Abstract
CodY is a global transcriptional regulator known to control expression of more than 100 genes and operons in Bacillus subtilis. Some of the most strongly repressed targets of CodY, the nupNOPQ (formerly, yufNOPQ) genes, were found to encode a guanosine transporter. Using DNase I footprinting experiments, we identified two high-affinity CodY-binding sites in the regulatory region of the nupN gene. The two sites are located 50 bp upstream and 163 bp downstream of the transcription start site. The downstream site was responsible for 6- to 8-fold nupN repression in the absence of the upstream site. When the upstream site was intact, however, only a minor contribution of the downstream site to nupN regulation could be detected under the conditions tested. Both sites contained 15-bp CodY-binding motifs with two mismatches each with respect to the consensus sequence, AATTTTCWGTTTTAA. However, the experimentally determined binding sites included additional sequences flanking the 15-bp CodY-binding motifs. An additional version of the 15-bp CodY-binding motif, with 5 mismatches with respect to the consensus but essential for efficient regulation by CodY, was found within the upstream site. The presence of multiple 15-bp motifs may be a common feature of CodY-binding sites.
INTRODUCTION
Bacillus subtilis cells are able to take up purine and pyrimidine nucleosides, allowing these compounds to serve as substrates for nucleotide biosynthesis via salvage pathways and as sources of carbon and nitrogen (40). Although two major transporters for pyrimidine and purine nucleosides, NupC and NupG, respectively, have been described in B. subtilis, mutants lacking NupC or NupG retain considerable ability to take up pyrimidine or purine nucleosides (19, 33). Thus, additional transporters for both pyrimidine and purine nucleosides remain to be identified (2, 19, 33).
The B. subtilis nupNOPQ (formerly, yufNOPQ) genes appear to encode the components of an ABC transport system of unknown specificity. The first gene of this apparent operon, nupN, codes for an apparent lipoprotein that could serve as the substrate-binding, specificity-determining subunit of the transporter. The putative product of nupN is homologous (43 to 50% identity) to several proteins that are involved in purine and pyrimidine nucleoside transport in Treponema pallidum, Streptococcus mutans, and Lactococcus lactis (9, 26, 41). The nupO and nupPQ genes appear to encode the ATP-binding protein and integral membrane proteins typical of ABC transporters, respectively.
The nupNOPQ genes were previously shown to be among the genes that are most highly repressed by CodY as detected by DNA microarray analysis and lacZ fusions (29). CodY is a global transcriptional regulator that controls expression of more than 100 genes and operons in B. subtilis.
Many of the CodY-regulated genes are involved in nitrogen or carbon metabolism (14, 29, 37–39). The main role of CodY in B. subtilis appears to be establishing the temporal hierarchy of utilization of various nitrogen (and carbon) compounds under conditions of nutrient excess. CodY homologs are also present in most other low-G+C, Gram-positive bacteria and in many species have been shown to coordinate expression of virulence-associated functions with expression of metabolic genes (see references 12, 21, 25, and 38 and references therein).
The activity of B. subtilis CodY as a DNA-binding protein (20, 23, 35) is increased by interaction with two types of effectors, branched-chain amino acids (isoleucine, leucine, and valine [ILV]) (10, 15, 22, 23, 30, 36) and GTP (17, 29, 31, 36), allowing the bacteria to change the pattern of CodY-dependent gene expression in response to the availability of nutrients in the growth medium.
We demonstrate here that the nupNOPQ genes encode a previously unidentified transporter for guanosine and, possibly, other nucleosides. Unexpected aspects of the mechanism by which CodY mediates repression of transcription from the nupN promoter are also described.
MATERIALS AND METHODS
Bacterial strains and culture media.
The B. subtilis strains constructed in this study were all derivatives of strain SMY (44) and are described in Table 1 and in the text. Escherichia coli strain JM107 (43) was used for isolation of plasmids. Bacterial growth in DS nutrient broth and minimal TSS medium was as described previously (5).
Table 1.
B. subtilis strains used in this study
| Strain | Genotype | Source or referencea |
|---|---|---|
| CU4121 | purM::Tn917 (erm) trpC2 | BGSC |
| PS251 | codY::(erm::spc) trpC2 | P. Serror |
| SMY | Prototroph | 44 |
| YXJAd | nupG::pMutin1 (erm) | NBRP |
| NUPNd | nupN::pMutin2 (erm) | NBRP |
| NUPOd | nupO::pMutin2 (erm) | NBRP |
| BB1888 | lacA::tet | 6 |
| BB2511 | ΔamyE::spc lacA::tet | 6 |
| BB2686 | ΔamyE::[erm Φ(nupN503p+-lacZ)]lacA::tet | BB2511 × pBB1474 |
| BB2809 | ΔamyE::[erm Φ(nupN276p+-lacZ)]lacA::tet | BB2511 × pBB1520 |
| BB2899 | ΔamyE::[erm Φ(nupN347p+-lacZ)]lacA::tet | BB2511 × pBB1521 |
| BB2900 | ΔamyE::[erm Φ(nupN120p+-lacZ)]lacA::tet | BB2511 × pBB1522 |
| BB2935 | ΔamyE::[erm Φ(nupN367p+-lacZ)]lacA::tet | BB2511 × pBB1534 |
| BB2936 | ΔamyE::[erm Φ(nupN140p+-lacZ)]lacA::tet | BB2511 × pBB1535 |
| BB2952 | ΔamyE::[erm Φ(nupN367p1-lacZ)] lacA::tet | BB2511 × pBB1542 |
| BB2953 | ΔamyE::[erm Φ(nupN380p+-lacZ)]lacA::tet | BB2511 × pBB1543 |
| BB2954 | ΔamyE::[erm Φ(nupN140p1-lacZ)] lacA::tet | BB2511 × pBB1544 |
| BB2955 | ΔamyE::[erm Φ(nupN153p+-lacZ)]lacA::tet | BB2511 × pBB1545 |
| BB2972 | ΔamyE::[erm Φ(nupN380p21-lacZ)] lacA::tet | BB2511 × pBB1554 |
| BB2973 | ΔamyE::[erm Φ(nupN380p22-lacZ)] lacA::tet | BB2511 × pBB1555 |
| BB2974 | ΔamyE::[erm Φ(nupN153p21-lacZ)] lacA::tet | BB2511 × pBB1556 |
| BB2975 | ΔamyE::[erm Φ(nupN153p22-lacZ)] lacA::tet | BB2511 × pBB1557 |
| BB3014 | ΔamyE::[erm Φ(nupN153p2-lacZ)] lacA::tet | BB2511 × pBB1574 |
| BB3015 | ΔamyE::[erm Φ(nupN153p3-lacZ)] lacA::tet | BB2511 × pBB1575 |
| BB3322 | ΔnupN amyE::spc lacA::tet | BB2511 × pBB1654 |
| BB3401 | ΔamyE::[erm Φ(nupN347p11-lacZ)] lacA::tet | BB2511 × pBB1676 |
| BB3402 | ΔamyE::[erm Φ(nupN503p12-lacZ)] lacA::tet | BB2511 × pBB1677 |
| BB3431 | ΔamyE::[erm Φ(nupN380p3/11-lacZ)] lacA::tet | BB2511 × pBB1691 |
| BB3484 | ΔnupN amyE::spc lacA::tet purM::Tn917 (erm) | BB3322 × CU4121 DNA |
| BB3486 | ΔnupN nupG::pMutin1 (erm) amyE::spc lacA::tet | BB3332 × YXJAd DNA |
| BB3491 | ΔamyE::[erm Φ(nupN380p3-lacZ)] lacA::tet | BB2511 × pBB1698 |
| BB3492 | ΔamyE::[erm Φ(nupN380p11-lacZ)] lacA::tet | BB2511 × pBB1699 |
| BB3493 | ΔamyE::[erm Φ(nupN347p12-lacZ)] lacA::tet | BB2511 × pBB1700 |
| BB3499 | ΔnupN nupG::pMutin1 (erm) amyE::spc lacA::tet purM::(Tn917::neo) | BB3485 × YXJAd DNA |
| BB3501 | nupG::pMutin1 (erm) lacA::tet | BB1888 × YXJAd DNA |
| BB3502 | nupG::pMutin1 (erm) lacA::tet purM::(Tn917::neo) | BB3501 × BB3499 DNA |
| BB3546 | purM::(Tn917::neo) | SMY × BB3485 DNA |
| BB3597 | ΔamyE::[erm Φ(nupN153p23-lacZ)]lacA::tet | BB2511 × pBB1724 |
| BB3598 | ΔamyE::[erm Φ(nupN153p24-lacZ)]lacA::tet | BB2511 × pBB1725 |
| BB3599 | ΔamyE::[erm Φ(nupN153p2/21-lacZ)]lacA::tet | BB2511 × pBB1726 |
BSCG, Bacillus Genetic Stock Center; NBRP, National BioResource Project (NIG, Japan): B. subtilis (http://www.shigen.nig.ac.jp/bsub/). × denotes transformation by plasmid or chromosomal DNA.
