Abstract
AdpA is a key regulator of morphological differentiation in Streptomyces. In contrast to Streptomyces griseus, relatively little is known about AdpA protein functions in Streptomyces coelicolor. Here, we report for the first time the translation accumulation profile of the S. coelicolor adpA (adpASc) gene; the level of S. coelicolor AdpA (AdpASc) increased, reaching a maximum in the early stage of aerial mycelium formation (after 36 h), and remained relatively stable for the next several hours (48 to 60 h), and then the signal intensity decreased considerably. AdpASc specifically binds the adpASc promoter region in vitro and in vivo, suggesting that its expression is autoregulated; surprisingly, in contrast to S. griseus, the protein presumably acts as a transcriptional activator. We also demonstrate a direct influence of AdpASc on the expression of several genes whose products play key roles in the differentiation of S. coelicolor: STI, a protease inhibitor; RamR, an atypical response regulator that itself activates expression of the genes for a small modified peptide that is required for aerial growth; and ClpP1, an ATP-dependent protease. The diverse influence of AdpASc protein on the expression of the analyzed genes presumably results mainly from different affinities of AdpASc protein to individual promoters.
INTRODUCTION
Streptomycetes, GC-rich Gram-positive soil bacteria known for their ability to produce many valuable antibiotics and other secondary metabolites, undergo complex morphological differentiation (4, 10). The genome of the model species Streptomyces coelicolor A3(2) was the first among the Streptomycetes to be completely sequenced (2).
Streptomyces bacteria grow by tip extension and hyphal branching to form a dense mycelial network of vegetative hyphae. In response to nutrient depletion and other signals, the vegetative mycelium is partially self-cannibalized by a nuclease(s) and protease(s) to supply nutrients for the growth of aerial hyphae, which subsequently transform into long chains of spores (3, 27). This morphological differentiation, which is usually accompanied by the production of secondary metabolite(s), is controlled by multilevel regulatory mechanisms. A key coordinating role in the regulation of morphological differentiation is played by the protein AdpA, which was originally discovered in Streptomyces griseus (37, 38). In all Streptomyces genomes sequenced so far, translation of adpA mRNA depends on a leucyl-tRNA for a rarely used TTA codon; the tRNAUAALeu is encoded by the bldA gene required for aerial mycelium formation. AdpA belongs to the AraC/XylS family of transcription regulators, whose members contain a dual helix-turn-helix (HTH) motif in the C-terminal DNA binding domain. In S. griseus, AdpA activates a number of genes whose products are required for morphological development and for secondary metabolites synthesis (e.g., streptomycin) (12, 28). During vegetative growth, the transcription of S. griseus adpA (adpASg) is repressed by ArpA, the receptor protein for the signaling γ-butyrolactone molecule A-factor; accumulation of A-factor and its interaction with ArpA cause derepression of adpASg expression and the subsequent activation of genes involved in differentiation and synthesis of secondary metabolites (12, 16) (for clarity, we used the subscripts Sg [for S. griseus] and Sc [for S. coelicolor] to distinguish between the adpA genes and AdpA proteins of these two organisms).
In Streptomyces coelicolor, somewhat less is known about AdpASc protein and its contribution to the regulatory events during differentiation. In contrast to S. griseus, a comparable γ-butyrolactone signaling system seems to have no marked effect on adpASc expression in S. coelicolor, and AdpASc does not appear to directly regulate any of the antibiotic biosynthetic gene sets analyzed so far (7, 40). In this paper, we characterize the AdpASc protein from S. coelicolor and its interaction with DNA. We also demonstrate a direct influence of AdpASc on the expression of several genes whose products play a key role in the differentiation of S. coelicolor: STI, a protease inhibitor (20); RamR, an atypical response regulator that itself activates expression of the genes for a small modified peptide that is required for aerial growth (17); and ClpP1, an ATP-dependent protease (9).
MATERIALS AND METHODS
DNA manipulation and bacterial strain growth conditions.
DNA manipulations were carried out by standard protocols (34). Enzymes were supplied by Fermentas, Promega, or Roche, and oligonucleotides were from Genomed (Poland). The Streptomyces coelicolor and Escherichia coli strains and plasmids used in this study are listed in Table 1. All the PCR-derived clones were analyzed by DNA sequencing to check their fidelity. Culture conditions, media, antibiotic concentrations, transformation and conjugation methods followed the general procedures for E. coli (34) and Streptomyces (18). S. coelicolor was cultivated in 79 liquid medium (32). For expression studies, the strains were cultivated on plates containing R2 or MM agar supplemented with 1% mannitol (18).
Table 1.
