Abstract
Establishing the risk of human infection is one of the goals of public health. For bacterial pathogens, the virulence and zoonotic potential can often be related to their host source. Escherichia coli bacteria are common contaminants of water associated with human recreation and consumption, and many strains are pathogenic. In this study, we analyzed three promoter-containing intergenic regions from 284 diverse E. coli isolates in an attempt to identify molecular signatures associated with specific host types. Promoter sequences controlling production of curli fimbriae, flagella, and nutrient import yielded a phylogenetic tree with isolates clustered by established phylogenetic grouping (A, B1, B2, and D) but not by host source. Virulence genes were more prevalent in groups B2 and D isolates and in human isolates. Group B1 isolates, primarily from nonhuman sources, were the most genetically similar, indicating that they lacked molecular adaptations to specific host environments and were likely host generalists. Conversely, B2 isolates, primarily from human sources, displayed greater genetic distances and were more likely to be host adapted. In agreement with these hypotheses, prevalence of σS activity and the rdar morphotype, phenotypes associated with environmental survival, were significantly higher in B1 isolates than in B2 isolates. Based on our findings, we speculate that E. coli host specificity is not defined by genome-wide sequence changes but, rather, by the presence or absence of specific genes and associated promoter elements. Furthermore, the requirements for colonization of the human gastrointestinal tract may lead to E. coli lifestyle changes along with selection for increased virulence.
INTRODUCTION
Pathogenic strains of Escherichia coli cause millions of cases of human infection each year (52) as well as severe problems in the livestock industry (78). Yet in other situations, many E. coli strains coexist peacefully as commensals in the intestinal tract of their warm-blooded hosts. A significant amount of research has sought to understand the relationship between pathogenic and commensal E. coli strains and determine if they can be distinguished from each other (18, 55, 72, 95). E. coli is also a common contaminant of water and various food sources (63). From a public health standpoint, it is important to establish the zoonotic and virulence potential of strains, which may be related to their natural lifestyle and host source.
E. coli has been proposed to have two principal habitats, the primary being the intestinal tract of mammals and birds and the secondary being water, sediment, and soil (58, 81). Survival and adaptation in both habitats are necessary for continued evolutionary success. For isolates that have evolved toward commensalism, signs of adaptation should be apparent, be it through altered regulatory sequences, gene expression, or metabolism (29). E. coli strains can be differentiated into four main phylogenetic groups (A, B1, B2, and D) along with two minor groups (C and E) (41). In general, these groups do not further divide into defined host lineages (23), except for the association of O157 enterohemorrhagic E. coli (EHEC; group E) with bovine sources (20, 66). Multilocus sequence typing (MLST) (39, 95) and more recent whole-genome comparisons (73, 85) have revealed that all E. coli strains share a highly similar core genome in addition to hot spots of recombination that result in a high percentage of unique or strain-specific genes. This characteristic makes phylogenetic analysis difficult but does not obscure the signal completely (55). Virulence genes can be scattered throughout different isolates, but groups B2 and D have an increased prevalence of extraintestinal virulence genes (8, 49, 69). Whether this is a requirement for or consequence of gastrointestinal (GI) tract colonization remains the subject of debate (21, 65).
Life in two different habitats requires E. coli to have a balanced strategy between faster growth rates in the primary habitat and increased stress resistance in the secondary habitat. σS (RpoS) is the central regulator of the general stress response in E. coli and helps direct transcription of genes essential for stress resistance (54, 91). There are complex feedback systems that regulate the levels of σS in the cell (36, 40, 75), which can shift the balance between stress resistance and the ability to utilize diverse nutrients (25, 26). Highly resistant E. coli isolates often have a reduced ability to compete for carbon sources due to low membrane permeability (53), whereas σS null mutants have increased scavenging capacity and growth advantages under certain conditions (64, 97). One of the key resistance phenotypes regulated by σS is the rdar (red, dry and rough) morphotype, a multicellular growth state that has been linked to long-term survival under harsh conditions (92). rdar cells produce an extracellular matrix comprised of curli fimbriae, cellulose, and other polysaccharides, resulting in colonies that have a distinctly patterned appearance and aggregative texture (14, 76, 77, 100). The matrix also provides increased resistance to disinfection (79, 86). Although there has never been an exhaustive examination of rdar morphotype prevalence in natural E. coli isolates, most, if not all, strains contain the genes necessary for production (4, 7). The relationship between σS activity, the rdar morphotype, and metabolic capacity is likely to reflect the evolutionary histories of different E. coli isolates.
Adaptation in bacteria occurs through genetic mutation and the acquisition and/or loss of genes. Techniques used to determine genetic relatedness, such as MLST and single nucleotide polymorphism (SNP) analysis (43, 55, 95), are usually based on the sequences of conserved genes, which are under mutational constraints. In contrast, noncoding, intergenic regions have not been used extensively for phylogenetic analysis, yet it is becoming clear that changes in promoter regions are key drivers of evolutionary adaptations (67, 98). In this study, we analyzed three E. coli intergenic regions from 284 diverse isolates to determine if there were molecular signatures associated with different host sources. Our analysis yielded a phylogenetic tree with isolates divided into the four defined E. coli phylogenetic groups but without any host-specific clustering. Phenotypic analysis for σS and rdar morphotype prevalence revealed that certain phylogenetic groups are exposed to different selection pressures, which is suggestive of a lifestyle shift. Human isolates were primarily in phylogenetic group B2 and had the lowest prevalence of σS activity and the rdar morphotype as well as the highest prevalence of virulence genes. In contrast, most nonhuman isolates, which were predominantly clustered into phylogenetic group B1, had the highest prevalence of σS activity and the rdar morphotype and the lowest prevalence of virulence genes. Despite the lack of clear host-specific molecular signatures, our results indicate that there is a correlation between E. coli host adaptation, increased virulence potential, and the loss of stress resistance pathways.
MATERIALS AND METHODS
E. coli isolation. (i) Animal isolates.
Isolates from a wide variety of animals were obtained from the Alberta Provincial Laboratory for Public Health (ProvLab) and originated from Calgary, Alberta, Canada. Fecal swabs were planted onto MacConkey agar and grown overnight at 37°C. Lactose-fermenting isolates from each specimen were swabbed to isolation on blood agar plates (BAP) and biochemically tested for reactivity with malonate (negative), citrate (negative), indole (positive), and oxidase (negative) (42). Presumptive animal isolates were confirmed as E. coli using API 20E strips (bioMérieux). Isolates from gulls, Canada geese, dogs, and cats were obtained from Environment Canada (Burlington, Ontario) and originated from the city of Toronto, Ontario, Canada. Fecal swabs were streaked onto mFC agar (Difco Inc.) and incubated at 44.5°C for 18 h. Isolates that were a typical dark blue on mFC agar were selected and streak-plated onto MacConkey agar for overnight growth at 37°C. Putative E. coli isolates on MacConkey plates were then tested for glucuronidase activity by growth and fluorescence in EC-MUG (EC broth with 4-methylumbelliferyl-β-d-glucuronide; Difco Inc.) and for indole production by growth in 1% (wt/vol) tryptone (Difco Inc.) and reaction with Kovac's reagent (Oxoid Inc.). E. coli ATCC 29194 and Klebsiella pneumoniae ATCC 33495 were used as a positive and negative controls, respectively, during confirmation tests.
(ii) Human isolates.
E. coli was isolated from fecal swabs submitted to the ProvLab for microbiological testing. Fecal swabs were plated onto MacConkey agar plates and incubated overnight at 35°C. Up to five mauve/pink presumptive E. coli colonies were picked from the MacConkey agar plates and plated to purity on BAP. Isolates were tested for reactivity to malonate (negative), citrate (negative), indole (positive), and oxidase (negative). Isolates confirmed as E. coli were swabbed to isolation on a BAP plate and frozen in skim milk at −70°C.
It is not known if the isolates selected for this study were clinically relevant or whether they simply represented the commensal E. coli from a patient suffering from enteric symptoms of other etiology. As part of the ethics submission for obtaining human samples, we delinked clinical laboratory information system numbers to our research samples, so we could not trace back to original submitter or trace back our samples to find out what the final clinical diagnosis was. We did not select for and isolate a “clinically relevant” pathotype of E. coli. In addition, 20 human isolates (17% of total) came from healthy donors and presumably would represent commensal E. coli.
(iii) Water isolates.
Environmental samples were collected as part of routine water testing surveillance programs at the ProvLab. Drinking water samples testing positive by enzyme substrate testing (Colilert; Idexx Laboratories) were inoculated (10-μl loops) onto 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc) plates and incubated overnight at 44.5°C. Dark blue colonies were isolated and swabbed to purity on BAP plates and tested as indicated above (for malonate, citrate, indole, and oxidase reactivity) or by API 20E strips (bioMérieux).
PCR.