DNA manipulations.
Methods for common DNA manipulations, transformation, primer extension, DNA sequencing, and sequence analysis were performed as previously described (5, 6, 32). Procedures for gel shift experiments and DNase I footprinting were described in detail previously (5, 6, 32). Chromosomal DNA of B. subtilis strain SMY or plasmids constructed in this work were used as templates for PCR. The oligonucleotides used in this work are described in Table 2. All cloned PCR-generated fragments were verified by sequencing.
Table 2.
Oligonucleotides used in this work
| Oligonucleotide type and name | Sequencea | Specificity |
|---|---|---|
| Flanking primers | ||
| Forward | ||
| oBB67 | 5′-GCTTCTAAGTCTTATTTCC | erm (pHK23) |
| oBB311 | 5′-CAATATCTAGATAAGAAAACGCACTGC | nupN503 |
| oBB365 | 5′-TTAAATCTAGATGCGAAATTCTATTTATTTC | nupN347, 5′ flank of nupN |
| oBB367 | 5′-CTAAATCTAGATAATTTTTAAAAAATTATGCG | nupN367 |
| oBB378 | 5′-CTAAATCTAGATAATTTTCAGAAAATTATGCG | nupNp1 |
| oBB379 | 5′-TTTTATCTAGATAATTCTAAAAATAGATAA | nupN380 |
| oBB390 | 5′-TTTTATCTAGATAATTCTGAAAATAGATAA | nupNp21 |
| oBB391 | 5′-TTTTATCTAGATAATTgTAAAAATAGATAA | nupNp22 |
| oBB400 | 5′-TTTTATCTAGATAATTCTAAAAATAGATAATTTTTAAcAAATTATGC | nupNp2 |
| oBB401 | 5′-TTTTATCTAGATAATTCTAAAAATAGATAAaTTTTAAAAAATTATG | nupNp3 |
| oBB412 | 5′-AAAAAGGTACCGTCTAGTTAATGTGTAAC | clpB (pMAD) |
| oBB437 | 5′-CCTACAAGCTTGATCAAGACGGAG | 3′ flank of nupN |
| oBB567 | 5′-GATCCTCTAGATAATTCTAcAAATAG | nupNp23 |
| oBB568 | 5′-GATCCTCTAGATAAaTCTAAAAATAG | nupNp24 |
| Reverse | ||
| oBB102 | 5′-CACCTTTTCCCTATATAAAAGC | lacZ (pHK23) |
| oBB253 | 5′-GGTTTTCCCGGTCGAC | lacZ (pHK23) |
| oBB312 | 5′-ACAAGAAGCTTCGCCAATCCGATTTTTC | nupN503, 5′ flank of nupN |
| oBB357 | 5′-CGGAAAAGCTTCCGGGCGGAAAACC | nupN276 |
| oBB408 | 5′-CGAGAGCCGGCTAAACCTTCCCGGCTTC | bgaB (pMAD) |
| oBB438 | 5′-TTTTCCTCGAGCAACGCGTGTATTTC | 3′ flank of nupN |
| Internal mutagenic primers | ||
| Forward | ||
| oBB502 | 5′-CCATTTGTAATTTTCAGAAAATTTTATC | nupNp11 |
| oBB504 | 5′-CCATTTGTTATTATCAcAAAATTTTATC | nupNp12 |
| Reverse | ||
| oBB501 | 5′-TTTCTGAAAATTACAAATGGAATGCGC | nupNp11 |
| oBB503 | 5′-TgTGATAATAACAAATGGAATG | nupNp12 |
The altered nucleotides are in boldface; those conferring up mutations in the CodY-binding motif are in uppercase, and those conferring down mutations are in lowercase. The restriction sites are underlined.
Construction of an nupN (yufN)-null mutant.
The 0.35-kb PCR fragments containing the 5′ or 3′ part of the nupN gene and the adjacent sequences, respectively, were cloned in two steps as contiguous inserts between the XbaI and HindIII or HindIII and XhoI sites of pBB544 (4). Then, the XbaI-XhoI fragment of the resulting plasmid, pBB1625, was recloned in the integrative plasmid pBB1579. pBB1579 (Neor) is a derivative of pBB544 that contains the clpB-bgaB construct expressing thermostable Bacillus stearothermophilus β-galactosidase from the constitutive clpB promoter of Staphylococcus aureus. The 2.1-kb clpP-bgaB fragment was synthesized by PCR using pMAD (1) as a template and cloned between the KpnI and NgoMI sites of pBB544.
The resulting plasmid, pBB1654, containing an in-frame deletion within the nupN gene was introduced by a single-crossover homologous recombination event into the nupN chromosomal locus of strain BB2511 (amyE lacA). White Neos colonies indicating excision of pBB1654 from the chromosome were searched for on plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), the colored substrate of β-galactosidase. The presence of the nupN deletion in strain BB3322 was confirmed by analyzing the size of the chromosomal nupN allele by PCR.
Construction of transcriptional lacZ fusions.
Plasmid pBB1474 containing a transcriptional nupN503-lacZ fusion was created by cloning in pHK23 (6) the XbaI- and HindIII-treated 0.52-kb nupN PCR product created with oligonucleotides oBB311 and oBB312 as the forward and reverse primers, respectively. pBB1474 contained a G-to-A substitution at position −176 with respect to the nupN transcription start point (see below) that was apparently introduced by PCR and was unlikely to affect nupN expression. (This mutation was later deleted when nupN380-lacZ and most other fusions were constructed.) Plasmids pBB1521 (nupN347p+-lacZ), pBB1534 (nupN367p+-lacZ), and pBB1543 (nupN380p+-lacZ) carrying a 347-bp, 367-bp, or 380-bp version of the nupN regulatory region truncated by 156, 136, or 123 bp at the 5′ end, respectively, were created in a manner similar to pBB1474, but with oligonucleotide oBB365 or oBB379 as the forward PCR primer, respectively.
Plasmids pBB1520 (nupN276p+-lacZ), pBB1522 (nupN120p+-lacZ), and pBB1545 (nupN153p+-lacZ), carrying 276-bp, 120-bp, or 153-bp versions of the nupN regulatory region truncated by 227 bp at the 3′ end were created in a manner similar to pBB1474, pBB1521, or pBB1543, respectively, but with oligonucleotide oBB357 as the reverse PCR primer.
Plasmid pBB1535 (nupN140p+-lacZ), carrying a 140-bp version of the nupN regulatory region truncated by 136 bp at the 5′ end and 227 bp at the 3′ end, was created using oligonucleotides oBB367 and oBB357 as forward and reverse PCR primers, respectively.
B. subtilis strains carrying various lacZ fusions at the amyE locus (Table 1) were isolated after transforming strain BB2511 (amyE::spc lacA) with the appropriate plasmids, by selecting for resistance to erythromycin, conferred by the plasmids, and screening for loss of the spectinomycin resistance marker, which indicated a double-crossover, homologous recombination event. Strain BB2511 and all of its derivatives have very low endogenous β-galactosidase activity due to a null mutation in the lacA gene (8a).
Mutations in CodY-binding sites.
Plasmids pBB1542 (nupN367p1-lacZ) and pBB1544 (nupN140p1-lacZ) were constructed as described above for pBB1534 (nupN367p+-lacZ) and pBB1535 (nupN140p+-lacZ), but using mutagenic oligonucleotide oBB378 as the forward PCR primer.
Plasmids pBB1554 (nupN380p21-lacZ), pBB1555 (nupN380p22-lacZ), pBB1556 (nupN153p21-lacZ), pBB1557 (nupN153p22-lacZ), pBB1574 (nupN153p2-lacZ), pBB1575 (nupN153p3-lacZ), pBB1698 (nupN380p3-lacZ), pBB1724 (nupN153p23-lacZ), and pBB1725 (nupN153p24-lacZ) were constructed as described above for pBB1543 (nupN380p+-lacZ) or pBB1545 (nupN153p+-lacZ), but using appropriate mutagenic oligonucleotides specified in Table 2 as forward PCR primers.
Plasmid pBB1726 (nupN153p2/21-lacZ) was constructed as described above for pBB1556 (nupN153p21-lacZ), but using pBB1574 (nupN153p2-lacZ) as the PCR template.