Bacterial strains and plasmids used in this study
| Bacterial strain or plasmid | Relevant genotype or phenotype or description | Source or reference(s) |
|---|---|---|
| E. coli strains | ||
| DH5α | φ80dlacZΔM15 recA1 endA1 gyrAB thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 phoA | Promega |
| BL21(DE3) | F−ompT hsdSB(rB− mB−) dcm gal λ(DE3) | Promega |
| BW25113/pIJ790 | Δ(araD-araB)567 ΔlacZ4787(::rrnB4) lacIp-40000(lacIq) λ−rpoS369(Am) rph-1 Δ(rhaD rhaB)568 hsdR514 on the bacterial chromosome; oriR101 repA1001(Ts) araBp-gam-be-exo on the pIJ790 plasmid | 11 |
| ET12567/pUZ8002 | dam dcm hsdS Cmr Tetr on the bacterial chromosome; tra KanrRP4 23 on pUZ8002 | 29 |
| US0 | ΔpyrF ΔhisB F′ | 25 |
| S. coelicolor strains | ||
| M145 | SCP1− SCP2− | 2 |
| M851 | M145 ΔadpASc | 36 |
| M145+pIJ6902-hyg | M145+pIJ6902-hyg | This study |
| M851+adpA(TTG) | M851+pIJ6902/2528-hyg | This study |
| M851+pIJ6902-hyg | M851+pIJ6902-hyg | This study |
| M851+LadpA(TTA) | M851+pHL71 | This study |
| M851+LadpA(TTG) | M851+pHL72 | This study |
| Plasmids | ||
| 2StC13 | Cosmid carrying adpASc gene | 33; http://strepdb.streptomyces.org.uk |
| pHZ2528 | pSET152 with 2.4-kb fragment harboring the adpASc gene (with TTG codon) | 36 |
| pHL71 | pSET152 with 3.2-kb fragment harboring the adpASc gene (with TTA codon) and surrounding genes SCO2791 and SCO2793 | 36 |
| pHL72 | pSET152 with 3.2-kb fragment harboring the adpASc gene (with TTG codon) and surounding genes SCO2791 and SCO2793 | 36 |
| pIJ6902/2528-hyg | pIJ6902-hyg containing adpASc under the control of the thiostrepton-inducible PtipA promoter | This study |
| pIJ6902-hyg | pIJ6902 derivative containing Hygr instead of Aprar | This study |
| pET-21a(+)adpAHis6 | pET-21a(+) harboring adpASc gene cloned into BamHI and XhoI sites | This study |
| pET-28a(+)His6-nuc-adpA(51) | pET28a(+) expressing His6-Nuc-AdpA(51) fusion protein cloned into BamHI and NotI restriction sites | This study |
| pB1H1-adpASc | pB1H1 vector harboring the adpASc gene cloned into restriction sites NotI and AvrII | This study |
| pH3U3-18random | pH3U3 vector harboring 18-bp randomized library cloned into NotI and SgsI restriction sites | This study |
qPCR.
For total RNA preparation, S. coelicolor mycelium grown on cellophane-covered MM agar containing 1% mannitol was harvested at different time points as described previously (18). RNA was prepared using TRI reagent (Sigma) according to the instructions of the manufacturer. RNA concentration was quantified using a Nanodrop ND-1000 spectrophotometer, and the quality of the RNA was analyzed on an agarose gel. cDNA was obtained after reverse transcription of 4 μg of DNase I-treated total RNA with MultiScribe murine leukemia virus (MuLV) reverse transcriptase and random hexamer primers (high-capacity cDNA reverse transcription kit; Applied Biosystems) following the procedures recommended by the manufacturer. Control reactions were carried out without reverse transcriptase. Expression of genes was analyzed by quantitative real-time PCR (qPCR) using ABI (StepOnePlus) and a real-time 2× PCR master Mix SYBR kit (A&A Biotechnology). The primers used for real-time PCR are listed in Table S1 in the supplemental material. All primers were designed using the PrimerExpress v. 3.0 program. The specificity of the PCR products was verified by performing melting curve analysis and running the DNA fragment on a gel at the end of each PCR. The reaction mixtures were heated at 95°C for 10 min before PCR cycling for 40 cycles, with 1 cycle consisting of 30 s at 95°C, 30 s at 60°C, and 30 s at 72°C. Fifty-picogram samples of cDNA from the wild type (WT) and mutants were used for analysis. The target gene transcript levels were normalized internally to the level of the hrdB gene. Each experiment was performed in duplicate, and each transcript level was measured in triplicate. With each set of primers, negative-control experiments, performed in the absence of reverse transcriptase, confirmed that PCR products were amplified on cDNA template (i.e., there was no significant contamination with chromosomal DNA).
Construction of mutant strains. (i) Construction of a strain overproducing AdpASc protein.
To construct a strain overproducing AdpASc protein, we used plasmid pHZ2528, which carries intact adpASc with a TTA→TTG mutation (36), and the PtipA (thiostrepton-inducible promoter) expression vector pIJ6902-hyg (hygromycin resistance instead of the apramycin resistance cassette; M. Wolanski, unpublished data), which integrates into the S. coelicolor chromosome at the φC31 attachment site attB (21). The PCR-amplified tetracycline resistance cassette fragment (pBR-tetF and pBR-tetR primers and pBR322 template) was used in the PCR targeting strategy to replace the original adpASc promoter in the pHZ2528 vector with the tetR gene and to create an NdeI restriction site in front of the start codon of the adpASc gene. The adpASc gene was subsequently cut out of the vector using NdeI and EcoRI and cloned into pIJ6902-hyg to give pIJ6902/2528-hyg. This construct was integrated into the attB site on the chromosome of S. coelicolor M851, giving M851+adpA(TTG).
(ii) Complementation of adpASc deletion mutant.
For complementation of the adpA deletion mutant, plasmid pHL71 or pHL72 (36) carrying the wild-type adpASc gene or a TTA-free version of the adpASc gene (TTA was replaced by TTG), respectively, was introduced into the M851 strain by conjugation selecting for apramycin-resistant colonies, yielding the M851+LadpA(TTA) or M851+LadpA(TTG) strain, respectively (Table 1). The presence of the TTA or TTG codon was confirmed by sequencing of PCR-amplified fragment of the complemented adpA gene.
SDS-PAGE and Western blotting.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% acrylamide) by the method of Laemmli (22) and transferred to a nitrocellulose membrane. The membrane was blocked for 1 h at room temperature with Tris-buffered saline with Tween 20 (TBST) (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) containing 3% bovine serum albumin (BSA) and subsequently incubated with anti-AdpASc polyclonal antibodies. The membrane was then washed and incubated with a goat anti-rabbit secondary antibody conjugated with alkaline phosphatase. The membrane was stained with 5-bromo-4-chloro-3-indolylphosphate (BCIP) and nitroblue tetrazolium (NBT).
EMSA.