Genomic DNA was prepared for PCR by boiling cells in a 5% solution of Chelex-100 (Bio-Rad Laboratories) as described by Walsh et al. (89). The phylogenetic grouping for each E. coli isolate was determined using a multiplex PCR assay for chuA, yjaA, and the TSPE4.C2 DNA fragment described by Clermont et al. (12). Isolates were termed unclassified if they were negative for all three reactions (33). To determine the prevalence of defined virulence genes, each E. coli isolate was tested using a multiplex PCR strategy, mostly adapted from Johnson and Stell (50); primers are listed in Table 1. PCR was conducted under standard conditions in 20-μl reaction volumes, containing 1× PCR buffer (Invitrogen), 7.5 ng/μl template, 1.5 mM MgCl2, 50 μM each deoxynucleoside triphosphate (dNTP), 0.2 μM primer, and 0.5 U of Taq polymerase. For multiplex reactions, PCR amplification conditions included an initial denaturation at 94°C for 2 min, 30 cycles of 94°C for 30 s, annealing at 63°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 7 min; multiplex reaction 8 used an annealing temperature of 55°C. The intergenic regions containing promoters for csgD and csgB, flhDC, and ompF were amplified individually under standard PCR conditions in 100-μl reaction mixtures containing 1× PCR buffer (Invitrogen), 2 mM MgCl2, 12.5 μM each dNTP, 0.5 μM each primer (Table 1), and 1.25 U of Taq polymerase. PCR amplification conditions included an initial denaturation at 94°C for 2 min, 30 cycles of 94°C for 30 s, annealing at 50°C for 30 s, and 72°C for 1 min, followed by a final extension at 72°C for 7 min; the only modification was for target flhDC, which required an annealing temperature of 53°C.
Table 1.
PCR primers used in this study
| Multiplex reaction no. | Target | Primera | Primer sequence (5′–3′) | Reference or source |
|---|---|---|---|---|
| 1 | aer | aer-F | TACCGGATTGTCATATGCAGACCGT | 83 |
| aer-R | AATATCTTCCTCCAGTCCGGAGAAG | |||
| papC | papC-F | GTGGCAGTATGAGTAATGACCGTTA | 50 | |
| papC-R | ATATCCTTTCTGCAGGGATGCAATA | |||
| traT | traT-F | GGTGTGGTGCGATGAGCACAG | 50 | |
| traT-R | CACGGTTCAGCCATCCCTGAG | |||
| 2 | iha | iha-F | CTGGCGGAGGCTCTGAGATCA | 83 |
| iha-R | TCCTTAAGCTCCCGCGGCTGA | |||
| usp | usp-F | CGGCTCTTACATCGGTGCGTTG | 83 | |
| usp-R | GACATATCCAGCCAGCGAGTTC | |||
| irp2 | irp2-F | AAGGATTCGCTGTTACCGGAC | 6 | |
| irp2-R | TCGTCGGGCAGCGTTTCTTCT | |||
| 3 | PAI | PAI-F | GGACATCCTGTTACAGCGCGCA | 50 |
| PAI-R | TCGCCACCAATCACAGCCGAAC | |||
| fimH | fimH-F | TGCAGAACGGATAAGCCGTGG | 50 | |
| fimH-R | GCAGTCACCTGCCCTCCGGTA | |||
| 4 | iroN | iroN-F | AAGTCAAAGCAGGGGTTGCCCG | 83 |
| iroN-R | GACGCCGACATTAAGACGCAG | |||
| iutA | iutA-F | GGCTGGACATCATGGGAACTGG | 50 | |
| iutA-R | CGTCGGGAACGGGTAGAATCG | |||
| ibeA | ibeA-F | AGGCAGGTGTGCGCCGCGTAC | 50 | |
| ibeA-R | TGGTGCTCCGGCAAACCATGC | |||
| 5 | cnfl | cnfl-F | AAGATGGAGTTTCCTATGCAGGAG | 83 |
| cnfl-R | CATTCAGAGTCCTGCCCTCATTATT | |||
| papGII | papGII-F | GGGATGAGCGGGCCTTTGAT | 50 | |
| papGII-R | CGGGCCCCCAAGTAACTCG | |||
| 6 | fuyA | fuyA-F | TGATTAACCCCGCGACGGGAA | 50 |
| fuyA-R | CGCAGTAGGCACGATGTTGTA | |||
| papGIII | papGIII-F | GGCCTGCAATGGATTTACCTGG | 50 | |
| papGIII-R | CCACCAAATGACCATGCCAGAC | |||
| 7 | sfa-foc | sfa/foc-F | CTCCGGAGAACTGGGTGCATCTTAC | 50 |
| sfa/foc-R | CGGAGGAGTAATTACAAACCTGGCA | |||
| hylA | hylA-F | AACAAGGATAAGCACTGTTCTGGCT | 50 | |
| hylA-R | ACCATATAAGCGGTCATTCCCGTCA | |||
| 8 | ompT | ompT-F | ATCTAGCCGAAGAAGGAGGC | 83 |
| ompT-R | CCCGGGTCATAGTGTTCATC | |||
| hra | hra-F | CAGAAAACAACCGGTATCAG | 6 | |
| hra-R | ACCAAGCATGATGTCATGAC | |||
| ompF | ompF_Ecnew1-F | TACGTGATGTGATTCCGTTC | This study | |
| ompF_Ecnew2-R | TGTTATAGATTTCTGCAGCG | |||
| csgD | agfD1 | GTGCTCGAGGGACTTCATTAAACATGATG | 92 | |
| agfD2 | GCCGGATCCTGTTTTTCATGCTGTCAC | |||
| flhDC | CG01-F | GCGGATCCGAGGTATGCATTATTCCCACCC | This study | |
| flhDC1-R | GCCCTCGAGTGGAGAAACGACGCAATC |
F indicates forward primers (5′ region of gene), and R indicates reverse primers (3′ region of gene).
Promoter sequence comparisons.
DNA sequencing was performed by Macrogen Inc. (Korea) and The University of Calgary Genetic Analysis laboratory (http://www.ucalgary.ca/dnalab/sequencing). Sequences were assembled into a concatenated, single file for each E. coli strain prior to alignment using the Clustal W algorithm (Vector NTI, version 11; Invitrogen). This Clustal W alignment was manually edited to trim the 5′ and 3′ regions of the alignment that contained missing data. The final alignment was 2,064 nucleotides (nt) in length. To obtain the most efficient parameters for the phylogenetic reconstruction, we used jModelTest (version 0.1) (70); using the Akaike information criterion, the TPM1uf+I+G model was selected. We used phyML (version 3.0) (38) to reconstruct the phylogeny. The initial topology was generated using the subtree pruning and regrafting (SPR) algorithm (44), and the branch support was calculated using Shimodaira-Hasegawa-like (SH-like) procedure (37). The default settings were used for the remaining SH-like parameters. In the resultant phylogenetic tree, internal nodes with less than SH-like support of <0.7 were collapsed. Branch lengths for this consensus tree were calculated using the baseml program from the PHYLIP software suite (version 4.3) (96). We used the interactive Tree Of Life (iTOL) online tool to map on the host-type and phylogenetic grouping data sets (56).
Reference E. coli isolates.
DNA sequences for intergenic regions containing csgD, csgB, flhDC, and ompF promoters were obtained from the GenBank from the following E. coli strains with fully sequenced genomes (accession numbers are listed in parentheses): 536 (CP000247), 53638 (AE014075), 55989 (CU928145), 101-1 (AAMK00000000), APEC01 (CP000468), B REL606 (CP000819), B171 (AAJX00000000), B7A (AAJT00000000), CFT073 (AE014075), E110019 (AAJW00000000), E22 (AAJV00000000), E24377A (CP000800), ED1a (CU928162), F11 (AAJU00000000), HS (CP000802), IAI1 (CU928160), IAI39 (CU928164), K-12 substrain MG1655 (U00096), K-12 substrain W3110 (AP009048), O157:H7 EDL933 (AE005174), O157:H7 Sakai (BA000007), S88 (CU928161), SMS-3-5 (CP000970), UMN026 (CU928163), and UT189 (CP000243).
Phenotypic testing for the rdar morphotype and σS activity.
Frozen E. coli isolates were recovered on LBns agar (LB without NaCl) and grown overnight at 37°C. Broth cultures were prepared in 100 μl of LBns broth in microtiter plates and grown overnight at 37°C. Spot colonies were prepared by inoculation of 1 μl of the overnight broth culture onto LBns agar or 1% tryptone (T) agar supplemented with 100 μg/ml Congo red. Colony morphologies were observed after incubation at 28°C for 48 to 72 h and were compared to known rdar-positive (rdar+) (Salmonella enterica serovar Typhimurium ATCC 14028), rdar-intermediate (S. Typhimurium ΔcsgA or ΔbcsA mutant) and rdar-negative (rdar−) isolates (S. Typhimurium ΔcsgD ΔrpoS) as outlined in White et al. (92). Because we wanted to make a distinction between nonaggregative and aggregative isolates in this study, isolates were recorded as rdar positive if they produced both curli and cellulose or either polymer alone. We did not test for rdar morphotype formation at 37°C. To our knowledge, all reported examples of E. coli strains that are rdar+ at 37°C are also positive at 28°C (7, 87).
For detection of σS activity, colonies grown on LBns at 28°C for 48 h were stained with iodine to detect glycogen production (described in reference35 and modified according to reference 94) and treated with hydrogen peroxide for detection of catalase activity (64). Results were compared to known σS-positive (S. Typhimurium ATCC 14028), σS-attenuated (S. Typhimurium ΔcsgD [17]), or σS-negative (S. Typhimurium ΔrpoS [93]) isolates. E. coli isolates were recorded as σS positive for full activity, whereas isolates with attenuated or null activity were classified as being σS impaired.