The p11 mutation in the nupN347 regulatory region was introduced by two-step overlapping PCR. In the first step, a product containing the 5′ part of the nupN regulatory region was synthesized by using oligonucleotide oBB365 as the forward primer and mutagenic oligonucleotide oBB501 as the reverse primer. A product containing the 3′ part of the nupN regulatory region was synthesized by using mutagenic oligonucleotide oBB502 as the forward primer and oBB312 as the reverse primer. The PCR products were used in a second, splicing step of mutagenesis as overlapping PCR templates to generate a modified fragment containing the entire nupN regulatory region; oligonucleotides oBB365 and oBB312 served as the forward and reverse PCR primers, respectively. The spliced PCR product was digested with XbaI and HindIII and cloned in pHK23, as described above, to create pBB1676 (nupN347p11-lacZ). Plasmid pBB1691 (nupN380p3/11-lacZ) was created in a similar manner by using pBB1698 (nupN380p3-lacZ) as the PCR template and oBB401 instead of oBB365. Plasmid pBB1699 (nupN380p11-lacZ) was created by using pBB1691 (nupN380p3/11-lacZ) as the PCR template and oBB379 and oBB312 as primers.
The p12 mutation in the nupN503 regulatory region [pBB1677 (nupN503p12-lacZ)] was introduced by two-step overlapping PCR, as described above, by using oligonucleotide pairs oBB67 and oBB503 or oBB504 and oBB312 in the first PCR step and oBB67 and oBB312 in the second PCR step. pBB1700 (nupN347p12-lacZ) was created by using pBB1677 (nupN503p12-lacZ) as the PCR template and oBB365 and oBB312 as primers.
Guanosine uptake.
Cells were grown at 37°C in TSS medium until mid-exponential phase, collected under vacuum on 0.45-μm-pore nitrocellulose filters, washed, and resuspended at an optical density at 600 nm (OD600) of ≈0.8 in the same medium without NH4Cl. Further incubation of cells was at 25°C. [3H]guanosine (Moravek Biochemicals) was added to 1 μCi/ml (0.14 μM), and 160-μl samples were taken at the indicated time, collected immediately under vacuum on 0.45-μm-pore nitrocellulose filters, washed with 5 ml of TSS without NH4Cl but containing 10 μg/ml guanosine, dried, and counted using Ecoscint H scintillation liquid (National Diagnostics). The protein concentration was determined in sonicated cell samples using Bio-Rad protein assay reagent. A 1-ml culture at an OD600 of 1 contained 127.5 μg of protein.
Labeling of DNA fragments.
The PCR products containing the regulatory region of the nupN gene were synthesized using vector-specific oligonucleotides oBB67 and oBB102 as the forward and reverse primers, respectively. oBB67 starts 96 bp upstream of the XbaI site used for cloning, and oBB102 starts 36 bp downstream of the HindIII site that serves as a junction between the promoters and the lacZ part of the nupN fusion. oBB102 (which would prime synthesis of the template strand of the PCR products) was labeled using T4 polynucleotide kinase and [γ-32P]ATP.
Purification of CodY.
Wild-type CodY was purified to near homogeneity as described previously (6).
Enzyme assays.
β-Galactosidase specific activity was determined as described previously (8).
RESULTS
Phenotype of a nupN (yufN)-null mutant.
A large DNA segment internal to the nupN gene was deleted without altering the reading frame of the distal part of the gene (see Materials and Methods). The deletion removed 84% of the nupN coding region. When introduced into the B. subtilis chromosome, the nupN mutation had no apparent effect on growth of cells in nutrient broth (DS) or in defined medium (TSS-glucose-ammonium). Moreover, the nupN-null mutation did not affect the ability of a purM strain, which is blocked in de novo purine biosynthesis, to use the nucleoside inosine, adenosine, guanosine, or xanthosine as a substrate for purine nucleotide biosynthesis by the salvage pathway.
B. subtilis NupG (YxjA) has been previously reported to be a major transporter of purine nucleosides inosine and guanosine. The nupG-null mutant apparently retained about 20% of its parent strain's ability to take up the nucleosides, indicating the existence of another transporter(s) with overlapping specificity (19). In our tests, the purM nupG double-null mutant (strain BB3502) was able to utilize inosine, adenosine, or guanosine to satisfy its auxotrophic requirement on solid minimal medium. (Addition of xanthosine allowed very slow growth.) However, the triple mutant strain BB3499 (purM nupG nupN) had a strong defect in the ability to grow with guanosine as the sole source of purines. No effect of the nupN mutation on the cell's ability to utilize inosine or adenosine or xanthosine as the sole purine source on solid medium was detected in the nupG purM genetic background (data not shown). In liquid minimal medium containing 0.2 mM guanosine, the purM and purM nupN mutants grew with a generation time of 63 to 65 min, the purM nupG strain had a generation time of 80 min, and the purM nupG nupN mutant grew extremely slowly, with a generation time of 5.5 h. All four strains grew at similar rates when provided with 0.2 mM adenosine or guanine (data not shown).
A similar phenotype of very slow growth with guanosine as the sole purine source was observed when a nupO-null mutation was combined with nupG and purM mutations (data not shown). Thus, nupN appears to encode a component, presumably the substrate-binding protein, of a guanosine uptake system that likely involves the products of the nupOPQ genes as well. The residual ability of the nupN nupG purM or nupO nupG purM triple-null mutants to grow with guanosine as the sole purine source, albeit very slowly, indicates that still another, minor transporter of guanosine is functional in B. subtilis cells.
Guanosine uptake.
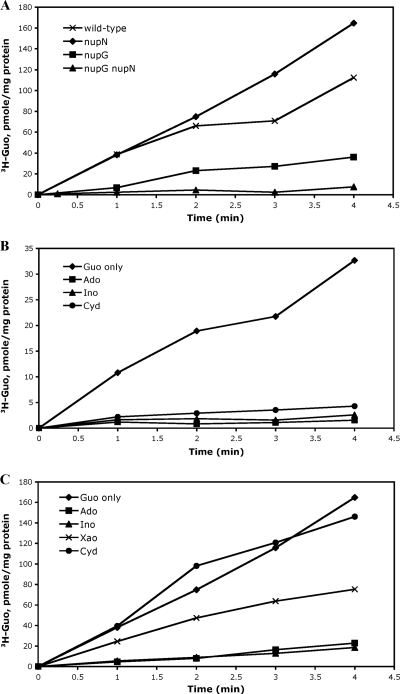
As shown previously (19), inactivation of the nupG gene led to a significant reduction in the rate of guanosine incorporation into resting cells (Fig. 1A). (Our uptake measurements do not preclude guanosine metabolism and incorporation of the guanosine derivatives into other low- and high-molecular-weight cellular compounds.) A null mutation in the nupN gene (strain BB3322) did not lead to any detectable defect in guanosine uptake. However, in the nupG nupN double-null mutant strain (BB3486), guanosine uptake was reduced to an almost undetectable level (Fig. 1A). Thus, NupN and NupG are together responsible for the bulk of guanosine uptake, though, as our growth experiments indicated, another low-activity guanosine transporter apparently exists in B. subtilis.
Fig. 1.
Preliminary characterization of the roles of NupNOPQ and NupG in nucleoside uptake. (A) Roles of NupNOPQ and NupG in guanosine uptake. Cells of strains BB2511 (wild-type), BB3322 (nupN), BB3501 (nupG), and BB3486 (nupG nupN) were assayed for guanosine incorporation as described in Materials and Methods. (B) Effect of nucleosides on guanosine uptake by NupNOPQ. Cells of strain BB3501 (nupG) were assayed for guanosine incorporation in the absence (Guo only) or presence of other nucleosides (at 50 μM) as described in Materials and Methods. Guo, guanosine; Ado, adenosine; Ino, inosine; Cyd, cytidine. (C) Effect of nucleosides on guanosine uptake by NupG. Cells of strains BB3322 (nupN) were assayed for guanosine incorporation in the absence (Guo only) or presence of other nucleosides (at 50 μM) as described in Materials and Methods. Xao, xanthosine.
Guanosine uptake in strain BB3501, provided mostly by NupN, was nearly abolished in the presence of 50 μM inosine or adenosine (357-fold excess over guanosine), suggesting that NupN binds these purine nucleosides and may also transport them (Fig. 1B). Xanthosine and the pyrimidine nucleosides cytidine, uridine, and thymidine (at 50 μM) reduced guanosine uptake partially (Fig. 1B) (data not shown), implying that NupN binds all of these nucleosides and suggesting that NupNOPQ may be a nucleoside transporter of broad specificity but with various affinities for different substrates.
The uptake of guanosine in strain BB3322, mediated mostly by NupG, was efficiently reduced by addition of inosine or adenosine, slightly reduced by xanthosine, and not affected by the presence of cytidine, uridine, or thymidine, indicating that NupG is a dedicated purine nucleoside transporter (Fig. 1C) (data not shown).