Electrophoretic mobility shift assay (EMSA) was carried out as described previously (42). Briefly, the 32P-labeled DNA fragment (or nonradioactive fragment) was incubated with increasing amounts of purified AdpASc protein with a six-His (His6) tag (AdpAScHis6) protein in the presence of a nonspecific competitor [poly(dI-dC)·poly(dI-dC)] in 1× Marians' buffer (1× Marians' buffer is 20 mM HEPES-KOH [pH 8.0], 5 mM magnesium acetate, 1 mM EDTA, 4 mM dithiothreitol, 0.2% Triton X-100, 3 mM ATP, 5 g liter−1 BSA, and 5% glycerol) for 30 min at room temperature. The nucleoprotein complexes were resolved in a 4 or 5% polyacrylamide gel in 0.25× Tris-borate-EDTA (TBE) buffer at 5 to 10 V cm−1. For the nonradioactive fragment, the gel was stained with ethidium bromide. The complexes were analyzed by a Typhoon 8600 variable-mode imager and ImageQuant software.
DNase I footprinting.
For DNase I footprinting experiments, a 289-bp-long PCR product obtained with primers padpA-pf and padpA-pr and encompassing the adpASc promoter region was used. The 5′-end-radiolabeled DNA fragments (∼10 fmol) were incubated with different amounts of AdpAScHis6 protein in 1× Marians' buffer at 25°C for 30 min. After DNase I digestion (24), the cleavage products were separated in an 8% polyacrylamide-urea sequencing gel. The gel was analyzed by a Typhoon 8600 variable-mode imager and ImageQuant software.
SPR analysis.
For the standard surface plasmon resonance (SPR) analysis, a promoter region of adpASc was PCR amplified with biotinylated oligonucleotide pb-AdpAEmr and nonbiotinylated oligonucleotide pAdpA-Emf and then immobilized on the chip surface (Sensor Chip SA; GE Healthcare) of the BIAcore 3000 apparatus; approximately 100 response units (RUs) of DNA were immobilized. A non-AdpA box DNA fragment was used as a negative control. DNA loosely attached to the surface of the chip was removed with a 20-μl pulse of 0.1% SDS (15 μl min−1). To exclude the effects of mass transport on the kinetics of the protein-DNA interactions, the measurements were performed at various AdpAScHis6 protein concentrations (2 to 100 nM) and at a continuous flow rate (15 μl min−1) for 120 s. The measurements were performed in HBS-200 buffer (10 mM HEPES [pH 7.4], 10 mM magnesium acetate, 200 mM NaCl, 3.4 mM EDTA, and 0.05% Tween 20). At the end of each cycle, bound AdpAScHis6 protein was removed by washing with a 20-μl pulse of 0.1% SDS (15 μl min−1). The results were plotted as sensograms after subtraction of the background response signal obtained in a control experiment. The BIAevaluation v. 4.1 program (Pharmacia Biosensor AB) was used for data analysis.
In vivo immunoprecipitation.
The immunoprecipitation assay was performed as described earlier (13, 35). Briefly, S. coelicolor strains cultivated for 24 h in 79 liquid medium were cross-linked by the addition of formaldehyde, and then the nucleoprotein complexes were immunoprecipitated with anti-AdpASc antibodies. Cells not subjected to cross-linking and the adpASc deletion mutant served as negative controls and were treated subsequently in the same way as the experimental samples. PCRs were carried out with primers flanking the promoter region of adpASc.
RESULTS AND DISCUSSION
AdpA reaches a maximum concentration at an early stage of aerial mycelium formation in S. coelicolor.
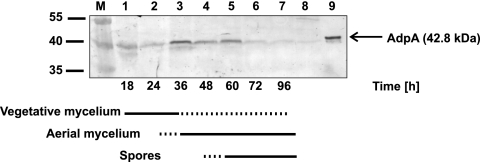
Until now, in both S. coelicolor and S. griseus, the expression of the adpA gene has been analyzed only on the transcript level. However, the level of the AdpA protein depends not only on transcriptional regulation but also on the availability of leucyl-tRNA corresponding to the rare TTA triplet in the adpA gene. In order to examine the expression profiles of the adpA gene during the differentiation of S. coelicolor, we performed quantitative real-time PCR (qPCR) (see Materials and Methods) and Western blotting. The profile of the adpASc mRNA level was similar to that obtained by Takano et al. (36); the transcript was at its highest levels before the formation of aerial hyphae (18 to 24 h), slightly diminished (36 h), and then returned to its high levels with the onset of sporulation (see Fig. S2 in the supplemental material). The abundance of AdpA in the S. coelicolor cell extracts was examined using purified rabbit antibodies raised against the protein (as described in Materials and Methods; see Fig. S1 in the supplemental material). A weak signal had already appeared at 18 h, while the maximum concentration of AdpA was reached at 36 h, i.e., in the early stage of aerial mycelium formation (Fig. 1). The level of AdpA protein dropped slightly and remained stable for the next several hours (48 to 60 h)—during aerial mycelium formation. The signal intensity then decreased significantly (72 h) and remained low until the end of the time course experiment. Surprisingly, the protein expression profile of the adpA mutant in which a rare TTA codon was replaced with TTG was similar to the protein expression profile of bacteria with the wild-type adpA gene (Fig. 1 and Fig. S1C). This is in agreement with the expression profiles of corresponding tRNAs; recently, Pettersson and Kirsebom (31) demonstrated that tRNAUAALeu and tRNACAALeu exhibited similar accumulation profiles and the levels of their accumulation increased after the onset of formation of aerial hyphae. Thus, our data showed that translation of the adpA gene is not limited by a rare tRNAUAALeu—the bldA tRNA. However, it has to be noted that AdpA is not produced in the bldA deletion mutant (11a, 22a, 26, 36).
Fig. 1.