PMs.
Phenotype microarrays (Biolog, Hayward CA) were performed to test the utilization of different carbon, nitrogen, phosphorus, and sulfur substrates by 43 selected E. coli isolates. Assays were conducted for phenotype microarray (PM) plates 1 through 4 as per the manufacturer's instructions with the following modifications. Frozen E. coli isolates were recovered on tryptic soy agar (TSA) and incubated overnight at 37°C. A single colony was further purified by TSA culture. Cell suspensions were concentrated to 48% transmittance (590 nm) before addition to the wells. PM plates were incubated at 37°C; absorbance at 600 nm was measured with a Wallac Victor2 plate reader (Perkin-Elmer Life Sciences, Boston, MA) after 24 and 48 h of growth. E. coli K-12 substrain MG1655 was used as the control strain.
Hierarchical clustering was performed using the heatmap.2 function in R, version 2.13.1 (74). Raw data were first normalized by subtraction of the background absorbance followed by a variance stabilization (vsn package). Clustering was performed using the McQuitty linkage method (61) and a Pearson correlation as the distance metric.
Statistical analysis of data.
Prevalence of virulence genes, rdar morphotype, or σS activity was compared between groups of isolates based on phylogenetic grouping (A, B1, B2, D, and unclassified) or host type (human, bovine, birds, water, and nonhuman mammals). Chi-square tests for heterogeneity or independence comparing all groups were performed, followed by Fisher exact tests between chosen groups. For each virulence gene, comparisons were made between phylogenetic groups with the highest and next highest prevalence; for host type, comparisons were made between the group of human isolates and the group of nonhuman isolates. For rdar and σS prevalence, comparisons were performed between all pairs of phylogenetic groups and host types.
RESULTS
Description of E. coli isolates and determination of phylogenetic groups.
A total of 284 E. coli isolates were obtained from a diverse collection of host types (Table 2). Humans were the most predominant host type, with 40% of isolates, each from separate individuals. The remaining isolates were obtained from fecal samples of nonhuman animals, with the exception of 23 isolates obtained from water samples. In almost all cases, each E. coli isolate corresponds to one animal, with the exception being 44 unique bovine isolates that came from a total of 29 animals (data not shown).
Table 2.
Summary of E. coli isolates from different host types
| Host typea | Total no. of isolates | No. of isolates by phylogenetic groupb |
||||
|---|---|---|---|---|---|---|
| A | B1 | B2 | D | Unclassifiedc | ||
| Human | 115 | 15 | 13 | 58 | 22 | 7 |
| Total nonhuman | 169 | 23 | 95 | 11 | 28 | 12 |
| Bovine | 44 | 15 | 15 | 12 | 2 | |
| Birds | ||||||
| Seagull | 20 | 2 | 14 | 1 | 2 | 1 |
| Goose | 21 | 18 | 1 | 2 | ||
| Chicken | 2 | 1 | 1 | |||
| Other nonhuman mammals | ||||||
| Dog | 28 | 2 | 15 | 4 | 6 | 1 |
| Cat | 21 | 16 | 3 | 2 | ||
| Pig | 9 | 3 | 1 | 1 | 4 | |
| Muskrat | 1 | 1 | ||||
| Water | 23 | 1 | 16 | 1 | 2 | 3 |
| Overall total | 284 | 38 | 108 | 69 | 50 | 19 |
Each E. coli isolate was cultured from fecal matter originating from the different host types, with the exception of water isolates, which were obtained from contaminated water samples.
E. coli genotype was determined using the triplex PCR classification scheme specific for ChuA, YjaA, and TspE4C2 described by Clermont et al. (12).
Unclassified isolates were negative for all three PCRs, as described by Gordon et al. (33).
For rapid determination of the E. coli phylogenetic group, all isolates were analyzed using the PCR-based method described by Clermont et al. (12). Overall, B1 was the most predominant phylogenetic group, followed by B2, D, and A; we were unable to classify 7% of isolates due to negative PCR results (Table 2) (33). Group B1 encompassed 56% of E. coli isolates from nonhuman sources; birds had the highest prevalence of B1 isolates at 74%. Fifty percent of human isolates were categorized into group B2, whereas only 11% were assigned to group B1.
Prevalence of virulence genes among E. coli isolates.
Many studies have been performed to determine the human virulence potential of E. coli isolates. Virulence of isolates in in vivo models is usually correlated with the presence of defined virulence factors (21, 48). These factors include adhesins (P, S, F1C, and type 1 fimbriae), toxins (hemolysin and cytotoxic necrotizing factor), iron acquisition systems (for aerobactin, yersiniabactin, and a catecholate siderophore), invasins (ibeA), and a variety of other genes including: traT (invasion), malX (pathogenicity island [PAI] marker), usp (bacteriocin), ompT (outer membrane endoprotease), iha (fimbriae or adhesin), and hra (heat-resistant agglutinin). To assess the virulence potential of E. coli isolates in our study, we used a multiplex PCR strategy to screen for the presence of 18 known virulence-associated genes (Table 3).
Table 3.
Prevalence of virulence genes among E. coli strains isolated from different hosts and environmental sources
| Isolate group or source | % of isolates carrying the indicated virulence geneb |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aer | cnf1 | fimH | fyuA | hlyA | hra | ibeA | iha | iroN | irp2 | ompT | PAI | papC | papG_II | papG_III | sfa-foc | traT | usp | |
| Phylogenetic groupa | ||||||||||||||||||
| A | 21.1 | 0.0 | 44.7 | 2.6 | 2.6 | 13.2 | 0.0 | 15.8 | 0.0 | 42.1 | 13.2 | 2.6 | 7.9 | 10.5 | 36.8 | 2.6 | 50.0 | 0.0 |
| B1 | 1.9 | 5.6 | 83.3 | 7.4 | 2.8 | 31.5 | 2.8 | 0.0 | 6.5 | 13.9 | 45.4 | 3.7 | 7.4 | 0.0 | 6.5 | 53.7* | 53.7 | 8.3 |
| B2 | 21.7 | 33.3* | 92.8 | 43.5* | 13.0 | 43.5 | 29.0* | 21.7 | 26.1 | 98.6* | 98.6* | 84.1* | 59.4* | 27.5* | 87.0* | 29.0 | 62.3 | 87.0* |
| D | 32.0 | 4.0 | 78.0 | 10.0 | 2.0 | 34.0 | 10.0 | 18.0 | 12.0 | 30.0 | 54.0 | 12.0 | 22.0 | 8.0 | 26.0 | 16.0 | 64.0 | 16.0 |
| Unclassified | 21.1 | 0.0 | 63.2 | 0.0 | 5.3 | 10.5 | 0.0 | 5.3 | 5.3 | 10.5 | 21.1 | 0.0 | 5.3 | 5.3 | 10.5 | 10.5 | 57.9 | 5.3 |
| Host type | ||||||||||||||||||
| Human | 26.1* | 13.0 | 88.7* | 19.1 | 5.2 | 24.3 | 12.2 | 24.3* | 15.7 | 71.3* | 69.6* | 48.7* | 36.5* | 22.6* | 64.3* | 13.0 | 55.7 | 51.3* |
| Nonhuman | 8.8 | 9.4 | 70.6 | 12.9 | 5.3 | 35.3 | 8.2 | 1.8 | 8.2 | 20.0 | 42.9 | 7.6 | 12.9 | 1.2 | 12.9 | 43.5* | 58.2 | 11.2 |
| Bovines | 18.2 | 0.0 | 56.8 | 2.3 | 9.1 | 40.9 | 0.0 | 0.0 | 0.0 | 13.6 | 34.1 | 0.0 | 9.1 | 2.3 | 6.8 | 20.5 | 79.5 | 0.0 |
| Birds | 7.0 | 11.6 | 95.3 | 16.3 | 0.0 | 37.2 | 9.3 | 2.3 | 16.3 | 20.9 | 55.8 | 9.3 | 14.0 | 0.0 | 11.6 | 74.4 | 44.2 | 20.9 |
| Other nonhuman mammals | 6.1 | 22.4 | 85.7 | 28.6 | 10.2 | 42.9 | 16.3 | 4.1 | 14.3 | 32.7 | 59.2 | 18.4 | 24.5 | 2.0 | 22.4 | 67.3 | 59.2 | 20.4 |
| Dogs | 10.7 | 25.0 | 75.0 | 32.1 | 10.7 | 39.3 | 17.9 | 7.1 | 10.7 | 35.7 | 57.1 | 21.4 | 25.0 | 3.6 | 21.4 | 46.4 | 50.0 | 28.6 |
| Cats | 0.0 | 14.3 | 100.0 | 19.0 | 9.5 | 38.1 | 9.5 | 0.0 | 19.0 | 19.0 | 57.1 | 14.3 | 19.0 | 0.0 | 14.3 | 95.2 | 47.6 | 9.5 |
| Pigs | 0.0 | 11.1 | 0.0 | 11.1 | 0.0 | 22.2 | 11.1 | 0.0 | 0.0 | 11.1 | 11.1 | 0.0 | 11.1 | 0.0 | 11.1 | 0.0 | 44.4 | 0.0 |
| Water | 4.3 | 0.0 | 52.2 | 0.0 | 0.0 | 21.7 | 8.7 | 0.0 | 0.0 | 13.0 | 21.7 | 0.0 | 0.0 | 0.0 | 13.0 | 0.0 | 69.6 | 0.0 |
The number of E. coli isolates corresponding to each phylogenetic group and host type is consistent with Table 2.