CodY-dependent regulation of the nupNOPQ genes.
The nupN gene appears to be the first gene of a four-gene (nupNOPQ) operon. We constructed a transcriptional fusion (nupN503-lacZ) containing a 503-bp DNA fragment that included the entire intergenic region upstream of the nupN gene (Fig. 2) and showed that the fusion was highly repressed by CodY. Under conditions of maximal CodY activity, in the glucose-ammonium minimal medium containing ILV and a mixture of 13 other amino acids (referred to here as the 16-amino-acid-containing medium), fusion activity in the codY-null mutant strain BB2691 was >100-fold higher (97.5 Miller units [MU]) than in wild-type strain BB2686 (Table 3).
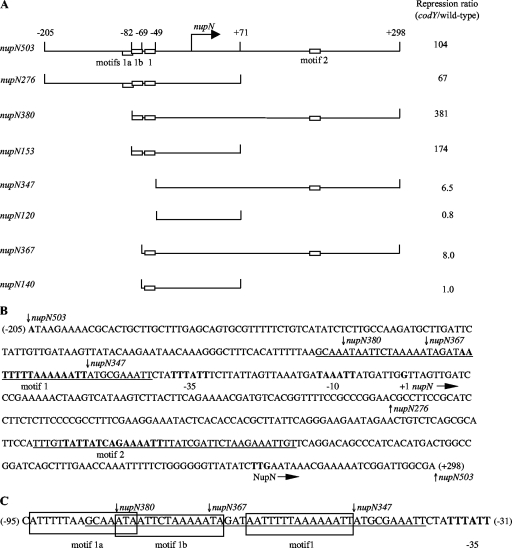
Fig. 2.
Plasmid maps and the sequence of the nupN regulatory region. (A) Schematic maps of the nupN inserts used to construct lacZ fusions. The location of the transcription start point is indicated by the bent arrow. CodY-binding motifs are shown as rectangles. The coordinates indicate the boundaries of different fusions with respect to the transcription start point. The repression ratio is the ratio of expression values for the corresponding fusions in the codY-null mutant and wild-type strain in the 16-amino-acid-containing medium. (B) Sequence of the coding (nontemplate) strand of the nupN regulatory region. The likely initiation codon, −10 and −35 promoter regions, transcription start site, and CodY-binding motifs 1 and 2 are in boldface. The direction of transcription and translation is indicated by the arrows. The sequences protected by CodY in DNase I footprinting experiments on the template strand of DNA are underlined. The boundaries of DNA fragments used to construct various lacZ fusions are indicated by vertical arrows. The coordinates of the 5′ and 3′ ends of the sequence with respect to the transcription start point are shown in parentheses. Note the incorrect annotation of the nupN initiation codon in older databases. (C) Motifs in the nupN CodY-binding site I. The sequences protected by CodY in DNase I footprinting experiments on the template strand of DNA are underlined. The sequences of motifs 1a, 1b, and 1 are boxed. The −35 promoter region is in boldface. The boundaries of DNA fragments used to construct various lacZ fusions are indicated by vertical arrows. The coordinates of the 5′ and 3′ ends of the sequence with respect to the transcription start point are shown in parentheses.
Table 3.
Expression of nupN-lacZ fusionsa
| Strain | Relevant genotype | Fusion genotypeb | Motif(s) presentc | Growth medium |
|||
|---|---|---|---|---|---|---|---|
| TSS + 16 amino acids |
TSS |
||||||
| β-Galactosidase activity (MU) | Repression ratiod | β-Galactosidase activity (MU) | Repression ratiod | ||||
| BB2686 | Wild type | nupN503p+ | 1a, 1b, 1, 2 | 0.94 | 104 | 138.0 | 1.3 |
| BB2691 | codY | 97.5 | 184.0 | ||||
| BB3402 | Wild type | nupN503p12 | 1a, 1b, 1, 2down | 2.73 | 64 | 178.0 | 1.3 |
| BB3417 | codY | 176.0 | 236.0 | ||||
| BB2809 | Wild type | nupN276p+ | 1a, 1b, 1 | 0.09 | 67 | 2.27 | 1.6 |
| BB2819 | codY | 6.04 | 3.54 | ||||
| BB2899 | Wild type | nupN347p+ | 2 | 1.61 | 6.5 | 16.1 | 0.99 |
| BB2906 | codY | 10.4 | 15.9 | ||||
| BB3401 | Wild type | nupN347p11 | 2up | 0.04 | 50 | 0.73 | 3.3 |
| BB3416 | codY | 2.01 | 2.42 | ||||
| BB3493 | Wild type | nupN347p12 | 2down | 10.8 | 1.3 | 20.3 | 0.93 |
| BB3497 | codY | 13.6 | 18.8 | ||||
| BB2900 | Wild type | nupN120p+ | None | 0.37 | 0.76 | ||
| BB2907 | codY | 0.28 | |||||
Cells were grown in TSS glucose-ammonium medium with or without a mixture of 16 amino acids. β-Galactosidase activity was assayed and expressed in Miller units (MU). All values are averages of at least two experiments, and the mean errors did not exceed 30%.
p+ indicates the presence of an unmodified promoter sequence within a fusion; for mutant promoters, the allele number is indicated.
The “up” and “down” notations indicate mutations that make CodY-binding motifs more or less similar to the consensus sequence.
The repression ratio is the ratio of expression values for the corresponding fusions in the codY-null mutant and wild-type strain in each medium.
In the wild-type strain, activity of the fusion was 15-fold derepressed (to 14.2 MU) in 13-amino-acid-containing medium (i.e., when isoleucine, leucine, and valine were omitted), and was further increased 10-fold in the absence of all amino acids. Addition of ILV alone to glucose-ammonium medium reduced expression from the nupN promoter 5-fold to 28.6 MU. This pattern of regulation is common for other CodY-dependent genes (5). Only a 1.9-fold effect of the medium composition on nupN expression was detected in cells defective for CodY, indicating that CodY itself or a CodY-dependent factor is the major regulator of the gene under the conditions tested (Table 3, strain BB2691). nupN expression in strain BB2686 was not affected by the presence of guanosine, inosine, adenosine, or uridine (200 μg/ml) or guanine or hypoxanthine (100 μg/ml) in the minimal glucose-ammonium medium (data not shown).
Transcription start point.
A primer extension experiment established that the 5′ end of nupN mRNA (corresponding to a likely transcription start point) is 270 bp upstream of the initiation codon (Fig. 2B and 3A). The sequences TTTATT and TAAATT, with 3 and 2 mismatches with respect to the −35 and −10 regions of σA-dependent promoters, and a 17-bp spacer region can be identified upstream of the apparent transcription start point (Fig. 2B).
Fig. 3.
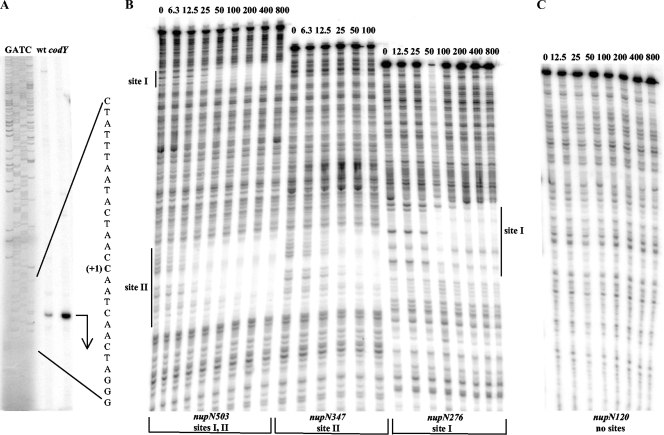
Determination of the nupN transcription start point and CodY-binding regions. (A) Primer extension analysis of the nupN mRNA. Primer oBB102 annealing to the lacZ gene of the nupN276-lacZ fusion was extended with reverse transcriptase using as the template total RNA from fusion-containing strains BB2809 (wt) and BB2819 (codY) grown in the 16-amino-acid-containing medium. The sequence of the template strand of pBB1520 determined from reactions primed with oBB102 is shown to the left. The apparent transcription start site of the nupN gene is in bold and marked by the +1 notation. A bent arrow indicates the direction of transcription. (B) DNase I footprinting analysis of CodY binding to the nupN regulatory region. The nupNp+ DNA fragments labeled on the template strand were incubated with increasing amounts of purified CodY in the presence of 10 mM ILV and 2 mM GTP and then with DNase I. The apparent transcription start site and direction of nupN transcription are shown by the bent arrow. The protected areas are indicated by the vertical lines. CodY concentrations used (nanomolar concentrations of monomers) are indicated above each lane. (C) Same as panel B, nupN120p+ fragment.