AdpA level in S. coelicolor during growth of hyphae. Western blot analysis of total S. coelicolor cell proteins using rabbit antibodies raised against the C terminus of recombinant AdpASc protein. Protein extracts were prepared from cultures growing on R2 medium at the corresponding time points (in hours) indicated below the figure. Lanes: M, molecular mass markers; 1 to 7, cell extracts of S. coelicolor M145, plate cultures (50 μg of total protein); 8, cell extract of strain M851 (adpA deletion mutant), 50-h plate culture (50 μg of total protein); 9, purified recombinant protein AdpAScHis6 (50 ng). The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel.
S. coelicolor AdpA protein binds in vitro and in vivo to its own promoter region.
So far, extensive studies regarding the interaction of AdpA protein with DNA have been focused on S. griseus AdpA. However, there are some known differences between S. griseus and the model organism S. coelicolor in the regulation and targets of AdpA; therefore, it was desirable to evaluate the details of the binding properties of AdpASc and target promoters in S. coelicolor (5, 36).
To elucidate the binding specificity of AdpASc, we applied the bacterial one-hybrid (B1H) system (for details, see Materials and Methods in the supplemental material). After screening a library of 18-bp randomized DNA sequences (encompassing putative AdpA binding sites), 10 different DNA fragments were selected. Sequence alignment of these fragments using the MEME algorithm (1) allowed us to determine the consensus binding site for the AdpASc protein (see Fig. S3 in the supplemental material), which as could be expected, is consistent with the consensus sequence for S. griseus AdpA protein (41) (both proteins exhibited high similarity, particularly within the DNA binding domain, which showed 100% identity). In particular, G, C, and A/T are highly conserved at positions 2, 4, and 8, respectively (Fig. S3).
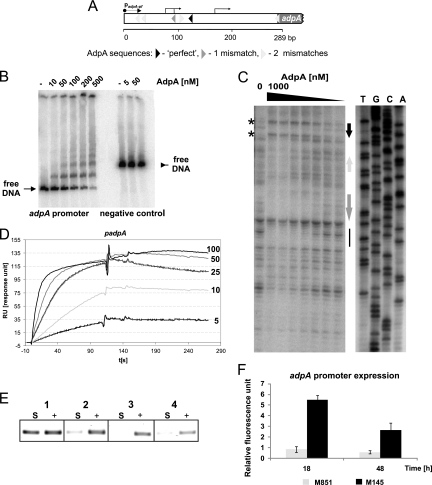
A genome search for the AdpA consensus sequence (5′ TGGCSNGWWY 3′ [41]) allowed us to identify 157 intergenic regions containing at least one consensus AdpA binding sequence in the S. coelicolor chromosome; usually the perfect binding site (0 mismatch) was accompanied by sequences with one or two mismatches. The adpASc promoter was found among the identified promoter regions; this region contains a single perfect sequence and five weak sequences (one with a single mismatch and four with two mismatches [Fig. 2 A]). To examine whether AdpASc interacts with its own promoter, the PCR-amplified adpASc gene was cloned into pET-21a(+) and then overexpressed as a C-terminal His-tagged protein in E. coli BL21 strain. The purified AdpAScHis6 protein was more than 95% pure (as judged by SDS-PAGE analysis [data not shown]) and formed a dimer in solution (per gel filtration column chromatography [data not shown]), similar to S. griseus AdpA [AdpASg] and other proteins belonging to the AraC/XylS protein family (41). To elucidate the interaction of the purified AdpAScHis6 protein with the adpASc promoter (PadpASc), we applied three independent methods: electrophoretic mobility shift assays (EMSAs), surface plasmon resonance (SPR), and DNase I footprinting. The EMSA analysis (Fig. 2B) demonstrated that the AdpAScHis6 protein binds specifically to its own promoter region: at low AdpAScHis6 concentrations, one nucleoprotein complex was observed, while increasing protein concentrations resulted in the formation of additional (one or two) higher-molecular-weight complexes. This suggests that at higher protein concentrations, AdpASc binds a weaker site(s). This is consistent with the DNase I footprint analysis; AdpAScHis6 protein did not protect the region with weak AdpASc sites at low protein concentrations (light-gray and medium-gray arrows in Fig. 2A and C), but the sites were not efficiently digested by the DNase I at high AdpAScHis6 concentrations. The weak sequences were bound by the AdpAScHis6 protein only if they were located in the vicinity of the strong AdpASc binding site(s); they did not form complexes with AdpAScHis6 protein when present alone even at a higher protein concentration (data not shown). Thus, AdpASc presumably interacts with weak target sequences via protein molecules anchored to the strong binding sequence. Binding to multiple sequences located in the adpASc promoter region leads to the formation of a large nucleoprotein complex; a long stretch of DNA was protected not only along AdpA boxes (see black and medium-gray arrows in Fig. 2C) but also in the region lacking in AdpA sequences (see the solid black vertical line in Fig. 2C). Binding of AdpAScHis6 also promoted many distortion points that were preferentially accessible to DNase I (see DNase I-hypersensitive sites in Fig. 2C). Moreover, in addition to the concentration-dependent association of the protein with the adpASc promoter, the SPR analysis (Fig. 2D) revealed the slow dissociation of the AdpAScHis6, suggesting that the complexes formed by AdpASc-PadpASc are fairly stable.
Fig. 2.