Virulence genes in each strain were detected by PCR using well-defined primer sets (Table 1). Numbers listed represent the percentage of isolates that carry the designated allele: aer, aerobactin; cnf1, cytotoxic necrotizing factor 1; fimH, d-mannose-specific adhesin; fyuA, ferric yersiniabactin receptor; hlyA, α-hemolysin; hra, heat-resistant agglutinin; ibeA, invasion of brain endothelium; iha, fimbriae or adhesin; iroN, novel catecholate siderophore receptor; irp2, yersiniabactin biosynthesis pathway; ompT, outer membrane endoprotease, PAI (malX), pathogenicity-associated island marker; papC, pilus assembly, central region of pap operon; papG, Gal(α1-4)Gal-specific pilus tip adhesin molecule; papGII, pyelonephritis-associated; papGIII; cystitis-associated; sfa-foc, central region of sfa (S fimbriae) and foc (F1C fimbriae) operons; traT, surface exclusion, serum survival (outer membrane protein); usp, uropathogenic-specific protein (bacteriocin). Statistical significance (*) for each gene was determined using a Chi-square test for heterogeneity or independence comparing all groups, followed by Fisher exact tests between two chosen groups: for phylogenetic groups, comparisons were made between group B2 and the group (usually D) with the next highest rate of prevalence for that particular gene; for host type, comparisons were made between the group of human isolates and the group of nonhuman isolates.
In agreement with previous studies, isolates in phylogenetic group B2 had a higher prevalence of virulence genes than isolates from the other phylogenetic groups. Ten of 18 genes were significantly more prevalent in group B2 isolates (Table 3). Among the other groups, group D isolates had a higher prevalence of virulence genes than group A isolates, while group B1 isolates had the lowest overall prevalence. When human and nonhuman isolates were compared to each other, 10 of 18 genes had significantly higher prevalence among human isolates (Table 3). Although these were not all the same 10 genes as those in group B2, the significance was correlated to the predominance of group B2 isolates from humans compared to group B1 isolates from nonhuman sources. All B1 isolates and all nonhuman isolates had significantly higher prevalence of sfa-foc fimbriae (Table 3). E. coli isolates from pigs and bovine sources had the lowest overall prevalence of virulence genes. There were no significant differences in prevalence of virulence-related genes when all host types were compared.
Phylogenetic analysis of E. coli strains based on intergenic sequence comparison.
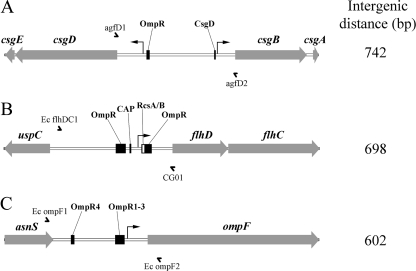
Although intergenic sequence and structural promoter analysis has been used to characterize genetic relatedness among species and strains of microbes (28, 30) or to examine the expression of virulence factors (67), little work has attempted to correlate this with host specificity. Three promoter-containing intergenic regions were sequenced from all E. coli isolates: between the divergent csgBAC and csgDEFG operons regulating the synthesis of curli fimbriae (76) (Fig. 1A), between uspC and flhDC, which code for the master regulator of flagellum biosynthesis (51) (Fig. 1B), and between asnS and ompF (Fig. 1C), which codes for an outer membrane porin expressed under low-osmolarity conditions (24). These intergenic regions were chosen because they were relatively large (>500 bp) and known to exhibit variation in E. coli and related organisms (15, 84, 87). In addition, because the associated gene products (i.e., curli, flagella, and OmpF) are conserved and found on the cell surface, their expression could be subject to selection pressure within host environments (95).
Fig. 1.
E. coli intergenic regions chosen for sequence comparisons. Intergenic regions are shown to scale from the genome of E. coli K-12 substrain MG1655 (Vector NTI, version 11; Invitrogen). Gray arrows represent genes; names are listed above in italics. The intergenic distances shown were measured between start or stop codons in genes flanking the region. Full arrowheads represent defined −35 and −10 promoter regions. Black or white boxes represent known operator binding sites that regulate promoter activity for csgD and csgB (9, 71), flhDC (1, 99), and ompF (46, 59). Names of transcription factors are displayed above each binding site. OmpR1-3 represents three adjacent binding sites near the ompF promoter. The PCR primers used for amplification and sequencing are represented by half arrows. CAP, catabolite activator protein.
Intergenic sequence comparison is effective for phylogenetic analysis in E. coli.
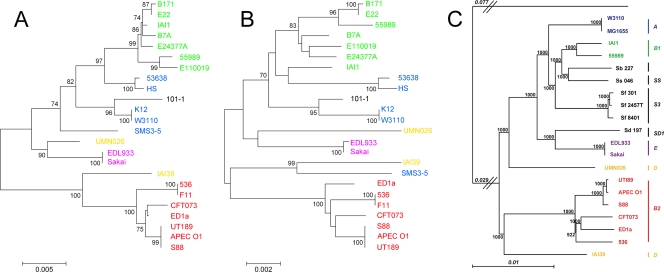
To determine if promoter sequence comparisons would provide adequate phylogenetic resolution to differentiate E. coli isolates, we performed in silico analysis on 24 E. coli strains with completely sequenced genomes. The phylogenetic tree reconstructed from promoter comparisons (Fig. 2A) was nearly identical to the tree generated by conventional MLST (Fig. 2B) (95). Despite a shorter overall sequence length (i.e., 2,064 nucleotides [nt] versus 3,423 nt), the intergenic sequence-based tree showed increased genetic distances for most isolates and higher bootstrap values at several nodes, demonstrating that this technique had more differentiating capacity than MLST, at least for the three intergenic regions analyzed. Four of the sequenced E. coli strains had insertion (IS)-like elements in the flhDC promoter region; these insertion sequences were removed prior to analysis. Both the intergenic sequence- and MLST-based trees had the same overall structures as the tree generated by comparison of 1,878 genes in the E. coli core genome (85) (Fig. 2C). The results of this in silico analysis indicated that comparison of promoter sequences could effectively be used to differentiate our 284 E. coli isolates.
Fig. 2.
Comparison of phylogenetic methods for differentiating E. coli isolates. Unrooted neighbor-joining trees were generated for 24 E. coli strains with completely sequenced genomes based on intergenic regions containing csgB-D, flhDC, and ompF promoter sequences (A) or conventional MLST of adk, fumC, gyrB, icd, mdh, purA, and recA sequences (B) (50) (http://mlst.ucc.ie/). Bootstrap values above 70%, based on 1,000 bootstraps, are displayed at nodes on each tree. Phylogenetic groups are represented by different colors: blue, A; green, B1; red, B2; yellow, D; purple, E; black, unclassified strain 101-1. (C) The maximum-likelihood tree generated for 14 E. coli and 6 Shigella strains is based on sequence comparison of all 1,878 genes in the Escherichia core genome (figure adapted from reference 85). Values at each node are based on 1,000 bootstraps, and the tree is rooted on Escherichia fergusonii. APEC, avian pathogenic E. coli.
Intergenic sequence comparisons can differentiate E. coli isolates into phylogenetic groups but cannot differentiate host sources.
In total, 284 of 296 E. coli isolates (96%) yielded PCR products for each of the three intergenic regions. IS-like insertion elements were detected in six isolates, with two having insertions in the csgB-csgD region and four in the asnS-ompF region; these insertion sequences were removed prior to analysis. The average pairwise identity for the final alignment of 2,064 nt was 97.6%. The csgB-csgD region was 736 nt in length with 98.6% pairwise identity, the uspC-flhDC region was 710 nt in length with 96.8% pairwise identity, and the asnS-ompF region was 618 nt in length with 97.2% pairwise identity.
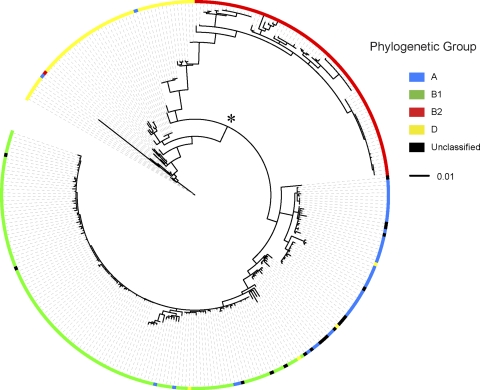
The 284 E. coli isolates clustered into their respective phylogroups, with only a few exceptions (Fig. 3). Phylogroup D was comprised of two distinct clusters of isolates (Fig. 3, star next to node). The D cluster adjacent to phylogroup B2 included reference E. coli strain IAI39 and is predicted to be ancestral to groups A and B1 and other group D isolates (55, 85). The second D cluster included reference E. coli strain UMN026 (85) and EHEC strains EDL933 and Sakai. EHEC isolates have previously been classified as phylogroup E (33); however, phylogroup E was not included in our analysis. The majority of unclassified or nontypeable isolates (33) were distributed within the phylogroup A cluster of isolates (Fig. 3). Phylogroup B2 isolates and the ancestral group D cluster of isolates displayed the largest genetic distances (distance from center of the tree), which reflected an increased number of sequence changes. From the point of view of host adaptation, these isolates would be the most likely to be adapted to their host environment. The phylogroup A and B1 isolates had intermediate branch lengths (Fig. 3). Within each phylogroup, the genetic similarity between isolates, as measured by average pairwise identity, ranged from 97.9% for group D, 98.5% for group A, and 98.9% for group B2 to a high of 99.6% for group B1.