CodY binding to the nupN gene.
Recently, we have shown that two B. subtilis genes, bcaP and ybgE, contain two CodY-binding sites each, as defined by gel mobility shift and DNase I footprinting assays. The upstream sites that overlap the corresponding promoters are apparently used for inhibiting transcription initiation, and binding of CodY to the downstream sites inhibits transcription elongation via a roadblock mechanism (5, 7). Sequence analysis revealed that the nupN regulatory region contains two sequences, called here motif 1 and motif 2, that strongly resemble the previously defined 15-bp CodY-binding consensus motif, AATTTTCWGAAAATT (6, 11, 16), and that are located at positions −64 to −50 and +169 to +183 with respect to the transcription start point (Table 5). (We define “motifs” as 15-bp sequences that are similar to the CodY-binding consensus and “sites” as experimentally determined regions of CodY binding.) In fact, we have already shown that the downstream motif 2 lies within a region where transcription elongation is blocked in vitro in the presence of CodY (7). Thus, it was important to assess the relationship between the motifs and CodY binding sites and the contribution of each motif to CodY-dependent regulation in vivo.
Table 5.
CodY-binding motifs of the nupN gene
| Motifa | Sequenceb | No. of mismatches | Scorec | Location with respect to transcription start point |
|---|---|---|---|---|
| Consensus | AATTTTCWGAAAATT | 0 | 13.8–14.1 | |
| 1a | AtTTTTaAGcAAATa | 4 | 4.0 | −94–−80 |
| 1b | AtaaTTCTaAAAATa | 5 | 4.6 | −82–−68 |
| 1b p21 | AtaaTTCTGAAAATa | 4 | 6.6 | |
| 1b p22 | AtaaTTgTaAAAATa | 6 | 1.9 | |
| 1b p23 | AtaaTTCTacAAATa | 6 | 0.85 | |
| 1b p24 | AtaaaTCTaAAAATa | 6 | 3.7 | |
| 1cd | tAaTTTtTaAAAAaT | 5 | 4.6 | −65–−51 |
| 1 p+ | AATTTTtAaAAAATT | 2 | 10.1 | −64–−50 |
| 1 p1 | AATTTTCAGAAAATT | 0 | 14.1 | |
| 1 p2 | AATTTTtAacAAATT | 3 | 6.4 | |
| 1 p3 | AAaTTTtAaAAAATT | 3 | 8.3 | |
| 2 p+ | tATTaTCAGAAAATT | 2 | 12.0 | +169–+183 |
| 2 p11 | AATTTTCAGAAAATT | 0 | 14.1 | |
| 2 p12 | tATTaTCAcAAAATT | 3 | 8.0 |
Shown are strong CodY-binding motifs 1 and 2 and weaker motifs found upstream of motif 1 and analyzed in this work. Additional weak motifs can be found downstream of motifs 1 and 2 (see Discussion).
Mismatches to the proposed CodY-binding consensus are indicated by lowercase letters. Mutations are in boldface. Overlapping parts of motifs 1a and 1b and the part of motif 1c that overlaps motif 1 are underlined.
The scores for individual CodY-binding motifs have been generated using the position-specific weight matrix, as described in reference 6.
Motif 1c almost completely overlaps strong motif 1 and therefore is unlikely to have a separate functional role. This is supported by the fact that the p1 mutation that increased CodY-mediated repression is a strong down mutation in motif 1c.
Using the entire 503-bp nupN regulatory region, two widely separated CodY binding sites were detected in a DNase I footprinting experiment in the presence of ILV and GTP (Fig. 3B). In order to establish more precisely the sites interacting with CodY, we performed separate DNase I footprinting experiments using two DNA fragments comprising the promoter-proximal and promoter-distal parts of the regulatory region. CodY protected with similarly high affinities two regions of the template DNA strand from positions −86 to −40 and from positions +164 to +204 with respect to the nupN transcription start point (Fig. 2 and 3B). The upstream (site I) and downstream (site II) regions of protection overlapped the previously recognized motifs 1 and 2, respectively (Fig. 2B).
The concentration dependence of CodY binding to site II was very similar whether this site was present on the same fragment of DNA with site I or on a separate fragment (Fig. 3B). Thus, binding of CodY to site II apparently occurs independently of its binding to site I. (The reverse experiment was difficult to perform because of the poor resolution of the site I footprint if present together with site II.)
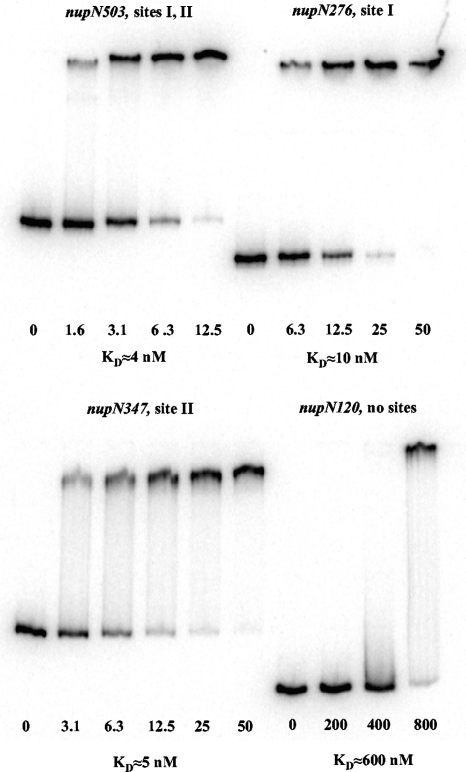
In gel shift experiments, CodY bound to DNA fragments containing only site I or site II with an apparent KD (equilibrium dissociation constant) of ≈10 or ≈5 nM, respectively, compared with ≈4 nM for the full-length fragment (Fig. 2A and 4). (We used the concentration of CodY at which 50% of the DNA molecules are bound as an approximation of KD.) No binding of CodY to the nupN120 fragment that lacked both CodY-binding sites was detected in a DNase I footprinting experiment (Fig. 2A and 3C). In gel-shift experiments, a similar nupN fragment lacking both CodY-binding sites was bound by CodY only at the concentration of 800 nM, apparently reflecting a nonspecific interaction between CodY and DNA (Fig. 4).
Fig. 4.
Gel shift assay of CodY affinity for nupN DNA fragments. Different labeled nupNp+ DNA fragments were incubated with increasing amounts of purified CodY in the presence of 10 mM ILV and 2 mM GTP. The CodY concentrations used (nanomolar concentrations of monomers) are indicated below each lane. KD, the apparent equilibrium dissociation constant, was estimated as the protein concentration needed to shift 50% of DNA fragments under conditions of vast protein excess over DNA.
Contribution of CodY-binding sites to nupN regulation.
To determine the relative contributions of the two CodY-binding sites to regulation of nupN, we used lacZ fusions containing truncated versions of the nupN regulatory region lacking either the downstream or the upstream CodY-binding site (Fig. 2). The resulting fusions, nupN276-lacZ (site I only) and nupN347-lacZ (site II only), were both still repressed by CodY 67- and 6.5-fold, respectively (Table 3; compare strains BB2809 and BB2899 and their codY derivatives). No repression by CodY was observed for the nupN120-lacZ fusion that lacked both CodY-binding sites (Fig. 2 and Table 3, strains BB2900 and BB2907).
Unexpectedly, CodY-dependent regulation of the nupN276-lacZ fusion lacking the downstream site II was almost as efficient as regulation of the full-length fusion nupN503-lacZ (67-fold versus 104-fold repression in 16-amino-acid-containing medium) (Table 3, strains BB2809, BB2686, and their codY derivatives). Therefore, although sites I and II were both able to confer CodY-mediated repression independently, it was not clear to what extent site II contributes to CodY-mediated repression when site I is present.
Interestingly, removal of the sequence upstream of position −49 in the nupN regulatory region reduced the maximal level of expression >10-fold (compare the activities of the nupN347-lacZ [−49 to +298] and nupN120-lacZ [−49 to +71)] fusions to those of their respective counterparts, nupN503-lacZ [−205 to +298] and nupN276-lacZ [−205 to +71], in the codY mutant strain (Table 3). Part of the deleted sequence may play a functional role similar to that of an UP promoter element (18, 28). The upstream boundary of this apparent activating sequence lies downstream of position −69 (see the results for the nupN367-lacZ [−69 to +298] and nupN140-lacZ [−69 to +71] fusions in Table 4). As shown below, some point mutations in this region also reduced expression of the nupN-lacZ fusions.
Table 4.