Interaction of AdpASc with the promoter region of its own gene. (A) Structure of the adpASc promoter region. Transcription start sites (36) are indicated by the bent arrows. padpA-pf is the primer that was labeled and used (together with the padpA-pr primer) for amplification of the adpASc promoter region and for sequencing. (B) EMSA. A 32P-labeled 289-bp DNA fragment (PCR amplified by the padpA-pf and padpA-pr primers [see Table S1 in the supplemental material]) was incubated with increasing amounts of AdpAScHis6, and the nucleoprotein complexes were analyzed in a 5% polyacrylamide gel. A DNA fragment amplified using the primer pair pH24parSfw and pH24parSrv served as a negative control for AdpASc binding. (C) DNase I footprinting. A 32P-labeled 289-bp DNA fragment was incubated with increasing amounts of AdpAScHis6 and then subjected to DNase I digestion. Black, medium-gray, and light-gray arrows mark the positions of AdpA sequences (see panel A). The asterisks indicate DNase I-hypersensitive sites. The solid black vertical line corresponds to the region lacking in AdpA boxes and protected from DNase I digestion. Lanes T, G, C, and A are sequencing reactions. (D) Surface plasmon resonance (SPR). Sensograms obtained at different concentrations of the AdpAScHis6 with biotinylated DNA immobilized on a streptavidin-coated chip of the BIAcore apparatus. AdpAScHis6 was used at concentrations of 5, 10, 25, 50, and 100 nM. (E) In vivo binding and PCR amplification of immunoprecipitated DNA fragment bound to AdpASc. Purified antibodies raised against AdpASc were used to immunoprecipitate AdpASc-DNA complexes after formaldehyde cross-linking. PCR was carried out with primers flanking the adpASc promoter region (Table S1). Lanes: S, sample of immunoprecipitated DNA; +, input (not immunoprecipitated) DNA (positive control); 1, S. coelicolor M851+adpA(TTG) strain, cross-linked; 2, M851+pIJ6902-hyg (deletion mutant) strain, cross-linked; 3, M851+adpA(TTG) strain, not cross-linked; 4, M851+pIJ6902-hyg (deletion mutant), not cross-linked. (F) Influence of AdpASc on its own expression. The bars represent adpASc promoter expression in M145 (wild-type strain) and M851 (deletion mutant) at the corresponding time points indicated below.
To address the question of whether AdpASc-PadpASc complexes exist in vivo as well, we performed formaldehyde cross-linking of proteins to DNA in intact cells followed by selective immunoprecipitation of protein-DNA complexes with antibodies raised against the AdpASc protein, and then the immunoprecipitated DNA fragments bound to AdpASc were detected by PCR (Fig. 2E). Since we had problems with the cross-linking reaction of cultures grown on solid medium, we decided to use the S. coelicolor M851+adpA(TTG) strain, which was able to produce the AdpASc protein (upon induction with thiostrepton) in liquid culture. The AdpASc-PadpASc complexes were indeed detected in the M851+adpA(TTG) strain, while no signal was observed in the M851+pIJ6902-hyg strain, which served as a negative control (Fig. 2E).
In summary, AdpASc specifically binds the adpASc promoter region in vitro and in vivo. We speculated that as in S. griseus, the transcription of adpASc is self-regulated (16). However, unlike the data presented for S. griseus, AdpASc acts as an activator rather than a repressor on its own gene transcription. Transcription analysis of the adpASc gene promoter indicates higher transcription activity for the M145 strain (wild-type strain) than for the M851 strain (adpASc deletion strain) (Fig. 2F). It has to be noted that Xu et al. (40) recently discovered an additional mechanism involved in the regulation of adpASc expression; they showed that RNase III (AbsB), which was initially discovered as a global regulator of antibiotic production, could cleave the adpASc mRNA. Thus, the expression of the adpASc gene appears to be regulated at three levels: transcription (autoactivation), posttranscription (RNase III), and translation (via the TTA codon). Moreover, both proteins, AdpA and RNase III, take part in a novel feedback control loop that reciprocally regulates their cellular levels (40).
AdpA directly switches on genes required for the morphological differentiation of S. coelicolor.
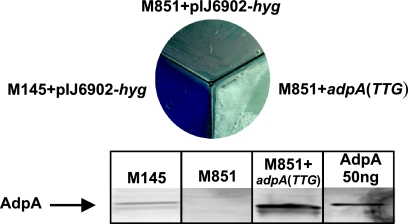
We found that strong AdpASc binding sites are present in promoters of several genes known to be implicated in the formation of aerial mycelium and spores, including sti1 (encoding Streptomyces trypsin inhibitor) (as previously reported [6, 20]), ramR (regulator of production of aerial mycelium-promoting peptide SapB [17, 23, 39]), and clpP1 (encoding ClpP1, an ATP-dependent protease that affects morphological and physiological differentiation [9]). We therefore examined the interaction of AdpASc with the promoters of these genes and the expression of sti1, ramR, and clpP1 in the adpASc deletion mutant (see Fig. 4) and in the strain overexpressing AdpASc protein in relation to the parental strain. For comparative expression studies, we constructed a strain, S. coelicolor M851+adpA(TTG) (Table 1) that upon induction with thiostrepton overexpresses the AdpASc protein (Fig. 3). The overexpression of AdpASc caused an acceleration in the formation of aerial hyphae which is consistent with the earlier observation reported by Nguyen et al. (26) (Fig. 3).
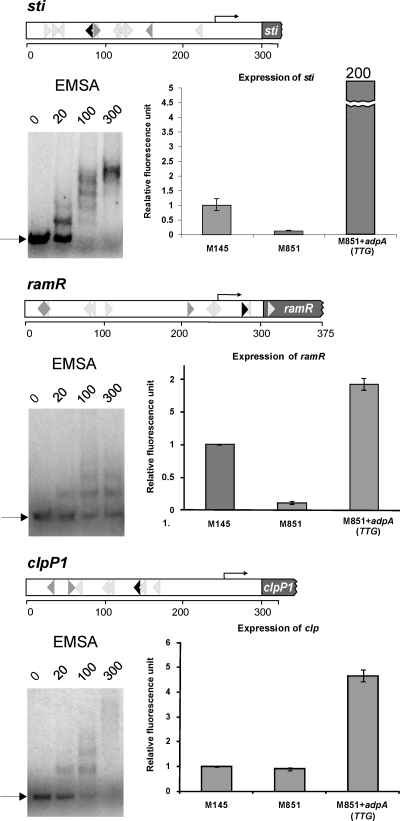
Fig. 4.