Fig. 3.
Phylogenetic analysis of E. coli isolates based on comparison of csgD-B, flhDC, and ompF promoter-containing intergenic sequences. An unrooted maximum-likelihood tree was based on comparison of intergenic regions containing csgB-D, flhDC, and ompF promoters from 284 E. coli isolates and 24 reference E. coli strains. Phylogenetic grouping was determined for each strain using multiplex PCR (12) and is matched with Table 1. This group information was overlaid on the phylogenetic tree using the interactive iTOL online tool. The node that divides group D isolates into two clusters is marked by the asterisk.
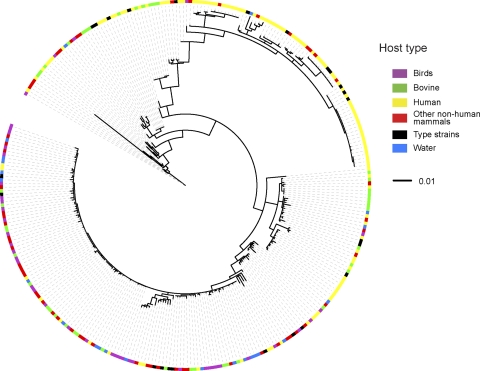
When the phylogenetic distribution of E. coli isolates was overlaid with the host source, a mosaic pattern was observed with no clear clustering of strains from different host types (Fig. 4). The only hosts displaying a trend were birds and humans, due to the predominance of bird isolates in phylogroup B1 and human isolates in phylogroup B2 (Table 2). The B1 phylogroup had an almost even distribution of isolates from different sources (Fig. 4 and Table 2). This diversity, coupled with the high similarity between isolates, suggested that phylogroup B1 isolates are host generalists with minimal adaptation to their host environments. Within phylogroup B2, 42 of 47 isolates that clustered with known urinary pathogenic E. coli (UPEC) strains (F11, 536, CFT073, APEC01, S88, and UT189) were of human origin, along with three isolates from a dog and two from cats. This suggested that humans and household pets are potential reservoirs for UPEC. Three of five isolates that clustered with reference EHEC strains EDL933 and Sakai were from bovine sources, along with one isolate from a pig and one isolate from a dog (data not shown). Overall, the lack of host type clustering revealed that there were no molecular signatures in the csgD-B, flhDC, and ompF intergenic regions associated with specific host types.
Fig. 4.
Phylogenetic tree for E. coli isolates based on intergenic sequence comparison matched to host source. The unrooted maximum-likelihood tree generated for our 284 E. coli isolates was overlaid with host or environmental source information from Table 1 using the interactive iTOL online tool.
The prevalence of the rdar morphotype and σS activity are not evenly distributed between E. coli phylogroups.
Host-adapted E. coli isolates are predicted to spend more time in their primary habitat, the intestinal tracts of mammals, where selection for a high growth rate would predominate (81). Host-generalist isolates, in contrast, would be predicted to spend more time in their secondary habitat, the environment, where selection for increased survival would predominate. Functional σS (RpoS) activity is essential for cells to adapt and survive in the face of a wide variety of environmental stresses (27, 68). σS controls formation of the rdar morphotype, a multicellular growth state that has been linked to long-term survival of E. coli and other enteric bacteria (7, 79, 86, 92).
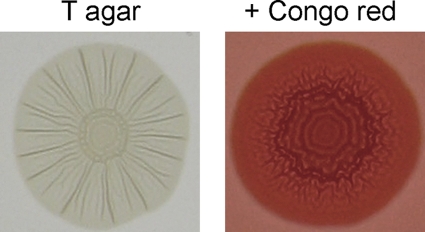
Phenotypic testing was performed on all E. coli isolates to determine the prevalence of the rdar morphotype and σS activity. Representative images of E. coli rdar morphotype colonies are displayed in Fig. 5. Overall, phylogroup B1 isolates had the highest rdar morphotype prevalence at 84.2%, followed by phylogroup D isolates at 58%, whereas <50% of isolates in phylogroup B2 and A were rdar+ (Table 4). This is consistent with the prediction that E. coli phylogroup B1 isolates are host generalists, whereas phylogroup B2 isolates are more host adapted. Within phylogroup D, there was a split between the ancestral cluster, with only 27% (4 of 15) rdar+ isolates, and the nonancestral cluster, with 71% (22 of 31) rdar+ isolates. For all phylogenetic groups, rdar morphotype prevalence was lower in human isolates than in nonhuman isolates (Table 4). Birds had the highest percentage of rdar+ E. coli isolates at 93%, whereas only 36.5% of human isolates were positive (Table 4).
Fig. 5.
Colony morphology of an E. coli isolate displaying the rdar morphotype. Colony morphology of an E. coli rdar+ isolate is shown after growth at 28°C for 72 h on tryptone agar or tryptone agar supplemented with Congo red. Note the distinctive, patterned appearance of the colony and deep red associated with extracellular matrix production and formation of the rdar morphotype.
Table 4.
Summary of phenotypic analysis of diverse E. coli isolates
| Isolate group or source | No. of isolatesa | Phenotype (% prevalence) by isolate sourced |
|||||
|---|---|---|---|---|---|---|---|
| Rdar morphotypeb |
σS activityc |
||||||
| All | Human | Nonhuman | All | Human | Nonhuman | ||
| Phylogenetic group | |||||||
| A | 38 (15, 23) | 34.2 | 26.7 | 39.1 | 71.1 | 53.3 | 82.6 |
| B1 | 108 (13, 95) | 84.2* | 61.5 | 87.4 | 88.9 | 69.2 | 91.6 |
| B2 | 69 (58, 11) | 40.5 | 36.2 | 63.6 | 42.0* | 32.7* | 90.9 |
| D | 50 (22, 28) | 58.0 | 36.4 | 75.0 | 78.0 | 63.6 | 89.3 |
| Ancestral D | 15 (9, 6) | 26.7 | 11.1 | 50.0 | 40.0 | 33.3 | 50.0 |
| Unclassified | 19 (7, 12) | 36.8 | 14.3 | 50.0 | 57.9 | 14.3 | 83.3 |
| Overall | 284 (115, 169) | 59.2 | 36.5 | 74.5 | 71.1 | 44.3 | 89.3 |
| Host type | |||||||
| Bovine | 44 | 59.0 | 77.3 | ||||
| Birds | 43 | 93.0* | 93.0 | ||||
| Other nonhuman mammals | 59 | 76.3 | 91.5 | ||||
| Human | 115 | 36.5* | 44.3* | ||||
| Water | 23 | 65.2 | 100 | ||||
For the phylogenetic groups, the numbers of human and nonhuman isolates, respectively, are shown in parentheses.
The ability to form rdar morphotype colonies was recorded after growth of isolates at 28°C on LBns agar supplemented with Congo Red.
σS activity was judged by glycogen production and catalase activity after growth of isolates at 28°C on LBns agar.
Prevalence was statistically different (*, P < 0.05) from all other phylogenetic groups or host types. Phylogroup D was evaluated as one group.
The prevalence of σS activity also varied greatly between different E. coli phylogroups, ranging from a low of 42% σS-positive for B2 isolates to a high of 88.9% for B1 isolates (Table 4). Like the rdar morphotype, there was a large discrepancy between human and nonhuman sources; nonhuman isolates had an overall σS-positive rate of 89% versus only 44% for human isolates (Table 4). Human group B2 isolates were by far the most likely to have impaired σS activity, with only 33% of isolates being σS positive (Table 4). With the exception of only 14 isolates, all σS-deficient isolates were negative for the rdar morphotype. The differences in prevalence of the rdar morphotype and σS activity between groups of E. coli isolates are thought to reflect lifestyle differences.
Very little metabolic differentiation between E. coli isolates.
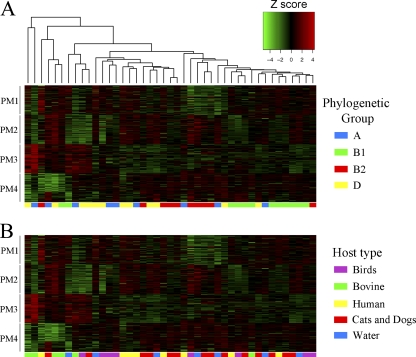
Although we did not identify sequence-based signatures that were host specific, we reasoned that host-adapted E. coli isolates could have elements of host-specific metabolism whereby their growth is adapted to carbon sources and nutrients that predominate in the mammalian intestine. In contrast, host-generalist isolates are hypothesized to retain maximum metabolic flexibility to give the best chance for survival in the environment. We analyzed 43 E. coli isolates from six different sources (gull, cow, cat, human, dog, and water) for their ability to utilize different carbon, nitrogen, phosphate, and sulfur sources for growth (Fig. 6). The chosen isolates were evenly distributed among the A, B1, B2, and D phylogenetic groups.