Expression of nupN-lacZ fusionsa
| Strain | Relevant genotype | Fusion genotype | Motifs present | Growth medium |
|||
|---|---|---|---|---|---|---|---|
| TSS + 16 amino acids |
TSS |
||||||
| β-Galactosidase activity (MU) | Repression ratio | β-Galactosidase activity (MU) | Repression ratio | ||||
| BB2953 | Wild type | nupN380p+ | 1b, 1, 2 | 0.91 | 381 | 171.0 | 2.7 |
| BB2960 | codY | 347.0 | 468.0 | ||||
| BB3491 | Wild type | nupN380p3 | 1b, 1down, 2 | 0.78 | 105 | 67.1 | 1.2 |
| BB3495 | codY | 81.8 | 77.4 | ||||
| BB3492 | Wild type | nupN380p11 | 1b, 1, 2up | 0.20 | 1,525 | 88.8 | 5.3 |
| BB3496 | codY | 305.0 | 471.0 | ||||
| BB3431 | Wild type | nupN380p3/11 | 1b, 1down, 2up | 0.77 | 326 | 166.0 | 2.7 |
| BB3435 | codY | 251.0 | 449.0 | ||||
| BB2972 | Wild type | nupN380p21 | 1bup, 1, 2 | 0.38 | 982 | 28.0 | 18 |
| BB2976 | codY | 373.0 | 492.0 | ||||
| BB2973 | Wild type | nupN380p22 | 1bdown, 1, 2 | 9.25 | 41 | 377.0 | 1.3 |
| BB2977 | codY | 379.0 | 496.0 | ||||
| BB2955 | Wild type | nupN153p+ | 1b, 1 | 0.09 | 174 | 4.01 | 2.4 |
| BB2962 | codY | 15.7 | 9.71 | ||||
| BB3014 | Wild type | nupN153p2 | 1b, 1down | 2.27 | 1.6 | 2.07 | 0.60 |
| BB3019 | codY | 3.54 | 1.24 | ||||
| BB3015 | Wild type | nupN153p3 | 1b, 1down | 2.41 | 6.8 | 12.5 | 0.81 |
| BB3020 | codY | 16.3 | 10.1 | ||||
| BB2974 | Wild type | nupN153p21 | 1bup, 1 | 0.04 | 423 | 0.27 | 47 |
| BB2978 | codY | 16.9 | 12.8 | ||||
| BB2975 | Wild type | nupN153p22 | 1bdown, 1 | 1.30 | 13 | 12.3 | 0.97 |
| BB2979 | codY | 17.0 | 11.9 | ||||
| BB3597 | Wild type | nupN153p23 | 1bdown, 1 | 9.30 | 1.5 | 8.96 | 0.82 |
| BB3600 | codY | 13.6 | 7.36 | ||||
| BB3598 | Wild type | nupN153p24 | 1bdown, 1 | 0.16 | 99 | 6.14 | 1.7 |
| BB3601 | codY | 15.9 | 10.3 | ||||
| BB3599 | Wild type | nupN153p2/21 | 1bup, 1down | 0.09 | 43 | 1.14 | 0.80 |
| BB3602 | codY | 3.91 | 1.42 | ||||
| BB2935 | Wild type | nupN367p+ | 1, 2 | 29.8 | 8.0 | 396.0 | 0.93 |
| BB2943 | codY | 238.0 | 368.0 | ||||
| BB2952 | Wild type | nupN367p1 | 1up, 2 | 0.31 | 623 | 30.6 | 8.2 |
| BB2959 | codY | 193.0 | 250.0 | ||||
| BB2936 | Wild type | nupN140p+ | 1 | 12.9 | 0.95 | 12.3 | 0.78 |
| BB2944 | codY | 12.3 | 9.64 | ||||
| BB2954 | Wild type | nupN140p1 | 1up | 0.05 | 172 | 0.37 | 16 |
| BB2961 | codY | 8.60 | 5.88 | ||||
Cells were grown, β-galactosidase activity was assayed, and repression ratios were calculated as described in Table 3.
It should be noted that truncation at position +71 also reduced the expression of the nupN-lacZ fusions (compare the nupN503-lacZ and nupN276-lacZ fusions or the nupN347-lacZ and nupN120-lacZ fusions in Table 3). Whereas this result might suggest the existence of an activating element downstream of position +71, it is well known that lacZ fusions with different junctions between the promoter-containing sequence and the reporter gene can yield very different activities of β-galactosidase, apparently due to differences in stability or translatability of the corresponding mRNA. To restrict our analysis to the role of CodY in nupN gene expression, we have compared the fold differences (repression ratio) between the activities of each fusion, rather than their absolute activities, in a codY mutant and wild-type cells under conditions in which CodY is maximally active (in the 16-amino-acid-containing glucose-ammonium medium).
The roles of multiple 15-bp CodY-binding motifs in nupN site I function.
Two CodY-binding motifs, 1a and 1b, with poor adherence to the consensus sequence (4 and 5 mismatches, respectively) could be recognized upstream of CodY-binding motif 1 (Fig. 2; Table 5). (We did not consider motifs with 6 mismatches that can be found very frequently in A+T-rich regulatory regions.) Moreover, the 22-bp region upstream of nupN motif 1 that was protected by CodY in DNase I footprinting experiments included the entire motif 1b and part of motif 1a (Fig. 2C). To analyze the potential roles of these sequences in CodY-mediated regulation, we constructed a series of lacZ fusions containing truncated versions of CodY-binding site I with or without site II. The nupN380-lacZ and nupN153-lacZ fusions that had most of motif 1a removed (Fig. 2) were repressed 170- to 380-fold by CodY in the 16-amino-acid-containing medium (Table 4, strains BB2953 and BB2955), i.e., to an even greater extent than the parent nupN503-lacZ and nupN276-lacZ fusions (Table 3, strains BB2686 and BB2809). However, the nupN367-lacZ fusion, in which all but 5 bp upstream of motif 1, including most of motif 1b, was removed, was repressed by CodY 13-fold less that the full-length fusion (Table 4, strain BB2935; and Table 3, strain BB2686) and at a level almost identical to that of the fusion that lacked site I entirely (Table 3, strain BB2899). When the nupN CodY-binding site II was deleted from the nupN367-lacZ fusion, the resulting nupN140-lacZ fusion (Fig. 2), retaining motif 1 but lacking motifs 1a, 1b, and 2, completely lost the ability to be regulated by CodY (Table 4, strain BB2936). Because a similar fusion, nupN153-lacZ, containing only 13 additional upstream nucleotides, was highly regulated by CodY, we conclude that the sequence within the nupN CodY-binding site I located upstream of motif 1 and overlapping motif 1b (from position −82 to position −70 with respect to the transcription start point) contains a region that is required for repression by CodY.
To analyze the role of motif 1b, we introduced an up mutation, p21, and down mutations, p22, p23, and p24, that made this motif more or less similar to the consensus sequence, respectively (Table 5). p21 increased the ability of CodY to repress the nupN380-lacZ or nupN153-lacZ fusions; all down mutations, including p24, which affects the least-conserved position of the consensus, had the opposite effect (Table 4, strains BB2972 to BB2975, BB3597, and BB3598). The p23 mutation affecting the most conserved position of the consensus motif almost completely eliminated regulation by CodY at site I. Taken together, these data strongly indicate that motif 1b, located 3 bp upstream of motif 1, is a functional regulatory element essential for CodY-dependent repression at site I.
To confirm our initial assumption that the 15-bp CodY-binding motif 1 is also important for the contribution of site I to nupN repression, we introduced single down mutations (p2 and p3) and a double up mutation (p1) into this sequence (Table 5). As expected, both down mutations greatly reduced the ability of CodY to repress, and the p1 up mutations increased the ability of CodY to repress (Table 4, strains BB2952, BB2954, BB3014, BB3015, and BB3491). The p1 mutation also eliminated the requirement for the motif 1b sequence for CodY-mediated repression (Table 4, strains BB2952 and BB2954). Conversely, the up mutation p21 in motif 1b was able to compensate partly for the defect in the adjacent motif 1 imposed by the down mutation p2 (Table 4, strains BB3014 and BB3599).
Mutational analysis of site II contribution to nupN regulation.