Influence of AdpASc on the expression of sti, ramR, and clpP1. For the three genes, each panel contains the structure of the analyzed promoter region (for details, see the legend to Fig. 2C), the interaction of AdpAScHis6 with the analyzed promoter region by EMSA (the arrow shows the position of free DNA; numbers indicate the nanomolar concentration of AdpAScHis6 protein), and quantitative real-time PCR analysis of the abundance of the analyzed mRNA. Total RNA was extracted from a culture growing on MM agar plates supplemented with 1% mannitol for 48 h. The primer pairs for the analyzed genes used in qPCR are listed in Table S1 in the supplemental material. cDNA levels of the target genes were normalized internally to hrdB cDNA, which is the control for a constitutively expressed gene. The error bars indicate the standard deviations from triplicate samples.
Fig. 3.
Phenotypes of S. coelicolor M851+pIJ6902-hyg (adpA deletion mutant) and M851+adpA(TTG) compared to the wild-type S. coelicolor M145+pIJ6902-hyg. Cultures were grown on R2 medium (supplemented with 2 μg ml−1 thiostrepton) for 48 h. The levels of expression of AdpA protein in the analyzed S. coelicolor strains are shown in the blot at the bottom of the figure. Western blot analysis of total S. coelicolor cell proteins (40 μg protein per lane) was performed using rabbit antibodies raised against the C terminus of recombinant AdpASc protein.
In EMSA experiments (Fig. 4), each of the PCR-amplified promoter regions was bound by AdpASc in a concentration-dependent manner. The protein showed the highest affinity toward the sti1 promoter region, presumably due to the presence of two closely spaced inverted AdpASc sequences: one perfect and the other containing a single mismatch. Moreover, the distance between the binding sites (2 bp) is supposed to be optimal for AdpASc binding, as was previously determined for S. griseus AdpA (41). The expression studies were performed when AdpASc was at its high level (at 48 h), and the S. coelicolor life cycle is associated with the formation of aerial hyphae. qPCR expression analysis showed a spectacular increase in sti1 transcript in the S. coelicolor M851+adpA(TTG) strain in relation to the parental strain (approximately 200 times), whereas the level of sti1 mRNA in the adpASc-null mutant was more than seven times lower than in the wild-type strain (Fig. 4). The data clearly demonstrated that the AdpASc protein is directly involved as a transcription activator in the expression of the sti1 gene: the binding of the AdpASc protein to the sti1 promoter region presumably recruits RNA polymerase and facilitates transcription initiation. Thus, the results confirm earlier suggestions regarding the involvement of AdpASc in sti1 gene regulation in S. coelicolor (20). It has been found that overproduction of STI (sti1 under the control of the strong constitutive ermEp promoter) has an inhibitory effect on the production of the blue polyketide antibiotic called actinorhodin (Act). We also found that the AdpASc overexpression resulted in significant inhibition of Act synthesis (Fig. 3). Interestingly, a red pigment antibiotic (undecylprodigiosin) is also not synthesized in the M851+adpA(TTG) strain (Fig. 3). STI is believed to act as an inhibitor of some extracellular trypsin-like protease(s) (15) and thus may play a role in aerial hyphal formation, since a relationship has been found between protease activity and the development of hyphae (19). The degradation of STI (presumably by a specific protease) releases trypsin-like protease(s), which then digests proteins of the old mycelium to supply nutrients for the growth of the reproductive aerial hyphae (3, 6).
In the case of the ramR gene, knowing that AdpASc exhibited a lower affinity toward its promoter than toward the sti1 promoter, we expected a weaker effect of AdpASc on ramR expression. Indeed, overexpression of AdpASc resulted in a slight increase in ramR mRNA level (nearly 2-fold) (Fig. 4). However, the expression of ramR in the adpASc deletion mutant was found to be considerably reduced relative to the wild-type strain (10-fold). These observations are consistent with other studies showing that sti and ramR expression are upregulated in strains that exhibit an elevated level of the AdpASc protein (26, 40). In the case of the clpP1 gene, overexpression of the AdpASc protein caused a moderate elevation of clpP1 mRNA level (4- to 5-fold), but the lack of AdpASc did not influence gene expression (Fig. 4).
Earlier studies suggested that in S. griseus, AdpASg protein regulates the expression of other genes essential for morphological differentiation: ssgA, which encodes a small acidic protein essential for sporulation (septum formation), and sgmA, which encodes a metalloendopeptidase probably involved in the apoptosis of substrate hyphae during the development of aerial hyphae. We decided to examine whether AdpASc can also regulate the expression of these genes in S. coelicolor. However, it has to be noted that in contrast to S. griseus, where each of these promoter regions contains two perfect AdpA binding sites (41), in S. coelicolor, neither sgmA nor ssgA possesses a perfect AdpASc binding site and their promoter regions were bound by AdpASc with rather weak affinity (data not shown) compared with the affinity of the protein to the sti1 or adpASc promoter. The results of qPCR analysis revealed that the levels of expression of ssgA and sgmA in the adpASc deletion mutant were 2- and 8-fold lower, respectively, than in the wild-type strain. Kato et al. (14) showed that no sgmA transcript production occurred in the adpASg deletion mutant of S. griseus. Surprisingly, overexpression of AdpASc reduced the expression of these genes in S. coelicolor (3 and 8 times for ssgA and sgmA, respectively [data not shown]), suggesting that at a higher protein concentration, AdpASc may somehow hinder the access of RNA polymerase to these promoter regions.