Fig. 6.
Hierarchical clustering of E. coli isolates based on their substrate utilization patterns. Phenotype microarray (PM) analysis (Biolog, Hayward, CA) was performed on 43 E. coli isolates to test their ability to metabolize different sources of carbon (PM1 and PM2), nitrogen (PM3), and phosphate and sulfate (PM4). Data were normalized by background subtraction and variance stabilization prior to clustering (see Materials and Methods). The Z-score metric corresponds to how many standard deviations above (red) or below (green) the mean the isolate was in comparison to other isolates. At the bottom of each cluster image, isolates were color coded by their phylogenetic group or host source.
Hierarchical cluster analysis revealed a slight correlation between metabolic profile and phylogenetic grouping (Fig. 6A) but no correlation with the host type (Fig. 6B). This was consistent with our sequence-based comparisons (Fig. 3 and 4). We hypothesized that host-generalist group B1 isolates would have more metabolic flexibility than isolates from other phylogenetic groups; however, this was observed for only one of the four array plates (PM1) (data not shown). The majority of group B1 isolates clustered together (Fig. 6A), indicating that they shared some common metabolic traits. Group A isolates, on the other hand, did not cluster together (Fig. 6A) and had reduced metabolic flexibility, with 12, 35, 53, and 31% reduced utilization on carbon, nitrogen, sulfate, and phosphate sources, respectively, compared to the other groups (data not shown). Our Biolog results might simply reflect the limited diversity of the isolates chosen or could be representative of a group-wide trend related to the degree of host adaptation. We concluded that only through analysis of a larger number of isolates would it be possible to identify trends in metabolic activity related to the host source or phylogenetic grouping of E. coli isolates.
DISCUSSION
The process of risk assessment for infectious disease includes determining the source of contamination and tracking the movement of causative organisms between animals and humans (10, 45). We reasoned that intergenic sequence comparisons could be a useful tool for microbial source tracking since promoter mutations can enable bacteria to adapt to changing environments (60, 67). We analyzed 284 E. coli isolates to see if host-specific molecular signatures could be detected within three large intergenic regions of the genome, controlling production of curli fimbriae, flagella, and an outer membrane porin. We hypothesized that this information, combined with the prevalence of virulence genes and other phenotypic traits, could be used to develop diagnostic tests to determine the host source of E. coli isolates and assess the risk of human infection.
Intergenic sequence alignment was able to differentiate the 284 E. coli isolates into their respective phylogroups (i.e., A, B1, B2, and D). This was expected because the E. coli phylogroups were originally identified using a sequence-based approach (41). The predictive power of the intergenic sequence analysis was equivalent to or greater than that of conventional MLST (95) despite being based on 40% less sequence information. Branch length differences in the intergenic sequence-based tree suggested there could be various degrees of host adaptation between different phylogenetic groups. Group B2 isolates and isolates in the ancestral group D (85) displayed the longest branch lengths. Because branch length is proportional to the number of sequence changes in the regions analyzed, which are assumed to be representative of other regions in the genome, we hypothesized that the B2 and ancestral group D isolates had the greatest probability of being host adapted. In contrast, group B1 isolates had shorter branch lengths and high genetic similarity and thus were predicted to be host generalists. Phylogroup A isolates and the remaining group D E. coli isolates had intermediate branch lengths. Gordon and Cowling (34) previously described A and B1 strains as generalists and B2 and D strains as specialists, confirming some of our predictions. In another large study examining E. coli diversity, Escobar-Paramo et al. (23) suggested that phylogroups A and B1 occupy similar commensal niches, distinct from niches occupied by phylogroups B2 and D. The main trend in our study was a predominance of B2 isolates from humans and B1 isolates from nonhuman sources, which would seem to agree with this.
Our analysis showed that E. coli as a group was too genetically similar to display large differences between isolates. No host-specific molecular signatures were identified in the intergenic regions analyzed. The average pairwise identity for the csgB-csgD intergenic region from 284 E. coli isolates was 98.6%. The −10 and −35 promoter regions, along with the key binding sites for OmpR in the csgD promoter (71) and CsgD in the csgB promoter (9), were nearly 100% conserved. By comparison, analysis of the same region among 26 diverse Salmonella isolates yielded an average pairwise identity of 89.2% (93). For the E. coli flhDC promoter region, we predicted there would be a lot of variability due to the presence of IS elements in laboratory strains (1) and the predominance of mutations in nonmotile, E. coli-related Shigella isolates (84). Although there were more mutations in operator binding sites within the flhDC promoter than in the other promoter regions analyzed, the overall mutation rate was still low (average pairwise identity of 96.8%). For ompF, promoter mutations have been shown to occur during in vitro evolution (3), and there is evidence that this gene is under selection in UPEC isolates (11). However, in our analysis, the ompF promoter region was also highly conserved (97.2% pairwise identity). Only one strain had a mutation in conserved nucleotides of the OmpR consensus binding site (46). In summary, we did not detect group-wide or host-specific changes predicted to inactivate the promoters analyzed. Given that recombination is predicted to occur frequently between E. coli isolates (39, 95), the high sequence identity within the three intergenic regions suggests that purifying selection is strong within the core genome of E. coli. It could be that noncore genes, which differ widely between isolates (85), determine E. coli host specificity. There is also the possibility that E. coli isolates are simply not differentiated into specific hosts (58).
The prevalence of the rdar morphotype and σS activity in E. coli isolates from different phylogroups and host sources could indicate potential lifestyle differences. It is hypothesized that true “commensal” isolates would have a reduced requirement for survival outside the host. These isolates would be predicted to be rdar negative, due to genetic drift and inactivity (i.e., genes are not needed and therefore are under reduced selection pressure) or because the rdar morphotype has a high energy cost (100). However, the evolutionary pressures could be considerably more complex if the extracellular matrix components curli and cellulose play a role in host colonization (31, 62, 80). Overall, 59% (168 of 284) of E. coli isolates analyzed were positive for rdar morphotype (curli and/or cellulose) production. This was consistent with Da Re et al. (16), who reported cellulose production by 53% (47 of 87) of natural E. coli isolates. In our collection, only 36.5% of human isolates were rdar positive. This value was lower than expected; Bokranz et al. (7) previously reported that 79% (41 of 52) of human fecal isolates and 67% (16 of 24) of urinary tract infection isolates were positive for curli and/or cellulose production. The discrepancy between our results may reflect geographical differences (19, 22). Among the phylogroups, B2, A, and the ancestral D isolates had reduced prevalence of the rdar morphotype. This can be explained for B2 and ancestral D isolates if they are host adapted. Phylogroup A isolates, however, appear to be somewhat of an anomaly; we are unsure how to explain the reduced rdar prevalence for this group of isolates. In contrast, group B1 isolates had by far the highest prevalence, which would make sense if these isolates are host generalists with an extended environmental phase in their life cycle.
For σS, this important sigma factor has been shown to regulate the expression of up to 10% of the genes in the E. coli genome (91). A robust stress response system is necessary to ensure E. coli survival under harsh conditions; however, it also associated with a decreased ability to utilize nutrients (53). In our study, nonhuman isolates in all phylogroups consistently had higher prevalence of σS activity than human isolates. This may indicate that nonhuman E. coli isolates spend more time in the environment than their human counterparts. Conversely, human E. coli isolates may have sacrificed their σS-mediated stress responses in order to maximize growth rate within the human GI tract (25). However, we did not detect any metabolic distinction between E. coli isolates from different host sources, in agreement with what has been reported previously (47). There was a slight correlation with different phylogenetic groups but not enough to identify clear metabolic trends. It is hypothesized that the cycling of E. coli isolates between the host and the environment will lead to a balance of intact and mutated rpoS alleles within the population (25). An overall rate of 20 to 30% rpoS-defective isolates has been detected previously in several natural populations (5, 26, 90), which is consistent with our analysis. The phylogroup B2 and ancestral group D isolates were by far the most likely to have attenuated σS activity in our study. In the trade-off between stress resistance and fast growth rates, this fits the hypothesis that these isolates have a reduced requirement for survival and persistence in the environment. There are likely two different scales of evolution occurring. Short-term adaptation within an individual host over time, as has been observed for Pseudomonas aeruginosa isolates from cystic fibrosis patients (82), could result in localized loss of σS activity and the rdar morphotype. A more specific, long-term “human” adaptation might be expected to result in loss of genes. However, this was not observed.