Since CodY binds to site II with higher affinity than to site I, it was surprising that deletion of site II, as in the nupN276-lacZ and nupN153-lacZ fusions (Fig. 2), affected the extent of nupN regulation by CodY only ≤2-fold. We were concerned that this result could be an artifact caused by extensive deletions used to construct the nupN276-lacZ and nupN153-lacZ fusions that led to their reduced expression. Therefore, to analyze further the actual role of site II in the regulation of nupN in the presence of site I, we introduced a single down mutation, p12, in the CodY-binding motif 2 (Table 5). As expected, in the context of a fusion that lacked site I (nupN347-lacZ), this mutation reduced CodY-dependent repression in the 16-amino-acid-containing medium >5-fold (Table 3, strains BB2899 and BB3493). However, the highly expressed nupN503p12-lacZ fusion, containing wild-type site I and a down mutation in site II, was repressed only 1.6-fold less strongly than the nupN503p+-lacZ fusion, consistent with the results of our deletion analysis and confirming that under the conditions tested site II does not contribute significantly to repression if site I is present (Table 3, strains BB3402 and BB2686).
To determine whether the strength of site II is limiting its ability to contribute to regulation, we introduced a two-nucleotide substitution (the up mutation p11) in motif 2 that made it a perfect match to the consensus. The efficiency of repression in the 16-amino-acid-containing medium of the nupN347p11-lacZ fusion, containing the improved site II as the only CodY-binding region, was increased 8-fold over that of the nupN347p+-lacZ fusion (Table 3, strains BB3401 and BB2899). Similarly, the nupN380p3/11-lacZ fusion, containing a partially inactive version of site I in addition to the improved site II, was repressed by CodY 4-fold more strongly than the nupN380p3-lacZ fusion (Table 4, strains BB3431 and BB3491). Finally, even the nupN380p11-lacZ fusion, containing both the intact site I and improved site II, was 4-fold more repressed by CodY in 16-amino-acid-containing medium than a similar fusion without the p11 mutation (Table 4, strains BB3431 and BB2953). We conclude that though site I is adequate for the full physiological extent of repression of nupN by CodY, increasing the similarity of motif 2 (within the CodY-binding site II) to the consensus sequence can cause more efficient repression.
Possible autoregulation of nupN.
It was previously suggested that the nupN promoter may be subject to positive autoregulation and cannot be transcribed if nupN or the downstream nupOPQ genes are not expressed (29). The suggestion was based on the lack of expression of an nupN::pMutin (lacZ) fusion when it was integrated at the nupN locus of the B. subtilis chromosome, thus disrupting the nupN gene and likely preventing expression of the downstream nupOPQ genes due to a polar effect on their transcription. However, expression of the ectopic nupN503-lacZ fusion in the strain containing a nupN in-frame deletion or the nupN::pMutin allele proceeded unimpeded (data not shown). We conclude that the nupN gene is unlikely to be subject to autoregulation.
DISCUSSION
Role of nupNOPQ in nucleoside uptake.
The results presented here show that the nupNOPQ genes encode an uptake system for guanosine and perhaps other nucleosides. At least two other transporters, NupG (19) and YutK (as mentioned in reference 27), have been found to have similar or overlapping specificities (none of them characterized in detail). NupG is the major adenosine and guanosine permease in cells grown in minimal medium (19). However, nupG expression is low in the presence of guanine or hypoxanthine in the medium (19) and is apparently subject to a guanine riboswitch-mediated control (25a). Thus, it is likely that the role of the NupNOPQ transporter in purine nucleoside uptake becomes more important in cells grown in the presence of guanine and related compounds, such as guanosine, hypoxanthine, or inosine. Our data indicate that NupNOPQ may also be a low-affinity transporter of pyrimidine nucleosides. NupC serves as the major transporter of pyrimidine nucleosides in B. subtilis (33); NupNOPQ may be the previously unidentified minor transporter of these nucleosides. NupNOPQ-like transporters in other bacteria take up both purine and pyrimidine nucleosides (9, 26, 41). A more detailed analysis of the contributions of NupG, NupNOPQ, YutK, and NupC to the uptake of different nucleosides in cells grown under different conditions was beyond the scope of this work.
Nucleosides, especially guanosine, are poor nitrogen and carbon sources (34, 40). Therefore, it makes physiological sense that their utilization under conditions of nutrient abundance is delayed by CodY until other, preferred sources of nitrogen and carbon have been consumed. Inhibition of guanosine uptake under growth conditions when CodY is highly active may also serve to keep the already high cellular pool of GTP, a positive effector of CodY, in check.
CodY-dependent regulation of the nupN promoter.
Two high-affinity CodY-binding sites were identified within the regulatory region of B. subtilis nupN. Each site was shown to bind CodY independently of the other.
CodY-binding site I, a major contributor to nupN repression, is located only 3 bp upstream of the −35 region of the promoter. Thus, CodY binding to this site is likely to prevent RNA polymerase binding and may specifically target binding of the σ-subunit or the α-subunit of RNA-polymerase; an A+T-rich sequence located downstream of position −69 and overlapping position −49 could potentially have a function similar to that of an α-interacting UP element (18, 28).
The downstream site II is, if anything, a more efficient CodY binder than site I in vitro, but only a minor role for this site in nupN regulation (≤2-fold) could be detected when the upstream site I was present. Nonetheless, site II serves as a roadblock for transcription elongation in in vitro transcription experiments (7) and negatively regulates expression in vivo of nupN-lacZ fusions that lack site I. Moreover, when we improved the similarity of motif 2 within site II to the CodY-binding consensus motif, the modified site II highly increased repression by CodY, even in the presence of site I. Given our inability to demonstrate a significant contribution of native site II to CodY-mediated repression in vivo, site II is likely to be a minor player in nupN regulation under most growth conditions. On the other hand, with its apparent higher affinity for CodY, site II could potentially contribute more prominently to nupN repression under growth conditions when CodY activity is low. Also, we cannot exclude the possibility that additional regulators operate at the nupN promoter and mask or modify effects of CodY in some of our experiments. The existence of such regulators could be consistent with our observation that some deletions and mutations affect nupN expression in a CodY-independent but medium-dependent and sequence context-specific manner.
In addition to nupN, the B. subtilis genes bcaP and ybgE were shown to possess two CodY-binding sites, of which one is located >60 bp downstream of the transcription start site. In all three cases, the downstream site has a higher affinity for CodY than does the upstream site and causes transcription elongation pausing or arrest due to a CodY-imposed roadblock in vitro (5, 7). However, the contributions of the downstream sites to CodY-mediated repression in vivo are drastically different: the ybgE downstream site is the major contributor to regulation (7), the bcaP downstream site contributes nearly as much as the upstream site (5), and the nupN downstream site is apparently a weak contributor. The efficiency of a transcription roadblock may depend both on the strength of the corresponding promoter and the sequence context of the roadblock site (13, 24).
The roles of multiple motifs within a single CodY-binding site.
To our surprise, we found that efficient CodY-mediated regulation at nupN site I requires the presence of two 15-bp motifs separated by 3 bp. This is reminiscent of the regulation of the B. subtilis putBCP operon (3). In the case of the putB promoter, the two motifs, separated by 2 bp, have 3 to 4 mismatches each with respect to the consensus sequence, and we surmised that it was their relative weakness that required the presence of both of them for efficient regulation by CodY. In two other cases, the dpp operon and the ybgE gene, strong CodY binding is associated with the presence of 3 to 4 adjacent or overlapping motifs (with 4 to 5 mismatches each), though their exact roles have not been characterized (6, 7). We presumed that weaker motifs are required to be in groups of at least two to provide effective binding and regulation by CodY. In the case of nupN, however, the native version of motif 1 has only 2 mismatches to the consensus, yet its contribution to regulation is fully dependent on the presence of the upstream motif 1b. The requirement for motif 1b in regulation is abolished if motif 1 is made stronger.
Binding of CodY to two adjacent motifs may be a more widespread phenomenon than currently assumed. Many strong motifs with 2 to 3 mismatches—the only ones that can be predicted to participate in CodY binding with some degree of confidence—are frequently associated with weaker motifs (with 4 to 5 mismatches) that are either adjacent to or overlap with the stronger motif (our unpublished analysis), consistent with the hypothesis that several nearby motifs can improve CodY binding to DNA or may be essential for such binding. Moreover, experimentally determined CodY-binding sites typically cover more extended regions than just a single 15-bp motif. Interestingly, additional 15-bp sequences with 5 mismatches to the consensus CodY-binding motif can be found within nupN site I (overlapping 14 or 4 promoter-proximal nucleotides of motif 1 from positions −63 to −49 and −53 to −39, respectively) and within nupN site II (5 bp downstream of motif 2 from positions +189 to +203). The roles of these sequences in regulation remain unknown. It should be noted that 15-bp sequences with 5 mismatches to the consensus CodY-binding motif are common in the (A+T)-rich B. subtilis genome and even more so in the (A+T)-enriched regulatory regions.
The data showing that efficient CodY-mediated regulation requires sequences outside a single 15-bp motif were also obtained for the bcaP gene (B. R. Belitsky and A. L. Sonenshein, unpublished data). Recently, the participation of two overlapping motifs was proposed to be a general rule for CodY binding (42). The accumulated results suggest a more complex mechanism of CodY-DNA interaction than previously thought.