In summary, the diverse influence of the AdpASc protein on the expression of the analyzed genes presumably results mainly from the different affinities of the AdpASc protein to individual promoters. However, the possibility that the spatial arrangement of AdpA sites and their location in respect to the promoter(s) may also influence gene expression cannot be excluded. In E. coli, DnaA protein (besides its primary function as an initiator) acts as a transcription factor that represses or activates several genes depending on the location and arrangement of DnaA boxes (25a).
Although the AdpASg and AdpASc proteins are highly conserved (84% identity) and adpA mutants are bald in both organisms (S. griseus and S. coelicolor), the AdpASg protein in S. griseus, in addition to morphological differentiation, is responsible for the positive regulation of secondary metabolites (including streptomycin) synthesis. The S. coelicolor adpA deletion strain retains the ability to synthesize red pigment antibiotic (undecylprodigiosin) (we observed premature and enhanced synthesis of red pigment), while in the S. coelicolor strain that overproduced AdpA protein, none of the two pigmented antibiotics were synthesized (Fig. 3). Knowledge regarding the involvement of the AdpA protein in the regulation of antibiotic production in S. coelicolor is rather obscure; AdpASc presumably acts directly (as in the case of STI) and/or indirectly at different levels of regulation. Moreover, there are also differences in the mechanisms regulating the adpA gene expression in both organisms (for details, see Fig. 5). In contrast to S. griseus, adpASc is not under the control of γ-butyrolactones, and its expression is presumably positively regulated by the AdpASc protein. Taking all the data together, it could be concluded that the AdpA-dependent regulation pathways in both organisms, despite some similarities, exhibit diversity (Fig. 5). It has to be emphasized that S. griseus and S. coelicolor are not closely related streptomycetes and the diversity in the AdpA-dependent pathways is presumably one of the important aspects of speciation which reflect differences in evolutionary adaptation to different soil microenvironments (5).
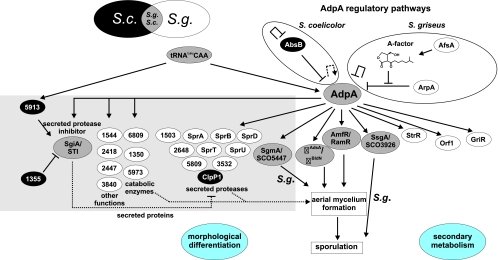
Fig. 5.
Comparison of functions dependent on AdpA protein in S. griseus (some data taken from reference 28) and S. coelicolor (some data taken from reference 40). Arrows indicate positive control. Perpendicular lines indicate negative control. S.c., S. coelicolor; S.g., S. griseus.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Keith Chater for valuable comments on the manuscript, Zhi-Xiong Deng for providing the S. coelicolor M851 strain, Scott Wolfe for plasmids required for bacterial one-hybrid system, Pawel Mackiewicz for help searching for AdpA binding motifs, and Olivier Bertrand for helpful comments regarding antibody preparation.
This work was supported by the Ministry of Science and Higher Education (grant N301 523838). M.W. was supported by a scholarship of the President of Polish Academy of Sciences. A.K.-O., R.D., P.M., and J. Z.-C. gratefully acknowledge financial support received from the Foundation for Polish Science (Start and MISTRZ Programmes).
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
Published ahead of print on 16 September 2011.
REFERENCES
- 1. Bailey T. L., Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 [PubMed] [Google Scholar]
- 2. Bentley S. D., et al. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147 [DOI] [PubMed] [Google Scholar]
- 3. Chater K. F., Losick R. 1997. Mycelial life style of Streptomyces coelicolor A3(2) and its relatives, p. 149–182 In Shapiro J. A., Dworkin M. (ed.), Bacteria as multicellular organisms. Oxford University Press, Oxford, England [Google Scholar]
- 4. Chater K. F. 2001. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr. Opin. Microbiol. 4:667–673 [DOI] [PubMed] [Google Scholar]
- 5. Chater K. F., Horinouchi S. 2003. Signalling early developmental events in two highly diverged Streptomyces species. Mol. Microbiol. 48:9–15 [DOI] [PubMed] [Google Scholar]
- 6. Chater K. F. 2006. Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chater K. F., Chandra G. 2008. The use of the rare UUA codon to define “expression space” for genes involved in secondary metabolism, development and environmental adaptation in Streptomyces. J. Microbiol. 46:1–11 [DOI] [PubMed] [Google Scholar]
- 8. Reference deleted.