One of the main questions in any phylogenetic analysis of E. coli is whether each isolate needs to be evaluated individually in terms of selection pressure and the environments that it is adapted to living in or whether there are group-wide trends within the population. Recent work by Luo et al. (58) has shown that E. coli diversity is greater than previously thought, with some isolates adapted to live almost exclusively in the environment. There is also a lot of variability in the pathogenic potential of isolates; although phylogroup B2 isolates consistently have the highest prevalence of extraintestinal virulence genes (21, 49, 50, 69), there are many examples of disease-causing E. coli strains within the normally nonvirulent phylogroup B1 cluster (88, 95). Another unknown in population-based studies is whether the E. coli isolates being evaluated are resident or transient strains. Short of analyzing gut biopsy samples, it may be impossible to differentiate these classes of isolates. Gordon and Cowling (34) stated that the likelihood of a host harboring E. coli depends on (i) the frequency with which a host individual is exposed to E. coli, (ii) the probability that an exposure event will result in the establishment of a population, and (iii) the mean length of time the E. coli population can persist in the host. Evolutionary success is likely dependent on the ability of an E. coli isolate to establish a population in the GI tract of the host (34). It has been suggested that acquisition of virulence genes may be advantageous for E. coli colonization of the human GI tract and represent a niche adaptation (55, 65). There has also been a human-specific lineage of E. coli phylogroup B2 recently described (13).
Many unresolved questions remain for understanding the complex population biology of E. coli. Our findings suggest that genome-wide, sequence-based approaches will not be successful in determining E. coli host specificity, at least within the core genome. However, phenotypic analysis revealed that there are different selection pressures acting on groups of E. coli isolates, which may be indicative of different lifestyles or ecological niches. This fits with the findings of Luo et al. (58), who recently questioned the dogma that the mammalian intestinal tract is the preferred niche of E. coli. Not every isolate survives an equal length of time in the environment (2, 32), and there is even the possibility of a dormant state (57). For some E. coli isolates, survival and persistence in the environment may be as important as the ability to colonize and grow within the GI tract of a host.
ACKNOWLEDGMENTS
A.P.W., K.A.S., M.G.S., and N.F.N. planned the experiments; A.P.W., K.A.S., C.D.S., and R.S. performed the experiments; A.P.W., K.A.S., R.S., T.A.E., and N.F.N. analyzed the data; J.D.W. curated the sequence alignments and generated the phylogenetic trees; and A.P.W., K.A.S., M.G.S., and N.F.N. wrote the paper.
Funding for this project was provided by the Alberta Water Research Institute.
We thank Brett Trost (University of Saskatchewan) for performing hierarchical cluster analysis of the Biolog data and Lorraine Ingham, Edie Ashton, Moira Dawson, Cheryl Hilner, and Sherilynn Fowler (Alberta Provincial Laboratory for Public Health) for laboratory isolation of E. coli from human and animal samples.
Footnotes
Published ahead of print on 9 September 2011.
REFERENCES
- 1. Barker C. S., Pruss B. M., Matsumura P. 2004. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J. Bacteriol. 186:7529–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Barnes B., Gordon D. M. 2004. Coliform dynamics and the implications for source tracking. Environ. Microbiol. 6:501–509 [DOI] [PubMed] [Google Scholar]
- 3. Barrick J. E., et al. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243–1247 [DOI] [PubMed] [Google Scholar]
- 4. Bauchart P., et al. 2010. Pathogenomic comparison of human extraintestinal and avian pathogenic Escherichia coli–search for factors involved in host specificity or zoonotic potential. Microb. Pathog. 49:105–115 [DOI] [PubMed] [Google Scholar]
- 5. Bhagwat A. A., et al. 2005. Characterization of enterohemorrhagic Escherichia coli strains based on acid resistance phenotypes. Infect. Immun. 73:4993–5003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bingen-Bidois M., et al. 2002. Phylogenetic analysis and prevalence of urosepsis strains of Escherichia coli bearing pathogenicity island-like domains. Infect. Immun. 70:3216–3226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bokranz W., Wang X., Tschape H., Romling U. 2005. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 54:1171–1182 [DOI] [PubMed] [Google Scholar]
- 8. Boyd E. F., Hartl D. L. 1998. Chromosomal regions specific to pathogenic isolates of Escherichia coli have a phylogenetically clustered distribution. J. Bacteriol. 180:1159–1165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brombacher E., Dorel C., Zehnder A. J., Landini P. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149:2847–2857 [DOI] [PubMed] [Google Scholar]
- 10. Caprioli A., Morabito S., Brugere H., Oswald E. 2005. Enterohaemorrhagic Escherichia coli: emerging issues on virulence and modes of transmission. Vet. Res. 36:289–311 [DOI] [PubMed] [Google Scholar]
- 11. Chen S. L., et al. 2006. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. U. S. A. 103:5977–5982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clermont O., Bonacorsi S., Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555–4558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Clermont O., et al. 2008. Evidence for a human-specific Escherichia coli clone. Environ. Microbiol. 10:1000–1006 [DOI] [PubMed] [Google Scholar]
- 14. Collinson S. K., Emody L., Trust T. J., Kay W. W. 1992. Thin aggregative fimbriae from diarrheagenic Escherichia coli. J. Bacteriol. 174:4490–4495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crozat E., et al. 2011. Altered regulation of the OmpF porin by Fis in Escherichia coli during an evolution experiment and between B and K-12 strains. J. Bacteriol. 193:429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Da Re S., Ghigo J. M. 2006. A CsgD-independent pathway for cellulose production and biofilm formation in Escherichia coli. J. Bacteriol. 188:3073–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Davidson C. J., White A. P., Surette M. G. 2008. Evolutionary loss of the rdar morphotype in Salmonella as a result of high mutation rates during laboratory passage. ISME J. 2:293–307 [DOI] [PubMed] [Google Scholar]
- 18. Dobrindt U., et al. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J. Bacteriol. 185:1831–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duriez P., et al. 2001. Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology 147:1671–1676 [DOI] [PubMed] [Google Scholar]
- 20. Elder R. O., et al. 2000. Correlation of enterohemorrhagic Escherichia coli O157 prevalence in feces, hides, and carcasses of beef cattle during processing. Proc. Natl. Acad. Sci. U. S. A. 97:2999–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Escobar-Paramo P., et al. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085–1094 [DOI] [PubMed] [Google Scholar]
- 22. Escobar-Paramo P., et al. 2004. Large-scale population structure of human commensal Escherichia coli isolates. Appl. Environ. Microbiol. 70:5698–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Escobar-Paramo P., et al. 2006. Identification of forces shaping the commensal Escherichia coli genetic structure by comparing animal and human isolates. Environ. Microbiol. 8:1975–1984 [DOI] [PubMed] [Google Scholar]
- 24. Ferenci T. 2005. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol. Microbiol. 57:1–8 [DOI] [PubMed] [Google Scholar]
- 25. Ferenci T. 2003. What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol. 11:457–461 [DOI] [PubMed] [Google Scholar]
- 26. Ferenci T., Galbiati H. F., Betteridge T., Phan K., Spira B. 2011. The constancy of global regulation across a species: the concentrations of ppGpp and RpoS are strain-specific in Escherichia coli. BMC Microbiol. 11:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferenci T., Spira B. 2007. Variation in stress responses within a bacterial species and the indirect costs of stress resistance. Ann. N. Y. Acad. Sci. 1113:105–113 [DOI] [PubMed] [Google Scholar]
- 28. Fournier P. E., Zhu Y., Ogata H., Raoult D. 2004. Use of highly variable intergenic spacer sequences for multispacer typing of Rickettsia conorii strains. J. Clin. Microbiol. 42:5757–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Futuyma D. J., Moreno G. 1988. The evolution of ecological specialization. Annu. Rev. Ecol. Evol. Syst. 19:207–233 [Google Scholar]
- 30. Glazunova O., et al. 2005. Coxiella burnetii genotyping. Emerg. Infect. Dis. 11:1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gophna U., et al. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69:2659–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gordon D. M. 2001. Geographical structure and host specificity in bacteria and the implications for tracing the source of coliform contamination. Microbiology 147:1079–1085 [DOI] [PubMed] [Google Scholar]
- 33. Gordon D. M., Clermont O., Tolley H., Denamur E. 2008. Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environ. Microbiol. 10:2484–2496 [DOI] [PubMed] [Google Scholar]
- 34. Gordon D. M., Cowling A. 2003. The distribution and genetic structure of Escherichia coli in Australian vertebrates: host and geographic effects. Microbiology 149:3575–3586 [DOI] [PubMed] [Google Scholar]
- 35. Govons S., Vinopal R., Ingraham J., Preiss J. 1969. Isolation of mutants of Escherichia coli B altered in their ability to synthesize glycogen. J. Bacteriol. 97:970–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gualdi L., Tagliabue L., Landini P. 2007. Biofilm formation-gene expression relay system in Escherichia coli: modulation of sigmaS-dependent gene expression by the CsgD regulatory protein via σS protein stabilization. J. Bacteriol. 189:8034–8043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guindon S., et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321 [DOI] [PubMed] [Google Scholar]
- 38. Guindon S., Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 39. Guttman D. S., Dykhuizen D. E. 1994. Clonal divergence in Escherichia coli as a result of recombination, not mutation. Science 266:1380–1383 [DOI] [PubMed] [Google Scholar]
- 40. Hengge R. 2008. The two-component network and the general stress sigma factor RpoS (sigma S) in Escherichia coli. Adv. Exp. Med. Biol. 631:40–53 [DOI] [PubMed] [Google Scholar]
