inputs to soil organic matter. Soil Biol. Biochem. 34:139–162 [Google Scholar]
- 32. Kopecký J., Novotná G., Ságová-Marečková M. 2009. Modification of the terminal restriction fragment length polymorphism analysis for assessment of a specific taxonomic group within a soil microbial community. Plant Soil Environ. 55:397–403 [Google Scholar]
- 33. Kopecký J., et al. 2011. Actinobacterial community dominated by a distinct clade in acidic soil of a waterlogged deciduous forest. FEMS Microbiol. Ecol. doi:10.1111/j.1574–6941.2011.01173.x [DOI] [PubMed] [Google Scholar]
- 34. Kropf S., Heuer H., Gruning M., Smalla K. 2004. Significance test for comparing complex microbial community fingerprints using pairwise similarity measures. J. Microbiol. Methods 57:187–195 [DOI] [PubMed] [Google Scholar]
- 35. Kumke M. U., Specht C. H., Brinkmann T., Frimmel F. H. 2001. Alkaline hydrolysis of humic substances—spectroscopic and chromatographic investigations. Chemosphere 45:1023–1031 [DOI] [PubMed] [Google Scholar]
- 36. Kyselkova M., et al. 2008. Development of a 16S rRNA gene-based prototype microarray for the detection of selected actinomycetes genera. Antonie Van Leeuwenhoek 94:439–453 [DOI] [PubMed] [Google Scholar]
- 37. Lane D. 1991. 16S/23S rRNA sequencing, p. 115–175 In Stackebrandt E., Goodfellow M. (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, West Sussex, United Kingdom [Google Scholar]
- 38. Lauber C. L., Hamady M., Knight R., Fierer N. 2009. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 74:5111–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Legendre P., Legendre L. 1998. Numerical ecology. Elsevier Science B.V., Amsterdam, The Netherlands [Google Scholar]
- 40. Lindedam J., Magid J., Poulsen P., Luxhøi J. 2009. Tissue architecture and soil fertility controls on decomposer communities and decomposition of roots. Soil Biol. Biochem. 41:1040–1049 [Google Scholar]
- 41. Liu Z., Liu G., Fu B., Zheng X. 2008. Relationship between plant species diversity and soil microbial functional diversity along a longitudinal gradient in temperate grasslands of Hulunbeir, Inner Mongolia, China. Ecol. Res. 23:511–518 [Google Scholar]
- 42. Manucharova N. A., Vlasenko A. N., Stepanov A. L. 2007. Temperature as an autoecological factor of chitinolytic microbial complex formation in soils. Biol. Bull. 34:163–169 [PubMed] [Google Scholar]
- 43. Muyzer G., de Waal E. C., Uitterlinden A. G. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Myers R. T., Zak D. R., White D. C., Peacock A. 2001. Landscape-level patterns of microbial community composition and substrate use in upland forest ecosystems. Soil Sci. Soc. Am. J. 65:359–367 [Google Scholar]
- 45. Paterson E., et al. 2008. Labile and recalcitrant plant fractions are utilised by distinct microbial communities in soil: independent of the presence of roots and mycorrhizal fungi. Soil Biol. Biochem. 40:1103–1113 [Google Scholar]
- 46. Patra A. K., Le Roux X., Grayston S. J., Loiseau P., Louault F. 2008. Unraveling the effects of management regime and plant species on soil organic carbon and microbial phospholipid fatty acid profiles in grassland soils. Bioresource Technol. 99:3545–3551 [DOI] [PubMed] [Google Scholar]
- 47. Ramirez K. S., Lauber C. L., Knight R., Bradford M. A., Fierer N. 2010. Consistent effects of nitrogen fertilization on soil bacterial communities in contrasting systems. Ecology 91:3463–3470 [DOI] [PubMed] [Google Scholar]
- 48. Rousk J., Brookes P. C., Bååth E. 2010. The microbial PLFA composition as affected by pH in an arable soil. Soil Biol. Biochem. 42:516–520 [Google Scholar]
- 49. Rousk J., Brookes P. C., Bååth E. 2010. Investigating the mechanisms for the opposing pH relationships of fungal and bacterial growth in soil. Soil Biol. Biochem. 42:926–934 [Google Scholar]
- 50. Rovira P., Vallejo V. R. 2002. Labile and recalcitrant pools of carbon and nitrogen in organic matter decomposing at different depths in soil: an acid hydrolysis approach. Geoderma 107:109–141 [Google Scholar]
- 51. Sagova-Mareckova M., et al. 2008. Innovative methods for soil DNA purification tested in soils with widely differing characteristics. Appl. Environ. Microbiol. 74:2902–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sakai M., Matsuka A., Komura T., Kanazawa S. 2004. Application of a new PCR primer for terminal restriction fragment length polymorphism analysis of the bacterial communities in plant roots. J. Microbiol. Methods 59:81–89 [DOI] [PubMed] [Google Scholar]
- 53. Schlatter D., et al. 2009. Resource amendments influence density and competitive phenotypes of Streptomyces in soil. Microb. Ecol. 57:413–420 [DOI] [PubMed] [Google Scholar]
- 54. Stach J. E. M., Maldonado L. A., Ward A. C., Goodfellow M., Bull A. T. 2003. New primers for the class Actinobacteria: application to marine and terrestrial environments. Environ. Microbiol. 5:828–841 [DOI] [PubMed] [Google Scholar]
- 55. Thoms C., Gattinger A., Jacob M., Thomas F. M., Gleixner G. 2010. Direct and indirect effects of tree diversity drive soil microbial diversity in temperate deciduous forest. Soil Biol. Biochem. 42:1558–1565 [Google Scholar]
- 56. Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. White T. J., Bruns T., Lee S., Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315–322 In Innis M. A., Gelfand D. H., Sninsky J. J., White T. J. (ed.), PCR protocols, a guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 58. Yergeau E., et al. 2010. Influences of space, soil, nematodes and plants on microbial community composition of chalk grassland soils. Environ. Microbiol. 12:2096–2106 [DOI] [PubMed] [Google Scholar]
- 59. Youssef N. H., Elshahed M. S. 2009. Diversity rankings among bacterial lineages in soil. ISME J. 3:305–313 [DOI] [PubMed] [Google Scholar]
- 60. Zak D. R., Kling G. W. 2006. Microbial community composition and function across an arctic tundra landscape. Ecology 87:1659–1670 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.