- 9. de Crécy-Lagard V., Servant-Moisson P., Viala J., Grandvalet C., Mazodier P. 1999. Alteration of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Mol. Microbiol. 32:505–517 [DOI] [PubMed] [Google Scholar]
- 10. Flärdh K., Buttner M. J. 2009. Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7:36–49 [DOI] [PubMed] [Google Scholar]
- 11. Gust B., Challis G. L., Fowler K., Kieser T., Chater K. F. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc. Natl. Acad. Sci. U. S. A. 100:1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11a. Higo A., Horinouchi S., Ohnishi Y. 2011. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Mol. Microbiol. 81:1607–1622 [DOI] [PubMed] [Google Scholar]
- 12. Horinouchi S. 2007. Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Biosci. Biotechnol. Biochem. 71:283–299 [DOI] [PubMed] [Google Scholar]
- 13. Jakimowicz D., Chater K., Zakrzewska-Czerwínska J. 2002. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol. Microbiol. 45:1365–1377 [DOI] [PubMed] [Google Scholar]
- 14. Kato J. Y., Suzuki A., Yamazaki H., Ohnishi Y., Horinouchi S. 2002. Control by A-factor of a metalloendopeptidase gene involved in aerial mycelium formation in Streptomyces griseus. J. Bacteriol. 184:6016–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kato J. Y., Hirano S., Ohnishi Y., Horinouchi S. 2005. The Streptomyces subtilisin inhibitor (SSI) gene in Streptomyces coelicolor A3(2). Biosci. Biotechnol. Biochem. 69:1624–1629 [DOI] [PubMed] [Google Scholar]
- 16. Kato J. Y., Ohnishi Y., Horinouchi S. 2005. Autorepression of AdpA of the AraC/XylS family, a key transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. J. Mol. Biol. 350:12–26 [DOI] [PubMed] [Google Scholar]
- 17. Keijser B. J., van Wezel G. P., Canters G. W., Kieser T., Vijgenboom E. 2000. The ram-dependence of Streptomyces lividans differentiation is bypassed by copper. J. Mol. Microbiol. Biotechnol. 2:565–574 [PubMed] [Google Scholar]
- 18. Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. 2000. Practical Streptomyces genetics. John Innes Foundation, Norwich, England [Google Scholar]
- 19. Kim I. S., Lee K. J. 1995. Physiological roles of leupeptin and extracellular proteases in mycelium development of Streptomyces exfoliatus SMF13. Microbiology 141:1017–1025 [DOI] [PubMed] [Google Scholar]
- 20. Kim D. W., Chater K., Lee K. J., Hesketh A. 2005. Changes in the extracellular proteome caused by the absence of the bldA gene product, a developmentally significant tRNA, reveal a new target for the pleiotropic regulator AdpA in Streptomyces coelicolor. J. Bacteriol. 187:2957–2966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuhstoss S., Rao R. N. 1991. Analysis of the integration function of the streptomycete bacteriophage φC31. J. Mol. Biol. 222:897–908 [DOI] [PubMed] [Google Scholar]
- 22. Laemmli U. K. 1970. Cleavage of structural proteins during assembly of the head of the bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 22a. Lopez-Garcia M. T., Santamarta I., Liras P. 2010. Morphological differentiation and clavulanic acid formation are affected in a Streptomyces clavuligerus adpA-deleted mutant. Microbiology 156:2354–2365 [DOI] [PubMed] [Google Scholar]
- 23. Ma H., Kendall K. 1994. Cloning and analysis of a gene cluster from Streptomyces coelicolor that causes accelerated aerial mycelium formation in Streptomyces lividans. J. Bacteriol. 176:3800–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Majka J., et al. 1999. Interactions of the Streptomyces lividans initiator protein DnaA with its target. Eur. J. Biochem. 260:325–335 [DOI] [PubMed] [Google Scholar]
- 25. Meng X., Brodsky M. H., Wolfe S. A. 2005. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 23:988–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a. Messer W., Weigel C. 1997. DnaA initiator-also a transcription factor. Mol. Microbiol. 24:1–6 [DOI] [PubMed] [Google Scholar]
- 26. Nguyen K. T., et al. 2003. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. J. Bacteriol. 185:7291–7296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nicieza R. G., Huergo J., Connolly B. A., Sanchez J. 1999. Purification, characterization, and role of nucleases and serine proteases in Streptomyces differentiation. Analogies with the biochemical processes described in late steps of eukaryotic apoptosis. J. Biol. Chem. 274:20366–20375 [DOI] [PubMed] [Google Scholar]
- 28. Ohnishi Y., Yamazaki H., Kato J. Y., Tomono A., Horinouchi S. 2005. AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci. Biotechnol. Biochem. 69:431–439 [DOI] [PubMed] [Google Scholar]
- 29. Paget M. S., Chamberlin L., Atrih A., Foster S. J., Buttner M. J. 1999. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2). J. Bacteriol. 181:204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Referece deleted.
- 31. Pettersson B. M., Kirsebom L. A. 2011. tRNA accumulation and suppression of the bldA phenotype during development in Streptomyces coelicolor. Mol. Microbiol. 79:1602–1614 [DOI] [PubMed] [Google Scholar]
- 32. Prauser H., Falta R. 1968. Phagensensibilitat, Zellwandzusammensetzung und Taxonomie von Actinomyceten. Z. Allg. Mikrobiol. 8:39–46 [DOI] [PubMed] [Google Scholar]
- 33. Redenbach M., et al. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77–96 [DOI] [PubMed] [Google Scholar]
- 34. Sambrook J., Fritsch E. F., Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 35. Solomon M. J., Varshavsky A. 1985. Formaldehyde-mediated DNA-protein crosslinking: a probe for in vivo chromatin structures. Proc. Natl. Acad. Sci. U. S. A. 82:6470–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Takano E., et al. 2003. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of S. coelicolor. Mol. Microbiol. 50:475–486 [DOI] [PubMed] [Google Scholar]
- 37. Vujaklija D., Ueda K., Hong S. K., Beppu T., Horinouchi S. 1991. Identification of an A-factor-dependent promoter in the streptomycin biosynthetic gene cluster of Streptomyces griseus. Mol. Gen. Genet. 229:119–128 [DOI] [PubMed] [Google Scholar]
- 38. Vujaklija D., Horinouchi S., Beppu T. 1993. Detection of an A-factor-responsive protein that binds to the upstream activation sequence of strR, a regulatory gene for streptomycin biosynthesis in Streptomyces griseus. J. Bacteriol. 175:2652–2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Willey J., Santamaria R., Guijarro J., Geistlich M., Losick R. 1991. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell 65:641–650 [DOI] [PubMed] [Google Scholar]
- 40. Xu W., Huang J., Lin R., Shi J., Cohen S. N. 2010. Regulation of morphological differentiation in S. coelicolor by RNase III (AbsB) cleavage of mRNA encoding the AdpA transcription factor. Mol. Microbiol. 75:781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yamazaki H., Tomono A., Ohnishi Y., Horinouchi S. 2004. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Mol. Microbiol. 53:555–572 [DOI] [PubMed] [Google Scholar]
- 42. Zawilak-Pawlik A., et al. 2005. Architecture of bacterial replication initiation complexes: orisomes from four unrelated bacteria. Biochem. J. 389:471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.