- 41. Herzer P. J., Inouye S., Inouye M., Whittam T. S. 1990. Phylogenetic distribution of branched RNA-linked multicopy single-stranded DNA among natural isolates of Escherichia coli. J. Bacteriol. 172:6175–6181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Holt J. G., Krieg N. R., Sneath P. H. A., Staley J. T., Williams S. T. (ed.). 1994. Bergey's manual of determinative bacteriology, 9th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 43. Hommais F., Pereira S., Acquaviva C., Escobar-Paramo P., Denamur E. 2005. Single-nucleotide polymorphism phylotyping of Escherichia coli. Appl. Environ. Microbiol. 71:4784–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hordijk W., Gascuel O. 2005. Improving the efficiency of SPR moves in phylogenetic tree search methods based on maximum likelihood. Bioinformatics 21:4338–4347 [DOI] [PubMed] [Google Scholar]
- 45. Hrudey S. E., Payment P., Huck P. M., Gillham R. W., Hrudey E. J. 2003. A fatal waterborne disease epidemic in Walkerton, Ontario: comparison with other waterborne outbreaks in the developed world. Water Sci. Technol. 47:7–14 [PubMed] [Google Scholar]
- 46. Huang K. J., Igo M. M. 1996. Identification of the bases in the ompF regulatory region, which interact with the transcription factor OmpR. J. Mol. Biol. 262:615–628 [DOI] [PubMed] [Google Scholar]
- 47. Ihssen J., et al. 2007. Comparative genomic hybridization and physiological characterization of environmental isolates indicate that significant (eco-)physiological properties are highly conserved in the species Escherichia coli. Microbiology 153:2052–2066 [DOI] [PubMed] [Google Scholar]
- 48. Johnson J. R., et al. 2006. Experimental mouse lethality of Escherichia coli isolates, in relation to accessory traits, phylogenetic group, and ecological source. J. Infect. Dis. 194:1141–1150 [DOI] [PubMed] [Google Scholar]
- 49. Johnson J. R., Delavari P., Kuskowski M., Stell A. L. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78–88 [DOI] [PubMed] [Google Scholar]
- 50. Johnson J. R., Stell A. L. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261–272 [DOI] [PubMed] [Google Scholar]
- 51. Kalir S., et al. 2001. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science 292:2080–2083 [DOI] [PubMed] [Google Scholar]
- 52. Kaper J. B., Nataro J. P., Mobley H. L. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 53. King T., Ishihama A., Kori A., Ferenci T. 2004. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J. Bacteriol. 186:5614–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lacour S., Landini P. 2004. SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186:7186–7195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Le Gall T., et al. 2007. Extraintestinal virulence is a coincidental by-product of commensalism in B2 phylogenetic group Escherichia coli strains. Mol. Biol. Evol. 24:2373–2384 [DOI] [PubMed] [Google Scholar]
- 56. Letunic I., Bork P. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127–128 [DOI] [PubMed] [Google Scholar]
- 57. Lim C. H., Flint K. P. 1989. The effects of nutrients on the survival of Escherichia coli in lake water. J. Appl. Bacteriol. 66:559–569 [DOI] [PubMed] [Google Scholar]
- 58. Luo C., et al. 2011. Genome sequencing of environmental Escherichia coli expands understanding of the ecology and speciation of the model bacterial species. Proc. Natl. Acad. Sci. U. S. A. 108:7200–7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mattison K., Oropeza R., Byers N., Kenney L. J. 2002. A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J. Mol. Biol. 315:497–511 [DOI] [PubMed] [Google Scholar]
- 60. McAdams H. H., Srinivasan B., Arkin A. P. 2004. The evolution of genetic regulatory systems in bacteria. Nat. Rev. Genet. 5:169–178 [DOI] [PubMed] [Google Scholar]
- 61. McQuitty L. L. 1966. Similarity analysis by reciprocal pairs for discrete and continuous data. Educ. Psychol. Meas. 26:825–831 [Google Scholar]
- 62. Monteiro C., et al. 2009. Characterization of cellulose production in Escherichia coli Nissle 1917 and its biological consequences. Environ. Microbiol. 11:1105–1116 [DOI] [PubMed] [Google Scholar]
- 63. Newell D. G., et al. 2010. Food-borne diseases—the challenges of 20 years ago still persist while new ones continue to emerge. Int. J. Food Microbiol. 139(Suppl. 1):S3–S15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Notley-McRobb L., King T., Ferenci T. 2002. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J. Bacteriol. 184:806–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nowrouzian F. L., Wold A. E., Adlerberth I. 2005. Escherichia coli strains belonging to phylogenetic group B2 have superior capacity to persist in the intestinal microflora of infants. J. Infect. Dis. 191:1078–1083 [DOI] [PubMed] [Google Scholar]
- 66. Ogura Y., et al. 2009. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 106:17939–17944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Osborne S. E., et al. 2009. Pathogenic adaptation of intracellular bacteria by rewiring a cis-regulatory input function. Proc. Natl. Acad. Sci. U. S. A. 106:3982–3987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Peterson C. N., Mandel M. J., Silhavy T. J. 2005. Escherichia coli starvation diets: essential nutrients weigh in distinctly. J. Bacteriol. 187:7549–7553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Picard B., et al. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Posada D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253–1256 [DOI] [PubMed] [Google Scholar]
- 71. Prigent-Combaret C., et al. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213–7223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Pupo G. M., Karaolis D. K., Lan R., Reeves P. R. 1997. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect. Immun. 65:2685–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rasko D. A., et al. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190:6881–6893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. R Development Core Team 2011. R: a language and environment for statistical computing, 2.13.1 ed. R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 75. Robbe-Saule V., et al. 2006. Crl activates transcription initiation of RpoS-regulated genes involved in the multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 188:3983–3994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Romling U., Bian Z., Hammar M., Sierralta W. D., Normark S. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Romling U., Sierralta W. D., Eriksson K., Normark S. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249–264 [DOI] [PubMed] [Google Scholar]
- 78. Ron E. Z. 2006. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr. Opin. Microbiol. 9:28–32 [DOI] [PubMed] [Google Scholar]
- 79. Ryu J. H., Beuchat L. R. 2005. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl. Environ. Microbiol. 71:247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Saldana Z., et al. 2009. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 11:992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Savageau M. A. 1983. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am. Nat. 122:732–744 [Google Scholar]
- 82. Smith E. E., et al. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Takahashi A., et al. 2006. Escherichia coli isolates associated with uncomplicated and complicated cystitis and asymptomatic bacteriuria possess similar phylogenies, virulence genes, and O-serogroup profiles. J. Clin. Microbiol. 44:4589–4592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tominaga A., Lan R., Reeves P. R. 2005. Evolutionary changes of the flhDC flagellar master operon in Shigella strains. J. Bacteriol. 187:4295–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Touchon M., et al. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Uhlich G. A., Cooke P. H., Solomon E. B. 2006. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl. Environ. Microbiol. 72:2564–2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Uhlich G. A., Keen J. E., Elder R. O. 2001. Mutations in the csgD promoter associated with variations in curli expression in certain strains of Escherichia coli O157:H7. Appl. Environ. Microbiol. 67:2367–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Viljanen M. K., et al. 1990. Outbreak of diarrhoea due to Escherichia coli O111:B4 in schoolchildren and adults: association of Vi antigen-like reactivity. Lancet 336:831–834 [DOI] [PubMed] [Google Scholar]
- 89. Walsh P. S., Metzger D. A., Higuchi R. 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10:506–513 [PubMed] [Google Scholar]
- 90. Waterman S. R., Small P. L. 1996. Characterization of the acid resistance phenotype and rpoS alleles of Shiga-like toxin-producing Escherichia coli. Infect. Immun. 64:2808–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Weber H., Polen T., Heuveling J., Wendisch V. F., Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. White A. P., Gibson D. L., Kim W., Kay W. W., Surette M. G. 2006. Thin aggregative fimbriae and cellulose enhance long-term survival and persistence of Salmonella. J. Bacteriol. 188:3219–3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. White A. P., Surette M. G. 2006. Comparative genetics of the rdar morphotype in Salmonella. J. Bacteriol. 188:8395–8406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. White A. P., et al. 2010. A global metabolic shift is linked to salmonella multicellular development. PLoS One 5:e11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Wirth T., et al. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol. Microbiol. 60:1136–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24:1586–1591 [DOI] [PubMed] [Google Scholar]
- 97. Zambrano M. M., Siegele D. A., Almiron M., Tormo A., Kolter R. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757–1760 [DOI] [PubMed] [Google Scholar]
- 98. Zaunbrecher M. A., Sikes R. D., Jr, Metchock B., Shinnick T. M., Posey J. E. 2009. Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U. S. A. 106:20004–20009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Zhao K., Liu M., Burgess R. R. 2010. Promoter and regulon analysis of nitrogen assimilation factor, sigma54, reveal alternative strategy for E. coli MG1655 flagellar biosynthesis. Nucleic Acids Res. 38:1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Zogaj X., Nimtz M., Rohde M., Bokranz W., Romling U. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452–1463 [DOI] [PubMed] [Google Scholar]