ACKNOWLEDGMENTS
We are grateful to the National BioResource Project (NIG, Japan): B. subtilis for the gift of strains and to L. V. Wray, Jr., and S. H. Fisher for sharing their results before publication.
This work was supported by a research grant (GM042219) from the U.S. Public Health Service.
Footnotes
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Arnaud M., Chastanet A., Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beaman T. C., et al. 1983. Specificity and control of uptake of purines and other compounds in Bacillus subtilis. J. Bacteriol. 156:1107–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Belitsky B. R. 2011. Indirect repression by Bacillus subtilis CodY via displacement of the activator of the proline utilization operon. J. Mol. Biol . 413:321–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Belitsky B. R., Gustafsson M. C., Sonenshein A. L., Von Wachenfeldt C. 1997. An lrp-like gene of Bacillus subtilis involved in branched-chain amino acid transport. J. Bacteriol. 179:5448–5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Belitsky B. R., Sonenshein A. L. 2011. Contributions of multiple binding sites and effector-independent binding to CodY-mediated regulation in Bacillus subtilis. J. Bacteriol. 193:473–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Belitsky B. R., Sonenshein A. L. 2008. Genetic and biochemical analysis of CodY-binding sites in Bacillus subtilis. J. Bacteriol. 190:1224–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Belitsky B. R., Sonenshein A. L. 2011. Roadblock repression of transcription by Bacillus subtilis CodY. J. Mol. Biol. 411:729–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Belitsky B. R., Sonenshein A. L. 1998. Role and regulation of Bacillus subtilis glutamate dehydrogenase genes. J. Bacteriol. 180:6298–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8a. Daniel R. A., Haiech J., Denizot F., Errington J. 1997. Isolation and characterization of the lacA gene encoding β-galactosidase in Bacillus subtilis and a regulator gene, lacR. J. Bacteriol . 179:5636–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deka R. K., et al. 2006. The PnrA (Tp0319; TmpC) lipoprotein represents a new family of bacterial purine nucleoside receptor encoded within an ATP-binding cassette (ABC)-like operon in Treponema pallidum. J. Biol. Chem. 281:8072–8081 [DOI] [PubMed] [Google Scholar]
- 10. den Hengst C. D., et al. 2005. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. J. Bacteriol. 187:512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. den Hengst C. D., et al. 2005. The Lactococcus lactis CodY regulon: identification of a conserved cis-regulatory element. J. Biol. Chem. 280:34332–34342 [DOI] [PubMed] [Google Scholar]
- 12. Dineen S. S., McBride S. M., Sonenshein A. L. 2010. Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 192:5350–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Epshtein V., Toulme F., Rahmouni A. R., Borukhov S., Nudler E. 2003. Transcription through the roadblocks: the role of RNA polymerase cooperation. EMBO J. 22:4719–4727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fisher S. H. 1999. Regulation of nitrogen metabolism in Bacillus subtilis: vive la difference! Mol. Microbiol. 32:223–232 [DOI] [PubMed] [Google Scholar]
- 15. Guedon E., Serror P., Ehrlich S. D., Renault P., Delorme C. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227–1239 [DOI] [PubMed] [Google Scholar]
- 16. Guedon E., Sperandio B., Pons N., Ehrlich S. D., Renault P. 2005. Overall control of nitrogen metabolism in Lactococcus lactis by CodY, and possible models for CodY regulation in Firmicutes. Microbiology 151:3895–3909 [DOI] [PubMed] [Google Scholar]
- 17. Handke L. D., Shivers R. P., Sonenshein A. L. 2008. Interaction of Bacillus subtilis CodY with GTP. J. Bacteriol. 190:798–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Helmann J. D. 1995. Compilation and analysis of Bacillus subtilis sigma A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johansen L. E., Nygaard P., Lassen C., Agerso Y., Saxild H. H. 2003. Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL). J. Bacteriol. 185:5200–5209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Joseph P., Ratnayake-Lecamwasam M., Sonenshein A. L. 2005. A region of Bacillus subtilis CodY protein required for interaction with DNA. J. Bacteriol. 187:4127–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kreth J., Chen Z., Ferretti J., Malke H. 2011. Counteractive balancing of transcriptome expression involving CodY and CovRS in Streptococcus pyogenes. J. Bacteriol. 193:4153–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Levdikov V. M., et al. 2009. Structural rearrangement accompanying ligand binding in the GAF domain of CodY from Bacillus subtilis. J. Mol. Biol. 390:1007–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Levdikov V. M., Blagova E., Joseph P., Sonenshein A. L., Wilkinson A. J. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria. J. Biol. Chem. 281:11366–11373 [DOI] [PubMed] [Google Scholar]
- 24. Lewis D. E., Komissarova N., Le P., Kashlev M., Adhya S. 2008. DNA sequences in gal operon override transcription elongation blocks. J. Mol. Biol. 382:843–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Majerczyk C. D., et al. 2010. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192:2861–2877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a. Mandal M., Boese B., Barrick J. E., Winkler W. C., Breaker R. R. 2003. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 13:577–586 [DOI] [PubMed] [Google Scholar]
- 26. Martinussen J., Sorensen C., Jendresen C. B., Kilstrup M. 2010. Two nucleoside transporters in Lactococcus lactis with different substrate specificities. Microbiology 156:3148–3157 [DOI] [PubMed] [Google Scholar]
- 27. Martinussen J., Wadskov-Hansen S. L., Hammer K. 2003. Two nucleoside uptake systems in Lactococcus lactis: competition between purine nucleosides and cytidine allows for modulation of intracellular nucleotide pools. J. Bacteriol. 185:1503–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meijer W. J. J., Salas M. 2004. Relevance of UP elements for three strong Bacillus subtilis phage phi29 promoters. Nucleic Acids Res. 32:1166–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Molle V., et al. 2003. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185:1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Petranovic D., et al. 2004. Intracellular effectors regulating the activity of the Lactococcus lactis CodY pleiotropic transcription regulator. Mol. Microbiol. 53:613–621 [DOI] [PubMed] [Google Scholar]
- 31. Ratnayake-Lecamwasam M., Serror P., Wong K. W., Sonenshein A. L. 2001. Bacillus subtilis CodY represses early-stationary-phase genes by sensing GTP levels. Genes Dev. 15:1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sambrook J., Fritsch E. F., Maniatis T. J. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 33. Saxild H. H., Andersen L. N., Hammer K. 1996. dra-nupC-pdp operon of Bacillus subtilis: nucleotide sequence, induction by deoxyribonucleosides, and transcriptional regulation by the deoR-encoded DeoR repressor protein. J. Bacteriol. 178:424–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schuch R., Garibian A., Saxild H. H., Piggot P. J., Nygaard P. 1999. Nucleosides as a carbon source in Bacillus subtilis: characterization of the drm-pupG operon. Microbiology 145:2957–2966 [DOI] [PubMed] [Google Scholar]
- 35. Serror P., Sonenshein A. L. 1996. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20:843–852 [DOI] [PubMed] [Google Scholar]
- 36. Shivers R. P., Sonenshein A. L. 2004. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol. Microbiol. 53:599–611 [DOI] [PubMed] [Google Scholar]
- 37. Slack F. J., Serror P., Joyce E., Sonenshein A. L. 1995. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol. Microbiol. 15:689–702 [DOI] [PubMed] [Google Scholar]
- 38. Sonenshein A. L. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8:203–207 [DOI] [PubMed] [Google Scholar]
- 39. Sonenshein A. L. 2007. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5:917–927 [DOI] [PubMed] [Google Scholar]
- 40. Switzer R. L., Zalkin H., Saxild H. H. 2002. Purine, pyrimidine, and pyridine nucleotide metabolism, p. 255–269In Sonenshein A. L., Hoch J. A., Losick R. (ed.), Bacillus subtilis and its closest relatives: from genes to cells. American Society for Microbiology, Washington, DC [Google Scholar]
- 41. Webb A. J., Hosie A. H. 2006. A member of the second carbohydrate uptake subfamily of ATP-binding cassette transporters is responsible for ribonucleoside uptake in Streptococcus mutans. J. Bacteriol. 188:8005–8012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wray L. V., Jr., Fisher S. H. 2011. Bacillus subtilis CodY operators contain overlapping CodY binding sites. J. Bacteriol. 193:4841–4848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yanisch-Perron C., Vieira J., Messing J. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103–119 [DOI] [PubMed] [Google Scholar]
- 44. Zeigler D. R., et al. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J. Bacteriol. 190:6983–6995 [DOI] [PMC free article] [PubMed] [Google Scholar]